Detection of Drug Resistance in Escherichia coli Isolated from Animals and ERIC Analysis of Multidrug-resistant Strains
2019-09-10XiaoxiaoDUANYuqianHEQizhengBINGShufengLIULinghongLIUWeishanCHANGYanLI
Xiaoxiao DUAN Yuqian HE Qizheng BING Shufeng LIU Linghong LIU Weishan CHANG Yan LI
Abstract The drug resistance of Escherichia coli from animals was detected in Shandong Province in 2016, the gene mapping of Enterobacterial Repetitive Intergenic Consensus PCR (ERIC PCR) of multidrug resistant isolates were analyzed, and then the relationship between ERIC PCR genotyping and drug resistance of highly resistant E. coli with multidrug resistance was discussed. A total of 110 E. coli isolates were separated and identified from diseased swine and avian, and their resistance to 10 kinds of antimicrobials was determined by the agar dilution method. Twenty highly resistant isolates with multidrug resistance were selected to carry on the ERIC PCR, followed by cluster genetic analysis according to the DNA fingerprints. The swine sourced E. coli isolates possessed serious resistance against several kinds of antimicrobial agents, for example, all isolates were tolerant to florfenicol, doxycycline and ampicillin (100%), but had a relative lower resistance rate to cefotaxime sodium (57.14%). The same situation was observed in the poultry sourced E. coli isolates, which had a resistance rate of 95.51% against florfenicol, while the lowest rate of 61.80% appeared on ciprofloxacin. Analysis of ERTIC showed dispersive fingerprint patterns of highly resistant and multidrug resistant isolates which represented a multiple clone resources, and there were certain correlation between the B genotype and the drug resistant characteristics. It indicated that the animal sourced E. coli isolates had a high level of drug resistance and were multidrug resistant, which meant there was severe antibiotic resistance against not only different kinds of antibiotics but also different drugs of the same kind. The highly resistant E. coli isolates with multidrug resistance had no apparent species preference, while their spread and pervasion posed a potential threat to the development of animal husbandry and the public health security.
Key words Escherichia coli; Drug resistance; ERIC PCR; Genotype
Escherichia coli is mostly a normal flora of humans and animals, but is a conditional pathogen under adverse conditions, and certain species are highly pathogenic and can cause severe diarrhea, septicemia, etc.[1]. In recent years, with the intensive development of aquaculture, the cultured animals are in a highly nutritious high density raising environment, so their autoimmunity is low. Plus various stress responses and vaccine protection are incomplete, infection is highly prone to occur. E. coli can become conditional pathogens easily as a common flora, causing E. coli infection and harm to the breeding industry. From the incidence of diseases, E. coli is the worlds first bacterial disease for animals, and has become one of the most common and difficult diseases in modern aquaculture, causing huge economic losses to the entire animal husbandry[2].
At present, the use of antibiotics is still the primary method to prevent E. coli disease in veterinary clinic. Numerous studies at home and abroad have shown that the large E. coli is highly prone to the formation of drug resistance. As a result, the drug resistance rate is gradually increasing; the drug resistance spectrum is rapidly broadened; and the formation and propagation speed of drug resistance is also increasing[3]. At the same time, the non standard use of antibacterial drugs not only exacerbates the residues in livestock and poultry, but also accelerates the spread of drug resistant strains or drug resistant gene elements among different species, thereby bringing hidden dangers and potential threats to the healthy and rapid development of Chinas animal husbandry and public health security[4]. In addition, the national management of antibiotics is in the process of exploration. For example, for different antimicrobial drugs of the same type, different use objects and scopes are currently divided, while whether this measure can effectively block the emergence and spread of drug resistance remains to be further studied.
Enterobacterial repetitive intergenic consensus (ERIC) PCR has attracted peoples attention because of its simple operation, easy implementation, high reproducibility and strong resolution[5]. In this study, the study swine and poultry sourced E. coli were collected from diseased animal samples and tested for their resistance to commonly used antibiotics in 2016; and meanwhile, the cross resistance between different drugs of the same category that differentiated between veterinary and medical use were investigated, and the relationship between the ERIC PCR map of the highly resistant E. coli isolates with multidrug resistance and their species specificity and transmission and between the their drug resistance and ERIC PCR genotyping were discussed by the ERIC PCR method. This study is of certain significance to the understanding of the antibiotic level in E. coli and the cross drug resistance between drugs, and the molecular biology research on the drug resistance mechanism of drug resistant isolates.
