Cry1Ah蛋白标准物质候选物的制备与纯化
2019-08-27郭林耿丽丽孙晓晓王美玲束长龙张杰
郭林,耿丽丽,孙晓晓,王美玲,束长龙,张杰
Cry1Ah蛋白标准物质候选物的制备与纯化
郭林1,2,耿丽丽2,孙晓晓2,王美玲2,束长龙2,张杰2
1 吉林农业大学 农学院,吉林 长春 130118 2 中国农业科学院植物保护研究所 植物病虫害生物学国家重点实验室,北京 100193
随着转基因技术的迅猛发展,转基因产品的安全性受到了广泛关注。转基因检测用有证标准物质在确保转基因产品定性、定量检测结果的可比性和可追溯性方面发挥着重要作用。但转基因蛋白质标准物质的开发相对缓慢,其中一个难点是制备高纯度的转基因蛋白质候选物。苏云金芽胞杆菌基因因其对亚洲玉米螟等鳞翅目害虫有很好的杀虫活性,已用于转基因抗虫作物的研制,并获得具有较好抗虫性状的转基因株系。为了研发Cry1Ah蛋白有证标准物质,亟需建立其制备及纯化体系。文中优化了利用Bt表达系统制备Cry1Ah蛋白的体系,利用离子交换色谱法和排阻色谱法逐级纯化的方法,获得了高纯度的Cry1Ah蛋白(排阻色谱纯度:99.6%)。生物活性测定结果表明,纯化的Cry1Ah蛋白与原毒素对小菜蛾的杀虫活性没有显著差异。最后使用Edman降解法和质谱法确定了Cry1Ah蛋白活化后的氨基酸序列。综上所述,获得的Cry1Ah纯蛋白可用于蛋白质标准物质的研制。
转基因生物,有证标准物质,苏云金芽胞杆菌,Cry1Ah蛋白
Since the United States developed the first transgenic plants in 1983, transgenic technology has received extensive attention due to the wide range of applications[1-2]. In the past 22 years of commercialization of biotech crops (1996 to 2017), the accumulated global area of biotech crops surged to a record 2.34 billion hectares, and the income generated by farmers increased by an additional US $204.3 billion. These results confirmed that biotech crops could yield huge economic, environmental and social benefits[3].
In the meantime, genetically modified (GM) product safety has been drawing increasing attention; therefore, qualitative and quantitative tests of GM products have become quite important[4-5]. The detection reagents for GM products are generally the nucleic acid sequence of the exogenously inserted genes or their encoded protein. The test results can be made more comparable and reliable by using genetically modified organism certified reference material (GMO CRM)[6-7]. Several GMO CRMs, including matrix reference material and DNA reference material, had been made, especially by the Institute for Reference Materials and Measurements and the American Oil Chemists Society; the transgenic crops involved were corn, soybeans, rape, cotton, etc.[8-13]. In the past 20 years, various national metrology institutes have been working on the development of protein reference material and have established an isotope dilution mass spectrometry method for the quantification of proteins[14-16]. At present, protein reference material, such as bovine serum albumin (7% solution), human angiotensinⅠ, human cardiac troponin complex, C-reactive protein solution and troponinⅠ, have been developed[17-21]. The development of these reference materials supports the accurate and comparable results of protein content determination in domestic, clinical, food and other fields. However, the development of GMO protein reference material remained absent. Protein with sufficient purity was the key element in the development of a protein reference material; therefore, it is especially necessary to establish a purification method for transgenic proteins in order to develop protein reference material.
(Bt) genes have been widely used in GM crops to control insect pests. Thegene cloned from Bt[22-23]encodes a protein with high insecticidal toxicity to several lepidopteran pests such as(LC50=0.05 μg/g),(LC50=1.48 μg/g) and(LC50=0.98 μg/g). At present, thegene has been transformed into maize, showing powerful resistance against some lepidopteran pests[24]. Moreover, risk assessment studies have been carried out on both Cry1Ah protein andtransgenic crops, and no adverse effects were found on the survival, development, behavior or bacterial diversity in the midgut of honeybees[25-28]. In this study, a purification protocol for Cry1Ah protein was established. In addition, the amino acid sequence was determined for subsequent preparation of the protein reference material using isotope dilution mass spectrometry.
