Construction of a replication-competent hepatitis B virus vector carrying secreted luciferase transgene and establishment of new hepatitis B virus replication and expression cell lines
2019-10-30JieRuanCaiYanPingShuoSunXinChengPengYuHanYinGeZhangDianXingSun
Jie Ruan, Cai-Yan Ping, Shuo Sun, Xin Cheng, Peng-Yu Han, Yin-Ge Zhang, Dian-Xing Sun
Abstract BACKGROUND Previously, we have successfully constructed replication-competent hepatitis B virus (HBV) vectors by uncoupling the P open reading frame (ORF) from the preC/C ORF to carefully design the transgene insertion site to overcome the compact organization of the HBV genome and maintain HBV replication competence. Consequently, the replication-competent HBV vectors carrying foreign genes, including pCH-BsdR, carrying blasticidin resistance gene (399 bp),and pCH-hrGFP, carrying humanized renilla green fluorescent protein gene (720 bp), were successfully obtained. However, the replication efficiency of the former is higher but it is tedious to use, while that of the latter is poor and cannot be quantified. Hence, we need to search for a new reporter gene that is convenient and quantifiable for further research.AIM To establish a helpful tool for intracellular HBV replication and anti-viral drugs screening studies.METHODS We utilized the replication-competent HBV viral vectors constructed by our laboratory, combined with the secreted luciferase reporter gene, to construct replication-competent HBV vectors expressing the reporter gene secretory Nanoluc Luciferase (SecNluc). HepG2.TA2-7 cells were transfected with this vector to obtain cell lines with stably secreted HBV particles carrying secNluc reporter gene.RESULTS The replication-competent HBV vector carrying the SecNluc reporter gene pCHsNLuc could produce all major viral RNAs and a full set of envelope proteins and achieve high-level secreted luciferase expression. HBV replication intermediates could be produced from this vector. Via transfection with pTRE-sNLuc and selection by hygromycin, we obtained isolated cell clones, named HBV-NLuc-35 cells, which could secrete secNLuc recombinant viruses, and were sensitive to existing anti-HBV drugs. Using differentiated HepaRG cells, it was verified that recombinant HBV possessed infectivity.CONCLUSION Our research demonstrated that a replication-competent HBV vector carrying a secreted luciferase transgene possesses replication and expression ability, and the established HBV replication and expression cell lines could stably secrete viral particles carrying secNluc reporter gene. More importantly, the cell line and the secreted recombinant viral particles could be used to trace HBV replication or infection.
Key words: Hepatitis B virus; Replication-competent hepatitis B virus vector; Secreted luciferase gene; Hepatitis B virus cell line
INTRODUCTION
There are many patients with hepatitis B virus (HBV) infection all over the world who carry a high risk of liver fibrosis, cirrhosis, and hepatocellular carcinoma[1]. Currently,antiviral therapy is critical for chronic infection with HBV, which includes type I interferon and nucleos(t)ide analogs. The former is only partially effective, and the latter is essential to long-term medication and susceptible to resistance[2]. Hence,exploring the mechanisms of HBV replication and developing new drugs are imperative. For this reason, it is absolutely necessary to establish suitable models of HBV infection and replication in vitro and in vivo. Unfortunately, due to species restriction and tissue tropism of HBV, the development of models is limited[3-5].
In the preliminary research, we have successfully constructed replicationcompetent HBV vectors carrying foreign genetic material[6]. Due to the characteristics of the HBV genome and replication strategy, interrupting any part of the genome by inserting a foreign sequence will inhibit HBV replication[7-9]. Hence, we uncoupled the P open reading frame (ORF) from the preC/C ORF to carefully design the transgene insertion site to overcome the compact organization of the HBV genome and maintain HBV replication competence, allowing it to carry nearly 400 bp (up to 720 bp) of foreign genetic information[6]. Consequently, we have successfully constructed the replication-competent HBV vectors pCH-BsdR, carrying blasticidin resistance gene(399 bp), and pCH-hrGFP, carrying humanized renilla green fluorescent protein gene(720 bp). The replication efficiency of the former is higher but it is tedious to use in that the expression of functional BsdR needs to be detected by blasticidin, which generates stable Bsd-resistant cell clones upon transfection of pCH-BsdR into cells.The latter could generate visible hrGFP expression by fluorescence microscopy, which is convenient for assessing the replication and expression of HBV; however,replication efficiency is poor and cannot be quantified.