Materials and Methods
Experiment materials
Susceptible standard E. coli strain ATCC 25922 was preserved by the Public Health Laboratory of the Institute of Animal Husbandry and Veterinary Medicine, Shandong Academy of Agricultural Sciences.
Medium: E. coli chromogenic medium ECC, purchased from Shanghai Central Bio engineering Co., Ltd.; MHA agar and MHB broth medium, purchased from Beijing Land Bridge Technology Co., Ltd.
Reagents: PCR buffer, dNTP and Taq DNA polymerase, purchased from Tiangen Biotech (Beijing) Co., Ltd.; DL5000DNA Marker, purchased from TaKaRa Biotechnology (Dalian) Co., Ltd.
Antibiotics: Aminoglycosides: gentamicin (CN); tetracyclines: doxycycline (DOX); chloramphenicols: florfenicol (FCC); β lactams: ampicillin (AMP), ceftriaxone (CRO), ceftiofur (CEF) and cefotaxime (CEQ); fluoroquinolones: enrofloxacin (EN), ciprofloxacin (CIP) and levofloxacin (LVX). The above reagents were purchased from Beijing Pu Boshin Biotechnology Co., Ltd.
Methods
E. coli isolation and identification
The diseased chicken and swine viscera and fecal swab samples collected from farms in different regions of Shandong Province in 2016 were streaked on E. coli chromogenic medium, and single blue suspicious colonies were picked after incubation at 37 ℃, and inoculated in non selective MHA agar medium. Then, the cultured bacteria were identified by MALDI TOF MS time of flight mass spectrometer (VETIK MS, France). The positive E. coli isolates were stored in 30% glycerin broth at -20 ℃ for later use.
E. coli drug sensitivity test
According to standards of the Clinical and Laboratory Standards Institute (CLSI), the isolates were tested for drug sensitivity to 10 drugs by the agar dilution method[6]. The drug resistance rate of each strain was determined, and the drug resistance of the poultry and swine sourced E. coli isolates and the multi drug resistance and cross resistance of each isolate were statistically analyzed.
ERIC Detection
Primers (F: 5′ ATGTAAGCTCCTGGGGATTCAC 3′, R: 5′ AAGTAAGTGACTGGGGTGAGCG 3′) were designed according to reference [7] and synthesized by Sangon Biotech (Shanghai) Co., Ltd.
According to the results of the drug sensitivity test, 10 poultry sourced highly resistant E. coli isolates and 10 swine sourced highly resistant E. coli isolates with multidrug resistance were picked and extracted for DNA, which was used for ERIC PCR amplification. The PCR was started with pre denaturation at 95 ℃ for 7 min, followed by 30 cycles of denaturation at 90 ℃ for 30 s, annealing at 52 ℃ for 1 min and extension at 65 ℃ for 8 min, and completely by extension at 65 ℃ for 16 min, obtaining the product which was preserved at 4 ℃. The ERIC fingerprints were recorded according to the electropherogram and analyzed for DNA polymorphism. Band was recorded as "1", and no band was recorded as "0". The DNA polymorphism data were formed into a data matrix, which was used for cluster genetic analysis. The similarity>0.75 indicated the same genotype, and the similarity<0.75 meant different genotypes[8].
Results and Analysis
E. coli isolation and identification
After isolation and identification on the collected samples, a total of 110 E. coli isolates were obtained, including 21 swine sourced E. coli isolates and 89 poultry sourced E. coli isolated.