1 Materials and methods
1.1 Materials
The Biot1Ah (HD73 strain transformed with pSXY422-1Ah vector containing thegene)[22]was stored by our lab. Trypsin (1:250) was purchased from Amresco (USA). Trifluoroacetic acid (TFA) and dithiothreitol (DTT) were obtained from Sigma-Aldrich (USA). High-performance liquid chromatography (HPLC) grade acetonitrile (Merck, Germany) was used for the mobile phase. HiTrap Q FF, Resource Q and HiLoad 26/600 Superdex 75 pg chromatography columns were purchased from GE Healthcare (Sweden). TSK gel G2000SWXL and Zorbax 300SB-C8 chromatography columns were purchased from Tosoh (Japan) and Agilent (USA), respectively. All other chemicals were of analytical grade.
1.2 Protein extraction and activation
Cry1Ah protein was extracted by the repeated crystal solubilization method[29]. The strain was grown in 1/2 LB medium at 30°C and 220 r/min until 50% of the crystals were released. After centrifugation at 8 200 ×for 10 min at 4 °C, the precipitate was collected and washed with 1.0 mol/L NaCl, and then washed with distilled water. The precipitate was resuspended in 50 mmol/L Na2CO3(pH 9.5) containing 20 mmol/L DTT and incubated on ice (with shaking at 85 r/min) for 4 h. The supernatant was collected by centrifugation at 18500 ×for 15 min at 4 °C, 3.0 mol/L NaAc-HAc (pH 4.5) was added until the pH of the solution reached 5, and the resulting solution was kept at 4 °C for at least 1 h. After centrifugation at 18 500 ×for 15 min at 4 °C, the precipitate was collected and washed twice with precooled distilled water, then dissolved in 50 mmol/L Na2CO3(pH 9.5). The final product was Cry1Ah protein. After analyzing the Cry1Ah protein by SDS-PAGE, the corresponding gel bands were excised and washed twice with purified water. Then, the gel bands were washed twice for 10 min each with 1.0 mL of the decolorizing solution (50% acetonitrile, 25 mmol/L ammonium bicarbonate). Dehydration was carried out by adding acetonitrile until the gel bands were completely whitened. Acetonitrile was removed by vacuum drying. Gel bands were completely absorbed by the addition of 10 mmol/L DTT and incubated for 1 h at 56 °C. Excess DTT was removed by pipette, and 55 mmol/L iodoacetamide was added and incubated for 45 min in the dark. Excess iodoacetamide was removed by pipette and the samples were washed with 25 mmol/L ammonium bicarbonate twice for 10 min each. The ammonium bicarbonate was removed and the samples were added to the decolorizing solution for 10 min, repeated once. The above dehydration step was repeated. Then, 0.1 mg/mL trypsin was added for digestion overnight at 37 °C. Finally, the digestion was terminated by the addition of formic acid at a final concentration of 0.1%. The instrument MicroTOF-QII was used for mass spectrometry detection of the treated gel bands, and Mascot software was used for data processing analysis.
The Cry1Ah protein was activated by trypsin (1:20, 1:10, 1:5, 1:2, 1:1, 2:1, 5:1, 10:1, and 20:1;/) at 37 °C for 2 h. Then, the Cry1Ah protein was activated under different levels of glycerol (0%, 5%, 10%and 15%). To compare the activation efficiency, all the samples were collected and analyzed by SDS-PAGE[30].
1.3 Protein purification
First, ion exchange chromatography (IEC) was performed using a HiTrap Q FF column. The activated Cry1Ah protein was centrifuged twice at 18 500 ×for 10 min and then loaded onto the column that had been pre-equilibrated with buffer containing 50 mmol/L Na2CO3(pH 9.5). Unbound impurities were removed by washing with 3 column volumes of the Na2CO3buffer. Afterwards, the protein was reverse eluted using a gradient of 0–1.0 mol/L NaCl. Peak fractions were collected based on UV absorbance at 280 nm. In this step, the flow rate was 2.0 mL/min.