For this reason, we need to search for a new reporter gene that is convenient and quantifiable for further research. Data from several preliminary studies indicated that some medium-sized transgenes approximately 500 bp are compatible with replication competence. The secreted luciferase (secNLuc) reporter gene (597 bp) can express luciferase protein that is secreted in culture supernatant and is beneficial for monitoring the transcriptional activation of the target gene, which is an extraordinarily helpful tool for molecular biology research[10]. In our study, we employed secNLuc reporter gene[10]as foreign genetic material inserted into the replication-competent HBV vector to achieve a high level of expression of HBV particles carrying secNluc (597 bp) reporter gene, which is slightly larger than the BsdR gene (400 bp). The results showed that the replication-competent HBV vector carrying secNluc reporter gene was able to replicate; however, replication efficiency was decreased compared to the wild-type vector.
Transient transfection is an available method to establish suitable cell models of HBV infection and replication[11,12]; however, transfection efficiency varies. As stable HBV-transfected human hepatoma cells, HepG2.2.15 cell lines are widely used in HBV molecular biology research[13-15], but HBV particle production efficiency is low.Previously, we established the tTA-expressing HepG2 TetOFF cell line HepG2.TA2-7 via a stably transfecting replication-competent HBV vector, and the HBV replication cell line HepG2.117 was successfully obtained, in which HBV particle production efficiency was ten times greater than HepG2.2.15 cell lines[16]. Now, we report the new cell lines, which have the same background as HepG2.117 cells and produce high titer secNluc recombinant HBV particles. To detect the infectivity of recombinant HBV particles, we employed the available HBV infectable cell line HepaRG[17]as the HBV infection model. Interestingly, these recombinant HBV particles are significantly infectious for HepaRG lines. Altogether, we describe a helpful tool for intracellular HBV replication and anti-viral drug screening studies.
MATERIALS AND METHODS
Plasmid construction
Construction of the vector was based on pCH-BsdR[6]arising from pCH-3093[16], which harbors HBV genotype D, subtype ayw (GenBank accession no. V01460.1). The secNluc vector was purchased from Promega Corporation. For construction of pCHsNLuc, the PCR product (template secNluc vector; amplification primer NLuc-S: TGT TGG TAA AGC CAC CAT GG and NLuc-AS: CGT AGA AGC TTA CGC CAG AAT GCG TTC G) was digested with Nco I and Hind III, and the resulting fragment was used to replace the Nco I-Hind III fragment of pCH-BsdR. The sequence was confirmed by DNA sequencing. Construction of pTRE-sNLuc was based on the pTREHBV-C7-5[16], which arises from pTRE-HBVT[16], a hygromycin resistance gene that serves as a selection marker. pCH-sNLuc and pTRE-HBV-C7-5 were digested with Sal I and Nhe I, respectively. Next, the HBV-sNLuc fragment of pCH-sNLuc was used to replace the Sal I-Nhe I fragment of pTRE-HBV-C7-5. The sequence was confirmed by DNA sequencing.
Cell culture, transfection, and clone selection
HepG2 and HepG2.TA2-7 cells were cultured as previously described[16]. HepG2 cells were transfected with pCH-3093, pCH-BsdR, and pCH-sNLuc, which was performed with Fugene HD reagent as recommended by the manufacturer (Roche). For clone selection, HepG2.TA2-7 cells were transfected with pTRE-sNLuc and cultured in medium supplemented with hygromycin. After several passages, the isolated clones that only grew in the presence of hygromycin were selected.
Southern blot, and Northern blot, and quantitative PCR
Isolation of intracellular and extracellular viral nucleocapsids was as previously described, and viral DNA was detected by Southern blot[16]. For Northern blot, RNA was isolated with the SV Total RNA Isolation System Kit (Promega). The HBV DNA in the culture supernatant was evaluated with a commercial HBV DNA kit (Kehua,Shanghai, China) on a SLANTM Real-Time PCR system (Hongshi, Shanghai, China).