Detection of drug resistance in E. coli
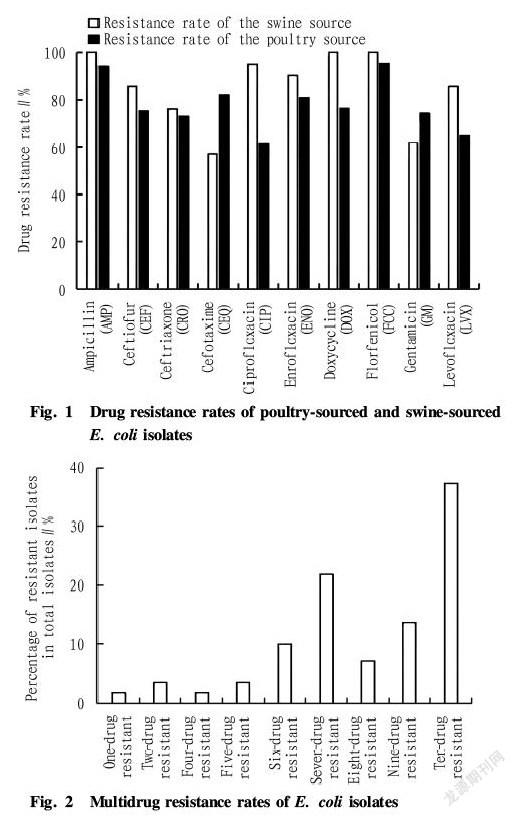
The drug sensitivity test was carried out on the isolated E. coli, and the results are shown in Fig. 1. It can be seen that the resistance rates of all strains were all higher, and swine sourced E. coli had the resistance rate to doxycycline, ampicillin and florfenicol reaching 100%, the resistance rate to ciprofloxacin and enrofloxacin higher than 90%, and relatively low resistance to cefotaxime, also at 57.14%. The poultry sourced E. coli had the resistance rate to florfenicol and ampicillin as high as 90%, and a lower resistance rate to ciprofloxacin, which was 61.80%.
Multidrug resistance in E. coli
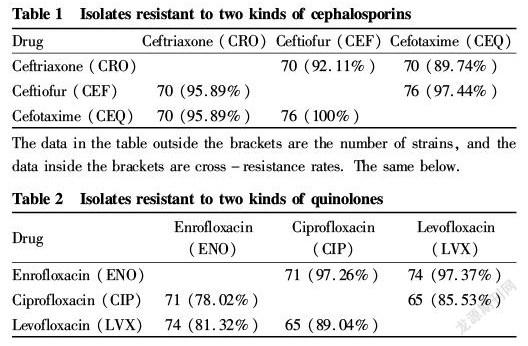
The results of the drug sensitivity test on the 110 E. coli isolates were statistically analyzed, and the multidrug resistance is shown in Fig. 2. It can be seen that no isolates were completely resistant to drugs, only one isolate was resistant to one antimicrobial agent, and other isolates showed resistance against two or multiple antimicrobial drugs, among which 41 isolates were resistant against 10 drugs (37.27%), 62 isolates (5.66%) were resistant against 6-9 antibiotics, and the isolates resistant to 1-5 drugs only accounted for 6.36% of the antimicrobial drugs.
Cross resistance against similar antibiotics in E. coli
In this study, three antibiotics belonging to the third generation cephalosporin, namely ceftiofur, cefotaxime and ceftriaxone were selected. Among them, the former is animal specific antibiotic, and the latter two are medical antibiotics.
According to the calculation formula of cross resistance rate[9]: Cross resistance rate of drug A to drug B (%) = Number of isolates resistant to drugs A and B/Total number of isolates resistant to drug B×100, the cross resistance rate between any two cephalosporins was shown in Table 1. There was very significant cross resistance between the cephalosporin antibiotics, and the cross drug resistance of any two of the antibiotics was mostly at 95%. The medical antibiotic and the animal antibiotics were not different in the resistance rate in E. coli isolates. Among the 110 E. coli isolates, there were 81 isolates resistant to one or more cephalosporin drugs, of which 70 isolates were resistant to all the three cephalosporins, and the rate was 86.4%. Similarly, three quinolone drugs were selected in the study, namely enrofloxacin, ciprofloxacin and levofloxacin. The former is animal specific and the latter two are medical. According to the calculation formula of cross resistance rate, the cross drug resistance rate between the quinolones was calculated, and the results are shown in Table 2. It is shown that the different quinolone antibiotics also had cross resistance. The cross resistance of any two of the antibiotics was higher than 80%. Among the 110 E. coli isolates, there were 95 isolates resistant to one or more quinolone drugs, of which 65 strains were resistant to all the three cephalosporin drugs, and the rate was 68.40%.