Second, a HiLoad 26/600 Superdex 75 pg column was used for size exclusion chromatography (SEC) to further purify proteins. Using a buffer containing 50 mmol/L Na2CO3(pH 9.5) at a flow rate of 2.6 mL/min, the target protein was collected and the small molecule impurities bound to the Q FF column in the previous step were removed.
Then, a Resource Q column for IEC was used to purify proteins. At a flow rate of 1.0 mL/min, the protein was loaded onto the pre-equilibrated column. Next, 20 column volumes were eluted with a gradient of 0−1.0 mol/L NaCl. Target proteins and nontarget proteins were effectively separated.
Finally, a HiLoad 26/600 Superdex 75 pg column for SEC was used to further purify proteins and exchange the buffer for 200 mmol/L NH4HCO3(pH 8.0).
1.4 Purity identification
To determine protein purity, the purified protein was analyzed by SDS-PAGE. Then, the purity of purified Cry1Ah protein was determined by HPLC. An Agilent Zorbax SB300-C8 column was used for reversed-phase chromatography (RPC) purity analysis. The flow rate was 1.0 mL/min and the injection volume was 20 μL. Mobile phase A and B were water and acetonitrile containing 0.1% TFA, respectively, and the gradient is shown in Table 1. Protein purity can be monitored using wavelengths of 210 nm, 254 nm and 280 nm, and it was found that the wavelength of 254 nm makes it easier to determine the amount of impurities in RPC. Therefore, a wavelength of 254 nm was used to monitor the purity and a column temperature of 40 °C was used.
In addition, a Tosoh TSK gel G2000SWXL column was used for SEC purity analysis. The flow rate was 0.45 mL/min and the injection volume was 20 μL. The mobile phase was a 3:7 mixture of water and acetonitrile containing 0.1% TFA. Of the three wavelengths, 210 nm makes it easier to determine the amount of impurities in SEC. A wavelength of 210 nm was used to monitor the purity and a column temperature of 40 °C was used.
1.5 Amino acid sequence determination
The activated and purified Cry1Ah protein was used as a sample. N-terminal sequence analysis was performed based on Edman degradation[31]. The gel was transferred to a PVDF membrane using semidry blotting after analysis by SDS-PAGE, and then the PVDF membrane test sample was placed into a PPSQ-31A (from SHIMADZU) that was set for sequencing 5 amino acids. PPSQ-30 software was used for data processing analysis. C-terminal sequence analysis was performed based on mass spectrometry. Trypsin, chymotrypsin and staphylococcal protease were used to treat the sample separately (method references protein extraction and activation) and the C-terminal sequence of the protein was obtained by mass spectrometry results of the three protease treatments. The N/C-terminal sequence analysis results were compared with the full-length amino acid sequence to obtain the activated Cry1Ah protein amino acid sequence.
1.6 Insect bioassay
Toxicity was analyzed by leaf-dip bioassay[32]. This assay included exposure of diamondback moth () larvae to fresh cabbage leaves that had been dipped in 7 concentrations of Cry1Ah protoxins and pure Cry1Ah protein (activated and purified). Thirty 2nd-instar larvae were placed on a leaf disk (cut into a circle with a diameter of 60 mm) and the number of surviving larvae was recorded after 2 days. The bioassays were repeated at least three times and 50% lethal concentration (LC50) values were calculated by probit analysis[33-34].
2 Results and discussion
2.1 Protein extraction and activation
The Cry1Ah protein with a size of 134 kDa was extracted by alkaline solubilization. Atomic force microscopy showed that the bipyramidal shape of the crystal surface contained regular horizontal stripes (data not shown). At the sametime, the expressed protein was identified by mass spectrometry, and the data were analyzed using Mascot software for proteomics identification. The corresponding Score value and sequence coverage were 26296 and 77%, respectively. The identification result was Cry1Ah.