Western blot and luciferase activity detection
HBV envelope proteins were detected with HBV human monoclonal antibody 4/7B[18], and isolation and Western blot analysis of intracellular envelope proteins were performed as previously described[16]. The luciferase activity of the culture supernatant was detected with the Nano-Glo luciferase assay reagent as recommended by the manufacturer (Promega).
Infection of HepaRG cells
HepaRG cells were purchased from Biopredic International (Rennes, France). Culture,differentiation, and infection of HepaRG cells were performed as previously described[5,6]. For infection, viral particles were collected from the culture supernatants of selected HBV-sNLuc cell clones by PEG 8000 precipitation and then incubated with HepaRG cells for 24 h. Next, the cells were washed, and the culture medium was changed every second day. For the negative control, HepaRG cells were incubated with free culture medium.
Drug susceptibility assay
Lamivudine (Xinjialin Biotech Ltd.) and IFN-α (Essex Pharma) were dissolved in double distilled water and stored at 4 °C. To observe lamivudine sensitivity, cells were seeded in 12-well plates and incubated with increasing concentrations of lamivudine (0, 0.1, 0.5, 2.5, 5.0, 12.5, and 25.0 µmol/L) for 72 h. Subsequently, the luciferase activity of the culture supernatant was detected with the Nano-Glo luciferase assay reagent, and HBV DNA in the culture supernatant was detected by qPCR. To observe the effect of IFN-α, cells were seeded in 6-well plates inoculated with IFN-α (3000 IU/mL) or free medium, and secreted luciferase expression was detected at the indicated time points (24, 48, 72, 96, and 120 h).
Statistical analysis
SPSS18.0 software was employed for statistical analyses, and one-way ANOVA was used to compare the difference among multiple groups. All data are expressed as the mean with standard deviation (SD), and P < 0.05 was regarded as statistically significant.
RESULTS
Construction of a replication-competent HBV vector carrying secreted luciferase reporter gene
Based on previous research, the secreted luciferase reporter gene sequence (597 bp)was inserted into a replication-competent HBV vector between the uncoupled P ORF and the preC/C ORF to construct the vector pCH-sNLuc (Figure 1A). The upstream and downstream portions of the reporter gene sequences were exactly connected by IRES elements. In addition, the size of the unit length HBV genome increased from 3182 bp to 4068 bp in pCH-sNLuc. For pTRE-sNLuc (Figure 1B), the sNLuc recombinant HBV genome was identical to pCH-sNLuc, and the difference in HBV pregenomic RNA (pgRNA) transcription was controlled by the TRE promoter, not the CMV promoter or the primitive HBV core promoter.
Replication activity of the replication-competent HBV vector carrying secreted luciferase reporter gene
To examine the replication activity of the replication-competent HBV vector, pCH-3093, pCH-BsdR, and pCH-sNLuc were transfected into HepG2 cells. Then,replication intermediates of the replication-competent HBV vector in intracellular viral particles were analyzed by Southern blot. The results (Figure 2A) showed that the RC-DNA and dsL-DNA signals from pCH-sNLuc (lane 3) were lower than those from pCH-3093 (lane 1) and pCH-BsdR (lane 2). These data demonstrated that pCHsNLuc was able to replicate; however, replication efficiency dropped compared with the wild-type HBV vector.
Detection of gene expression of the replication-competent HBV vector carrying secreted luciferase reporter gene

Figure 1 Genomic organization of wild-type hepatitis B virus vector pCH-3093 and replication-competent hepatitis B virus vectors.