Analysis of ERIC fingerprints
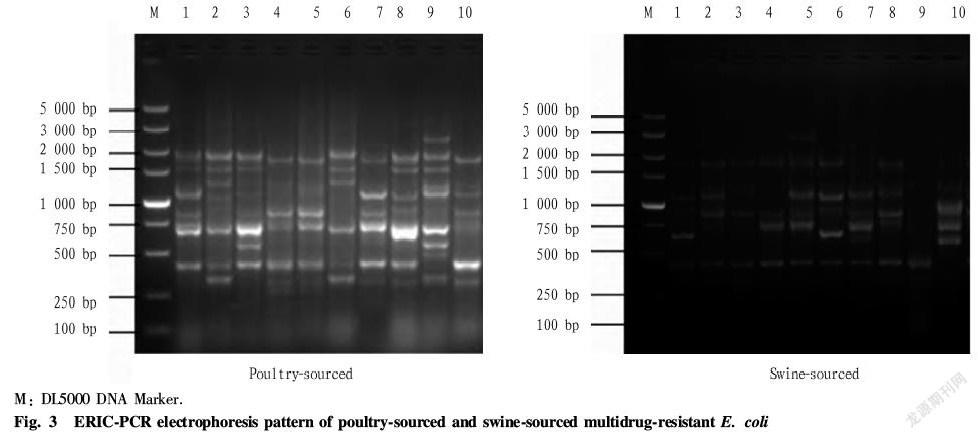
According to the comprehensive evaluation of the results of the drug susceptibility test, 10 poultry sourced and 10 swine sourced highly resistant isolates with multidrug resistance were selected, and subjected to multiple times of ERIC PCR, thereby obtaining stable fingerprints. There were about 1-8 bands of ERIC PCR fingerprints, which mainly ranged from 200 to 3 000 bp, among which those at about 400 and 1 800 bp were the main bands; and the swine sourced and poultry sourced fingerprints shared no significant homology. The fingerprints of the 4 and 5 lanes of the poultry source were almost identical, indicating that the 4 and 5 isolates may belong to the same strain; no obvious patterns were found in the swine sourced fingerprints; and the poultry and swine sourced DNA fingerprints showed a total of 19 ERIC patterns, as shown in the Fig. 3.
Cluster analysis of DNA polymorphism
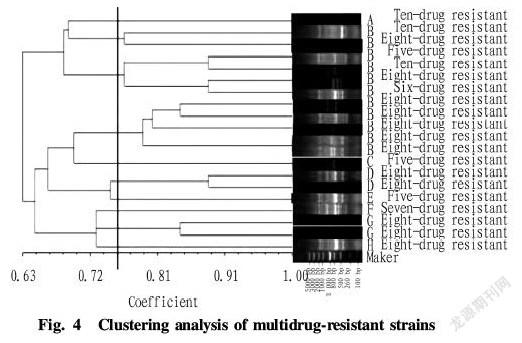
According to the cluster analysis tree (Fig. 4), the 20 resistant isolates included eight ERIC PCR genotypes and were not associated with the poultry or swine source. Among them, 55% of the resistant isolates showed the same ERIC PCR genotype, i.e., type B, and 63.64% of these isolates showed the same drug resistance pattern, that is, resistant to all of the 10 antibiotics. A total of 10 isolates of the selected isolates were resistant to the 10 antimicrobial agents, among which 7 were of the B ERIC PCR genotype.
Discussion and Conclusions
In recent years, the number of resistant E. coli isolates has increased, and a large number of bacteria with multidrug resistance have appeared. Studies at home and abroad have shown that drug resistant E. coli is distributed worldwide[10-11]. Lei[12], Yu[13] and He et al.[14] all reported that the drug resistance of E. coli from animal sources was dominated by multidrug resistance. The results of this study found that only one isolate of the 110 E. coli isolates was a single drug resistant isolate, and the rest were multidrug resistant isolates. The multidrug resistant isolates in this study were highly concentrated, which might be due to the large scale use of antimicrobial drugs in the veterinary clinic to treat bacterial infections, or the addition of antibiotics in the feed to promote growth and prevent diseases, resulting in the long term emergence of more and more significant resistance in E. coli under drug selection pressure[15]. At the same time, the emergence of a large number of drug resistant bacteria might be caused by the drug resistant genes spreading between bacteria which carry plasmids that mediate drug resistance. According to research reports, the emergence of multidrug resistant bacteria may be the result of changes in cell membrane permeability and increased expression of the active efflux system in bacteria[16-17].