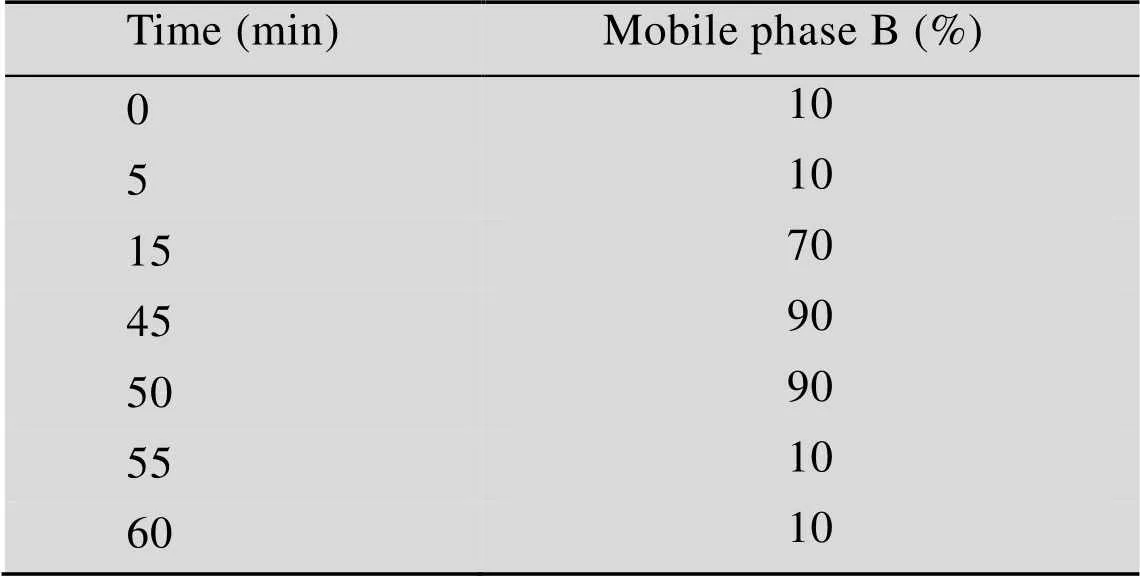
Table 1 The mobile phase gradient for Cry1Ah protein by RPC
Mobile phase B was acetonitrile containing 0.1% TFA.
The Cry1Ah protein was then treated with different amounts of trypsin (1:20, 1:10, 1:5, 1:2, 1:1, 2:1, 5:1, 10:1 and 20:1;/) at 37 °C for 2 h to form a fragment with a size of approximately 63 kDa. To obtain a single fragment for subsequent purification, a mass ratio of 10:1 was selected as the best activation condition (Fig. 1A). However, during the activation of Cry1Ah protein, most proteins precipitatedout (data not shown). To change this phenomenon, we optimized the activation conditions and added glycerol to promote the dissolution of the protein. Under these activation conditions, different proportions of glycerol (0%, 5%, 10% and 15%) were added. Compared with water, glycerol has a lower polarity and characteristic strong hydrophilic properties and hydrogen bond-forming ability. The addition of glycerol can reduce the polarity of the buffer, thereby preventing hydrophobic aggregation of the protein and increasing protein stability. At the same time, it can also reduce nonspecific adsorption during protein sample processing and improve the recovery rate. In the case of supplementation with 15% glycerol, the activated protein was effectively dissolved (Fig. 1B). The Cry1Ah protein activation conditions were determined as follows: the protein was activated by trypsin (protein:trypsin=10:1,/) with 15% glycerol at 37 °C for 2 h.
2.2 Protein purification and purity identification
The target proteins and impurities could not be effectively separated when the activated protein was directly purified by the IEC (data not shown). However, after removing most of the impurities, this situation was resolved. As shown in Fig. 2A, the HiTrap Q FF column was used to enrich the target protein and remove impurities that were not bound to the column. The HiLoad 26/600 Superdex 75 pg column was then used to remove impurities with molecular weights that are much larger or much smaller than the target protein (Fig. 2B). At this point, most of the impurities had been removed; on this basis, a Resource Q column with high resolution was used to achieve baseline separation of the target protein and impurities (Fig. 2C). Finally, a HiLoad 26/600 Superdex 75 pg column was used for further purification and replacement of the protein into the desired buffer (Fig. 2D). Compared to recombinant labeled proteins using affinity chromatography, this method results in high purification of natural proteins. After removing most of the impurities, a high-purity ion exchange resin can be used to obtain a highly pure target protein. This purification protocol promises to provide a novel method for the purification of natural proteins.