To evaluate the transcription level of recombinant RNAs, we transfected pCH-sNLuc into HepG2 cells, controlled with the wild-type HBV expression vector pCH-3093 and the replication-competent HBV vector pCH-BsdR. The results of Northern blot showed that the three vectors could transcribe pgRNA and subgenomic RNA(sgRNA), and all RNA levels were similar (Figure 2B). The size of pgRNA derived from pCH-sNLuc (lane 3) was increased compared to that from pCH-3093 (lane 1) and was similar to that from pCH-BsdR (lane 2). This phenomenon indirectly implied that the presence of the transgene in the recombinant pgRNA comes from pCH-sNLuc. To investigate whether the transgene interferes with the expression of viral structural proteins, we employed Western blot to directly detect HBV envelope proteins (Figure 2C). The test revealed that the levels of envelope proteins L, M, and S in differentially glycosylated forms (gp42/p39, gp36/gp33, and gp27/p24, respectively) from the replication-competent HBV vector pCH-sNLuc (lane 3) were similar to those from the wild-type HBV vector pCH-3093 (lane 1) and the other replication-competent HBV vector pCH-BsdR (lane 2). Subsequently, to detect the transgene expression level of pCH-sNLuc, we transfected pCH-sNLuc into HepG2 cells to observe luciferase activity in the cell supernatant with the Nano-Glo luciferase assay reagent at the indicated time points. The test confirmed high-level luciferase expression up to 2 × 109RLU (relative light unit) at 72 h after transfection (Figure 2D).
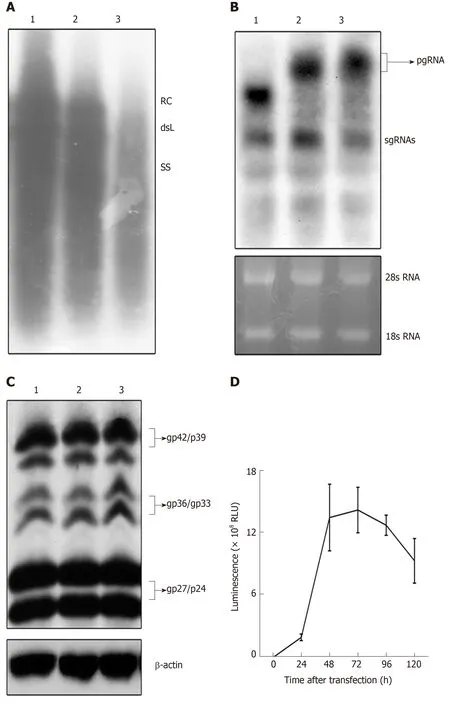
Figure 2 Detection of expression and replication of the recombinant hepatitis B virus vectors.
Established cell lines with stable secretion of HBV particles carrying secNluc reporter gene
To obtain cell lines with stable secretion of HBV particles carrying secNluc reporter gene, HepG2 TA-7 cells were transfected with pTRE-sNLuc. Isolated cell clones were selected in the presence of hygromycin and allowed to grow to confluence. After several passages, we cultured the cell clones Nos. 17, 22, 23, and 35. Among them, the luciferase activity of Nos. 23 and 35 was significantly superior to that of other cell clones (Figure 3A). Next, we assessed the HBV replication levels of superiority cell lines by analyzing extracellular viral particles by Southern blot. As shown in Figure 3B, the RC-DNA and dsL-DNA signals from No. 35 were more dominant than those from No. 7 and No. 23. There was evidence that cell line No. 35 possessed a greater ability to secrete HBV particles carrying secNLuc reporter gene, which are HBVNLuc-35 cells. Together, these data suggested that we have successfully established HBV-NLuc-35 cell lines that could stably form HBV particles carrying secNluc reporter gene.
Infectivity of secNluc recombinant HBV particles
To test the infectivity of recombinant virions carrying secNLuc reporter gene, we collected culture supernatants of HBV-NLuc-35 cells and infected HepaRG cells after concentrating and then monitored the luciferase level. The results of the infection experiment are shown in Figure 3C. The luciferase level in supernatants increased on day 2 post inoculation and gradually increased with time, up to 106RLU on day 6, and gradually decreased on day 10 compared with the lower 103RLU in the negative control group. These data indicated that secNLuc recombinant HBV particles possess infectivity that could successfully infect HepaRG cells.