This study showed that the isolated E. coli not only developed resistance to various antibiotics, but also had serious cross resistance to different drugs of the same type of antibiotics, such as cephalosporins and quinolones, and the isolates were highly resistant to medical antimicrobials. Although this phenomenon may be related to nonstandard use of drugs in the farm, the same type of antimicrobial drugs with similar action mechanism and therapeutic effect cannot be used in large quantities and repeatedly. Therefore, the phenomenon of high cross resistance should be highly valued. A drug produces drug resistance, and other drugs of the same type may cause cross resistance. Therefore, it is impossible to prevent the spread and pervasion of drug resistant strains by only separating animal specific antimicrobial drugs from the medical antimicrobial drugs, regardless of the cross resistance problem. The conclusion of this study that cross resistance exists between similar antibiotics agrees with the result reported by Li et al.[18] that there is cross resistance between quinolones. The in vitro induced resistance tests of quinolone antimicrobial agents prove that the cross resistance between these drugs is due to the same parent structure. Similarly, the resistance between cephalosporin antibiotics is due to the same cyclic structure[19]. The detection of cross resistance can reflect the actual situation of veterinary clinical drug resistance and lay a foundation for veterinary clinical guidance medication, and meanwhile, cross resistance brings challenges and potential threats to the public health field.
Pathogens bring high risks to the breeding industry, and especially, the multi resistant highly resistant strains bring great harm to the actual production. This paper discussed the relationship between the phenotypes of multi resistant highly resistant isolates and their DNA fingerprints, analyzed the relationship between the homology of drug resistant isolates and their drug resistance phenotype, and the ERIC PCR genotype, and explored the similarities between the resistance mechanisms of drug resistant isolates. It was found from this study that the patterns of the tested isolates were very dispersive, and there was no obvious species specificity between the poultry and swine sourced E. coli, which indicates that the isolates can spread between different species easily, and thus increase the possibility of diseasing different species including humans. The results also showed a certain degree of correlation between strain resistance and specific ERIC PCR genotypes, specifically the maximum overlap and coverage of the multi resistant high resistant isolates resistant to all the 10 drugs with the B type ERIC PCR genotype. It indicates that E. coli of this genotype is easy to form multidrug resistance, or that this genotype of E. coli has strong ability to spread and pervade, posing a potential threat to the aquaculture and the world public health security.
The statistical analysis of the relationship between the drug resistance in animal sourced E. coli and ERIC genotypes in resistant strains helps to detect E. coli resistance from multiple aspects including E. coli phenotype and molecular level. In this study, DNA fingerprinting was correlated with drug resistance and homology, laying a foundation for the study on the molecular mechanism of the drug resistance in drug resistant strains. The use of ERIC PCR technology to study the relationship between DNA fingerprint and drug resistance phenotype requires more data support, and further research is needed.
References
[1] ORTH D, GRIF K, ZIMMERHACKL LB, et al. Prevention and treatment of enterohemorrhagic Escherichia coli infections in humans[J]. Expert Rev. Anti. Infect. Ther., 2008, 6: 101-108.
[2] AN WEI, ZHANG XY, LI R, et al. Advances in research on pathogenic Escherichia coli virulence factors and drug resistance[J]. Animal Husbandry and Veterinary Medicine, 2013, 45(8): 106-110. (in Chinese)
[3] TITILAWO Y, SIBANDA T, OBI L, et al. Multiple antibiotic resistance indexing of Escherichia coli to identify high risk sources of faecal contamination of water[J]. Environ. Sci. Pollut. Res. Int., 2015, 22: 10969-10980.