The purified protein was analyzed by SDS-PAGE. From the results of SDS-PAGE (Fig. 3A), Cry1Ah showed only one band, which proved that the obtained Bt protein achieved a high purity. The average purity of the Cry1Ah protein was95.5% with a standard deviation of 0.4% as determined by reverse-phase HPLC purity analysis (Fig. 3B). The purity of the Cry1Ah protein was further verified by size exclusion HPLC to check for the presence of degraded fragments in the macromolecule protein. The average purity was 99.6% and the standard deviation was 0.2% (Fig. 3C). This high-purity Cry1Ah protein sample can be used as a candidate for the development of protein reference materials.
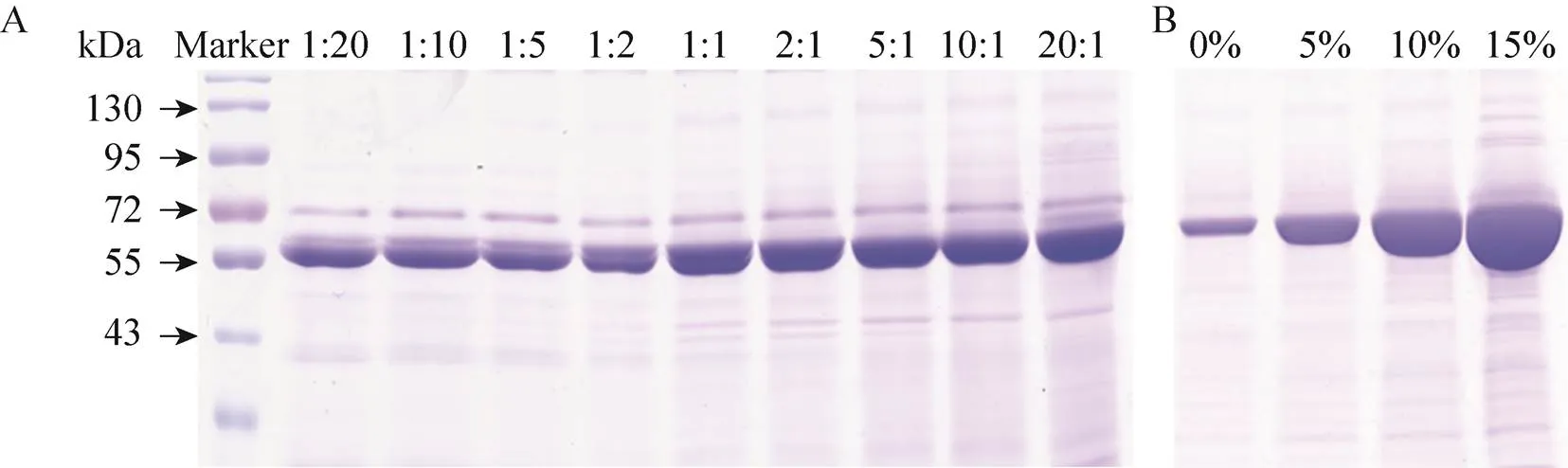
Fig. 1 Effects of different concentrations of trypsin and glycerol on the activation of Cry1Ah protein. (A) Effect of the amount of trypsin on activation results (protein trypsin = 1:20, 1:10, 1:5, 1:2, 1:1, 2:1, 5:1, 10:1 and 20:1; W/W). (B) Effect of the amount of glycerol on the dissolution rate of activated protein (0%, 5%, 10% and 15%).
The protein purity determined by reverse-phase HPLC was lower than that of SDS-PAGE and size exclusion HPLC which may be caused by a small amount of excessive activated proteins produced during the activation process. These proteins were close to the size of the target protein, and not completely separated using a Resource Q column.