Drug susceptibility testing for the new cell lines
In addition, we observed the effects of lamivudine and IFN-α on HBV-NLuc-35 cells.The luciferase level and the amount of viral genomes in the culture supernatants were determined by Nano-Glo luciferase assay and quantitative PCR (qPCR) after treatment with lamivudine. It was revealed that the copies of secreted viral DNA gradually decreased in a dose-dependent manner (Figure 4B). When the HBV-NLuc-35 cells were lamivudine-free, they produced as many as 5 × 105copies/mL of virions;however, the concentration of lamivudine up to 25 µmol/L reduced the copy numbers of HBV below the detection lower limit compared with the luciferase level above 3 × 106RLU despite the increased concentration of lamivudine (Figure 4A). In addition, the results showed that the luciferase level peaked at 5 × 106RLU at 72 h and then gradually decreased to the original level at 120 h in the cell supernatant after treatment with IFN-α compared with the control group, in which the luciferase level was above 1 × 107RLU (Figure 4C).
DISCUSSION
Replication-competent virus vectors carrying foreign reporter genes have become helpful tools in virus molecular biology research and anti-viral drug screening in various diseases, such as infections with HCV[19,20], human immunodeficiency virus(HIV)[21,22], and influenza virus[23,24]. Various replication-defective HBV vectors were constructed by substitution of the S gene[25-27]or the Core gene[28]with the gene of interest. We reported the construction of double shRNA expression in HBV Core and S regions[29]. But these HBV vectors require complementation in trans by a helper virus genome, which provides the essential functional proteins. Cotransfection of the chimeric genome of HBV vector and helper genome into permissive cultured hepatoma cells results in the release of enveloped infectious chimeric virions. Several published studies report attempts to generate replication-competent HBV vectors.Chaisomchit[30]demonstrated that a functional HIV tat was expressed before the preS1. Except that the 276 bp small insertion almost completely abolished generation of recombinant HBV, infectivity of its particles remained uncertain and was most probably lost because the expression of preS1 might be missed. Hanafusa[31]showed that the HBV could carry 63 bp of extra DNA by destroying the DR2 region, which should be a critical cis element of HBV replication. The only defined data is that a little of HBV DNA was observed by Southern blot in the cell line HuH-7 but not HepG2.Bai et al[32]constructed a new kind of HBV vector by inserting transgene at the spliced HBV polymerase spacer region and proved that it could replicate in hepatocytes. But as the PreS1 region was replaced by the transgene, it should have lost the infection ability.
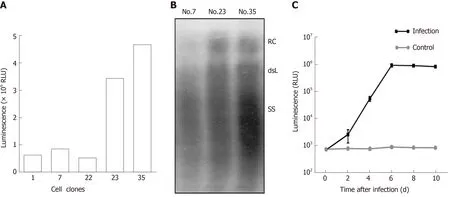
Figure 3 Establishment of secretory Nanoluc Luciferase reporter gene cell lines carrying stably secreted hepatitis B virus particles.
Previously, we successfully constructed replication-competent HBV vectors by inserting two 22 nt Rbm3 IRES sequences and transgene in between the overlap region of Core and Polymerase (Pol) genes to fully maintain HBV genetic information and minimal impact on HBV replication competence[6]. These vectors were able to replicate and express and even generate infectious recombinant virions. However,after transgene size increased from 400 bp to 720 bp, the replication efficiency of vectors decreased from approximately 40% to 1% compared with the level of wildtype HBV[6]. Some medium-sized transgenes of approximately 500 bp are compatible with replication competence.
The secNLuc reporter gene (597 bp) is a convenient and quantifiable tool for target gene molecular biology research[10]. Hence, we utilized the secNLuc reporter gene as a foreign gene to construct the neo replication-competent HBV vector, pCH-sNLuc. This vector could produce all major viral RNAs and a full set of envelope proteins and achieve high levels of secreted luciferase expression (Figure 2). The size of pgRNAs stemming from pCH-sNLuc was larger than pCH-3093 and similar to pCH-BsdR,confirming that the transcription product of the secNLuc gene was fused into recombinant HBV pgRNA. Importantly, the level of transcription was stronger. Pol was translated from HBV pgRNA and necessary for reverse transcription to convert into RC-DNA[33]. Southern blot revealed that HBV replication intermediates could be produced from the pCH-sNLuc vector, implying that Pol could be translated from recombinant HBV pgRNA. However, the replication activity of pCH-sNLuc was lower than those of pCH-BsdR and pCH-3093. The plausible explanation is that the size of the secNLuc reporter gene (597 bp) is large enough to interfere with the reverse transcription of recombinant HBV pgRNA.