[4] LIN JC, ZHUO JZ, JIANG HX, et al. Surveillance of antimicrobial resistance among Escherichia coli isolates from swine and poultry in different regions[J]. Journal of South China Agricultural University, 2009, 30(1): 86-88. (in Chinese)
[5] HULTON CS, HIGGNSC F. ERIC sequences: a novel family of repetitive elements in the genomes of Escherichia coli, Salmonella typhimurium and other enterobacteria[J]. Journal of Molecular Microbiology and Biotechnology, 1991, 5: 825-830.
[6] Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing: 18th informational supplement [S]. CLSI Document M100 S18, 2008.
[7] VERSALOVIC J, KOEUTH T, LUPSKI JR. Distribution of repetitive DNA sequences in eubacteria and application to finerpriting of bacterial genomes [J]. Nucleic Acids Research, 1991, 19 (24): 6823-6831.
[8] SANDVANG D, AARESTRUP FM, JENSEN LB. Characterisation of integrons and antibiotic resistance genes in Danish multiresistant Salmonella enterica Typhimutium DT104 [J]. FEMS Microbiology Letters, 1998, 160(1): 37-41.
[9] ZHANG JF, LI YL, CHEN XH, et al. Studies on cross resistance of avain Escherichia coli to fluoroquinolones[J]. Guangdong Agricultural Sciences, 2005(2): 74-76. (in Chinese)
[10] CHAIEB K, ABBASSI MS, TOUATI A, et al. Molecular characterization of Staphylococcus epidermidis isolated from biomaterials in a dialysis service[J]. Annals of Microbiology, 2005, 55: 307-312.
[11] MATHAI E, CHANDY S, THOMAS K, et al. Antimicrobial resistance surveillance among commensal Escherichia coli in rural and urban areas in Southern India[J]. Trop. Med. Int. Health, 2008, 13(1): 41-45.
[12] LEI LC, ZHENG D, HAN WY, et al. Antibiotic sensitivity detection and analysis of Escherichia coli isolated clinically in China[J]. Chinese Journal of Veterinary Science, 2005, 25(5): 470-473. (in Chinese)
[13] SUI H, YANG JS. Isolation and identification of animal derived multidrug resistant Escherichia coli[J]. Chinese Journal of Veterinary Medicine, 2013, 49(6): 48-50. (in Chinese)
[14] HE DD, HUANG LZ, CHEN XJ, et al. Investigation of antibiotic resistance among Escherichia coli isolated from different animals[J]. China Animal Husbandry and Veterinary Medicine, 2013, 40(10) : 211-215. (in Chinese)
[15] ZHANG CP, NING YB, SONG L. Resistance to tetracycline and distribution of tetracycline resistance determinants in commensal Escherichia coli isolated from clinically healthy chickens and pigs[J]. Scientia Agricultura Sinica, 2010, 43(12): 2578-2583. (in Chinese)
[16] ZHAO J, YANG HC, ZHA XL. Monitoring of Escherichia coli resistance in large scale pig farms[J]. Chinese Journal of Veterinary Medicine, 1998, 24(11): 12-13. (in Chinese)
[17] ZHANG XL, LI JT. Distribution and expression of active efflux system in Escherichia coli[J]. The Chinese Journal of Clinical Pharmacology, 1999, 15(3): 171-174. (in Chinese)
[18] LI YH, QU F, BAO CM, et al. Resistance and cross resistance of intestinal bacteria to fluoroquinolones[J]. Chinese Journal of Antibiotics, 2009(4): 251-253. (in Chinese)
[19] BARRY AL, JONES RN. Cross resistance among cinoxacin, ciprofloxacin, DJ 6783, enoxacin, nalidixic acid, no rfloxacin, and oxolinic acid after in vitro selection of resistant populations[J]. Antimicrob. Agents Chemo. Ther., 1984, 25: 775-777.
杂志排行
农业生物技术(英文版)的其它文章
- Study on Chemical Composition of the Ethyl Acetate Extract of Pratia
- Effects of Different Selenium Fertilizer Types on Selenium Content and Quality of “Lingfeng” Grapes
- Protoplast Culture and Its Application in Fruit Breeding
- Indexes of Tree Structure of Cylindrical Pear Orchards at the Sapling Stage
- Carbon Storage and Distribution of the Mature Pinus massoniana Plantation in Northwest Guangxi
- Pollution and Quality Control of Mycotoxins in Foods and Feeds