2.3 Amino acid sequence determination
The advantages of isotope dilution mass spectrometry make it widely used in organic compound quantification and CRM development for goals including absolute protein quantification[35-37]. Accurate calculation of the protein content using the amino acid content determined by isotope dilution mass spectrometry requires identification of the amino acid sequence of the protein. In this study, we chose to separately analyze the N/C-terminus of the activated protein to obtain its amino acid sequence. The first five amino acids of the N-terminal sequence of Cry1Ah protein were determined using the Edman degradation method and the result was ISVGN (Fig. 4A). Simultaneously,the last enzymatic peptide(trypsin- or chymotrypsin-treated) of the C-terminal sequence of the Cry1Ah protein was determinedbymassspectrometry to be NFSGTAGVIIDR (Fig. 4B). The determined N/C-terminus sequence was compared with the full-length amino acid sequence to obtain an activated Cry1Ah protein sequence between amino acids 50I to 622R. The resulting sequence contains 573 amino acids with a molecular weight of 63962.73 Da, which is consistent with the approximately 63 kDa band observed by SDS-PAGE.
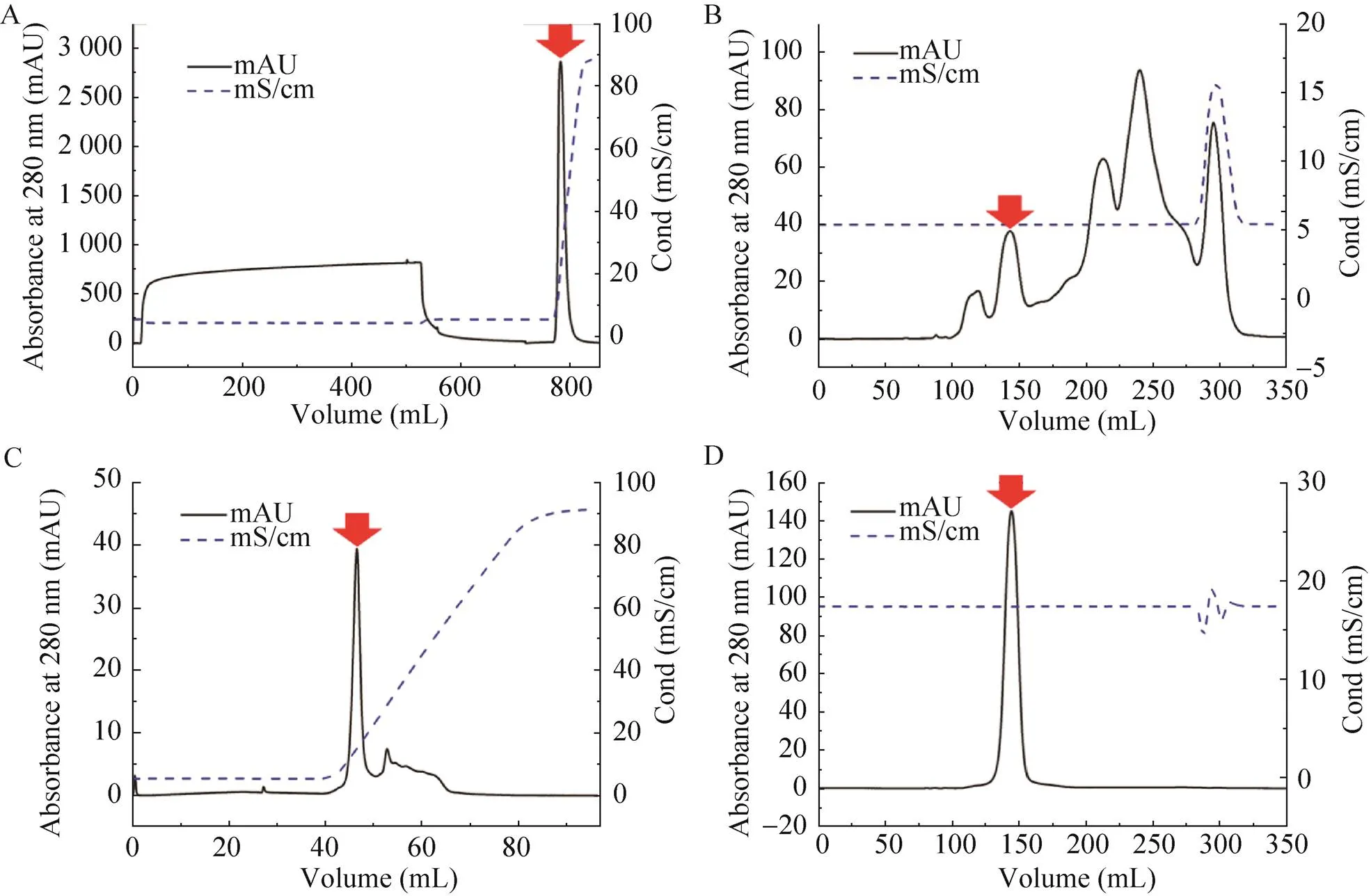
Fig. 2 Purification process of Cry1Ah protein. (A) The HiTrap Q FF column was used to enrich the target protein and remove impurities that were not bound to the column. (B) The HiLoad 26/600 Superdex 75 pg column was then used to remove impurities with molecular weights that are much larger or much smaller than the target protein. (C) The Resource Q column with high resolution was used to achieve baseline separation of the target protein and impurities. (D) The HiLoad 26/600 Superdex 75 pg column was used for further purification and replacement of the protein into the desired buffer. The arrows mark the target protein.
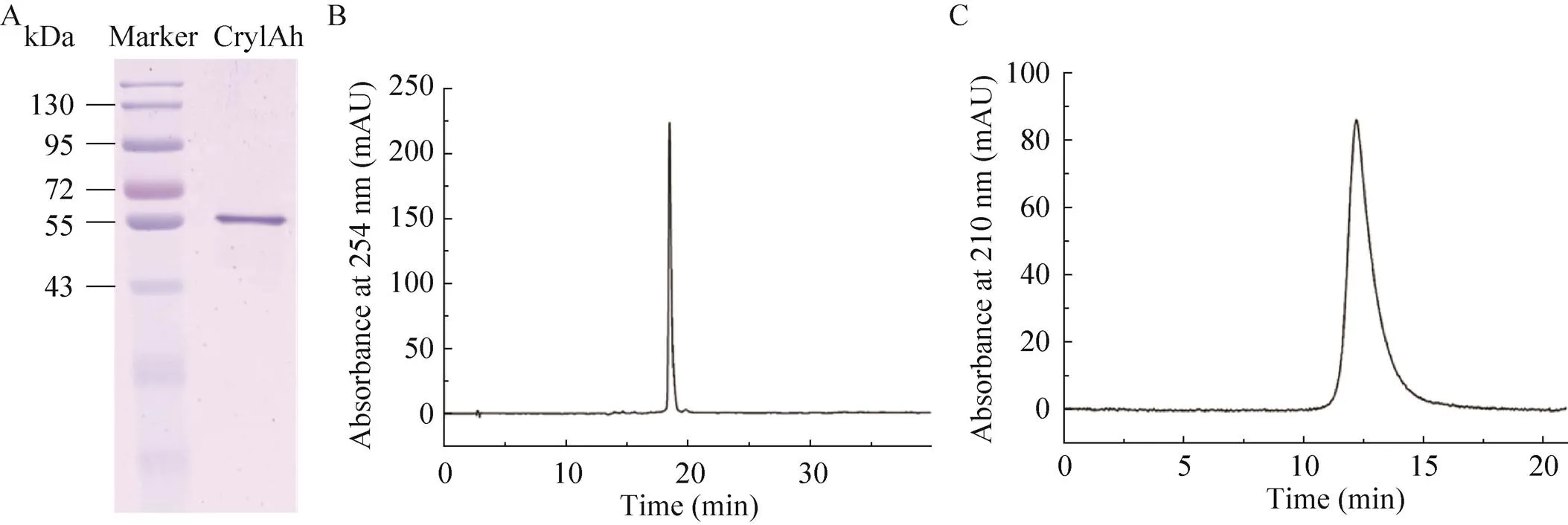
Fig. 3 Purity identification of Cry1Ah protein. (A) Gel electrophoresis result. (B) Reverse-phase HPLC diagram. (C) Size exclusion HPLC diagram.