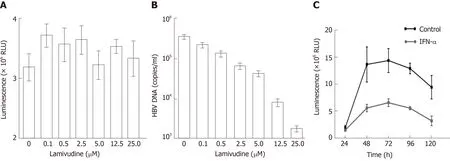
Figure 4 Drug susceptibility testing for the new hepatitis B virus -NLuc-35 cell lines.
For simplified detection of HBV replication, it is necessary to structure quantifiable and standardized cell lines stably transfected with a replication-competent HBV vector[16], which could stably secrete HBV particles carrying foreign genes, such as secNLuc report gene[10]. The tetracycline(Tet)OFF and TetON are notably regulatable systems[34]; if Tet transactivator (tTA) is present, Tet responsive promoter (TRE) could control HBV pgRNA transcription. In terms of the TetOFF system, transcription could be regulated by tTA when Tet was lost. Based on this, we established the tTAexpressing HepG2 TetOFF line HepG2.TA2-7, which efficiently regulated HBV replication and expression[16]. Consequently, via transfection of the TRE-controlled wild-type HBV genome, the high-level HBV replication cell line HepG2.117 was obtained[16]. Similarly, we utilized the same background to establish new HBV replication cell lines. For this purpose, the new replication-competent HBV vector pTRE-sNLuc resulted in the replacement of the HBV core promoter with the TRE promoter to initiate HBV pgRNA transcription. Next, via transfection with pTREsNLuc and selection with hygromycin, we obtained several isolated cell clones,named HBV-NLuc cells (Figure 3). The levels of HBV replication in all selected clones were detected by Southern blot and secNLuc activity by Nano-Glo luciferase assay.When comparing among different clones, it was found that secNLuc activity was closely related to the level of HBV replication; the No. 35 clone possessed stronger HBV replication competence and better secNLuc activity up to 5×, implying that the new cell lines were able to form secreted recombinant viruses. Using differentiated HepaRG[5,35]cells, it was verified that recombinant HBV possessed infectivity. The secNLuc reporter gene inserted among the uncoupled P ORF and preC/C ORF and the recombinant virus could transcribe recombinant HBV pgRNA, which plays an important role in the HBV life cycle, including encoding Core protein and Pol and acting as HBV reverse transcription template for forming novel cccDNA[9], and most importantly, the level of secNluc expression was highly concordant with HBV replication, implying that secNLuc expression could be used to evaluate the level of HBV replication. The increasing secretion levels of secNLuc in supernatant from the secNLuc recombinant virus-infected HepaRG cells suggest that efficient infection can be achieved.
Drug susceptibility of HBV-NLuc-35 cells was detected by the response of cell lines encoding recombinant viruses to LAM and IFN-α. The LAM EC50value was recommended by previous research[16,36]. Following the increasing LAM concentration,the cell line-borne recombinant viruses were strongly inhibited; however, this phenomenon was not observed in luciferase expression, which cannot be affected by LAM. This finding is in line with the theory that LAM could inhibit HBV pgRNA reverse transcription to reduce HBV replication but did not inhibit transcription of cccDNA and translation of pgRNA[37]. pgRNA contains all HBV genetic information as a template for HBV reverse transcription, and as C/P mRNA for encoding core protein and Pol[9]. HBV-NLuc-35 cells could form secNLuc recombinant pgRNA.Regarding IFN-α, the susceptibility assays show that it could reduce secNLuc expression in HBV-NLuc-35 cells. Because IFN-α could inhibit HBV replication,transcription, and expression[38], and cccDNA was an exclusive template for virus transcription, this process depended on the presence of HBx[37,39], implying that secNLuc recombinant cccDNA may be present in HBV-NLuc-35 cells. Altogether,these data demonstrated that new cell lines could secrete secNLuc recombinant viruses and were sensitive to existing anti-HBV drugs. The cell line and the secreted recombinant viral particle could trace HBV replication or infection.
ARTICLE HIGHLIGHTS
Research background
Replication-competent viral vectors carrying additional genetic information have become invaluable tools for virus molecular biology research and anti-viral drug screening, such as those for infections with HCV, HIV, and influenza virus. Due to the extremely compact organization of the hepatitis B virus (HBV) genome, HBV-based vectors had met with very limited success. In our previously study, via inserting two 22 nt Rbm3 IRES sequences and transgene in between the overlap region of Core and Polymerase genes, the replication-competent HBV vectors can be successfully constructed, which allowed production of HBV vectors carrying at least around 400 bp and possibly up to 720 bp of foreign genetic information yet maintaining replication competence and even infectivity.
Research motivation
Regarding the replication-competent HBV vectors, the pCH-BsdR carries blasticidin resistance gene (399 bp), the replication efficiency is higher, but it is tedious to use. The pCH-hrGFP carries humanized renilla green fluorescent protein gene (720 bp), but the replication efficiency is poor and could not be quantified. Hence, we tried to use the secreted luciferase (secNLuc) report gene(597bp) as foreign genetic inserted to the replication-competent HBV vector, which is convenient and quantifiable for the further research.
Research objectives
The secNLuc report gene can express luciferase protein that is secreted in culture supernatant,which is beneficial for monitoring transcriptional activation of target gene. We utilized this report gene to construct the other replication-competent HBV vector which can generate secNLuc recombinant HBV particles, and to establish quantifiable and standardized HBV replication cell lines that can stably secret recombinant HBV particles.
Research methods
We utilized the replication-competent HBV viral vectors constructed by our laboratory,combined with secreted luciferase reporter gene, to construct replication-competent HBV vectors expressing the reporter gene secretory Nanoluc Luciferase (SecNluc). HepG2.TA2-7 cells were transfected with this vector, to obtain cell lines that can stably secret HBV particles carrying secNluc report gene.
Research results
We successfully constructed a replication-competent HBV vector carrying SecNluc reporter gene,pCH-sNLuc and pTRE-sNLuc, and successfully obtained quantifiable and standardized HBV replication cell lines, HBV-NLuc-35 cells. The former could produce all major viral RNAs and full set of envelope proteins, and achieve high level expression of secreted luciferase. The latter could secret secNLuc recombinant viruses that are sensitive for existing anti-HBV drugs. Using differentiated HepaRG cells, it was verified that recombinant HBV possessed infectivity.
Research conclusions
Despite that the organization of HBV genomes is extremely compact, the replication-competent HBV vector can be successfully constructed by redesigning HBV genomes to fully maintain the HBV genetic information and creating transgene insertion site to have a minimal impact on HBV replication competence. The recombinant HBV pgRNA carrying small-to-medium-sized transgenes can be packaged. By carefully redesigning its intricate genome organization, HBV can be harnessed into a replication-competent infectious vector bearing substantial additional genetic information. HBV genomes can be reformed to have a minimal impact on HBV replication competence and expression. Numerous available reporter and effector genes meet the apparent size limit of 500-700 bp. In addition, viral-based vectors could be highly used for drug screening.With the increase in transgene size, the replication efficiency of HBV vectors gradually decreases compared with that of wild-type HBV. Some medium-sized transgenes about 500 bp are compatible with replication competence. The replication-competent HBV vectors carrying appropriate transgenes can be expected to find numerous applications, from further unraveling the molecular mechanism of HBV infections, including the involved host factors, to the identification of infectable cells and new antiviral drugs.
Research perspectives
Given strict hepatocyte tropism, the convenient HBV infection model has met with very limited success. The replication-competent HBV vectors carrying transgenes will provide a helpful tool for this research. The replication-competent HBV vectors carrying transgenes could be utilized to establish animal models of HBV infection. HBV genomes could be reformed to overcome species specificity.
杂志排行
World Journal of Gastroenterology的其它文章
- Gut-liver axis signaling in portal hypertension
- Role of tristetraprolin phosphorylation in paediatric patients with inflammatory bowel disease
- STW 5 is effective against nonsteroidal anti-inflammatory drugs induced gastro-duodenal lesions in rats
- Superior gallstone dissolubility and safety of tert-amyl ethyl ether over methyl-tertiary butyl ether
- Gender differences in vascular reactivity of mesenteric arterioles in portal hypertensive and non-portal hypertensive rats
- Differentially expressed long noncoding RNAs and regulatory mechanism of LINC02407 in human gastric adenocarcinoma