Fig. 4 Sequence analysis of activated Cry1Ah protein. (A) N-terminal sequence analysis by Edman degradation. (B) C-terminal sequence analysis by mass spectrometry.
High-purity Cry1Ah protein was obtained, and its amino acid sequence was determined. Based on this finding, isotope dilution mass spectrometry can be used to prepare Cry1Ah protein reference material.
2.4 Insect bioassay
Cry1Ah protoxin and pure Cry1Ah protein (activated and purified) from the Biot1Ah were tested for insecticidal activities against. Both proteins were quantified by SDS-PAGE using BSA standards. The LC50values for Cry1Ah protoxin and pure Cry1Ah protein were 7.40 μg/mL and 3.86 μg/mL respectively; 95% confidence intervals were 4.48–22.82 and 2.89–5.59 respectively. Bioassay results showed that the 95% confidence limits overlapped for the two proteins. Thus, no significant difference in the insecticidal activity of the protoxins and purified Cry1Ah toxin againstwas observed. The process of activation and purification does not affect the insecticidal activity of the Cry1Ah protein against. In Xue’s study, it was confirmed that the minimal active fragment of the Cry1Ah toxin was located between amino acid residues 50I and 639E[38], and the results here validate this conclusion.
3 Conclusion
In this study, the Cry1Ah protein extracted from Bt was activated and purified, and its purity was determined by HPLC. At the same time, the amino acid sequence of the protein was determined. This purified Cry1Ah protein can be used to prepare a protein reference material.
[1] Zambryski P, Joos H, Genetello C, et al. Ti plasmid vector for the introduction of DNA into plant cells without alteration of their normal regeneration capacity. EMBO J, 1983, 2(12): 2143–2150.
[2] James C, Krattiger AF. Global Review of the Field Testing and Commercialization of Transgenic Plants, 1986 to 1995: The First Decade of Crop Biotechnology. Ithaca: ISAAA, 1996.
[3] ISAAA. Global Status of Commercialized Biotech/GM Crops in 2017: Biotech Crop Adoption Surges as Economic Benefits Accumulate in 22 years. Ithaca: ISAAA, 2017.
[4] Ciabatti I, Marchesi U, Froiio A, et al. Role of the “national reference centre for genetically modified organisms (GMO) detection” in the official control of food and feed. Vet Res Commun, 2005, 29(2): 31–34.
[5] Devos Y, Aguilera J, Diveki Z, et al. EFSA’s scientific activities and achievements on the risk assessment of genetically modified organisms (GMOs) during its first decade of existence: looking back and ahead. Transgenic Res, 2014, 23(1): 1–25.
[6] Ahmed FE. Detection of genetically modified organisms in foods. Trends Biotechnol, 2002, 20(5): 215–223.
[7] Elenis DS, Kalogianni DP, Glynou K, et al. Advances in molecular techniques for the detection and quantification of genetically modified organisms. Anal Bioanal Chem, 2008, 392(3): 347–354.
[8] Maria KA, Blagica D, Hanne L, et al. The certification of different mass fractions of DAS-81910–7 in cotton powder: Certified reference materials ERM®-440a, ERM®-440b, ERM®-440c, ERM®-440d and ERM®-440e. Geel, Belgium: ERM, 2018.
[9] Maria KA, Blagica D, John S, et al. The certification of different mass fractions of Bt11 in maize powder: Certified reference materials: ERM®-BF412ak, ERM®-BF412bk, ERM®-BF412ck, ERM®-BF412dk and ERM®-BF412ek. Geel, Belgium: ERM, 2017.
[10] Workman L, Ludwig C, Atkinson C, et al. Report of the certification process for GT73/RT73 canola certified reference materials first batch. Urbana, IL: AOCS, 2016.
[11] Miller A, Bloomer S. Report of the certification process for T25 maize certified reference materials ninth batch. Urbana, IL: AOCS, 2017.
[12] Clapper G, Cantrill R. The certification of conventional, roundup ready®, bollgard®, and bollgard II®cottonseed reference materials AOCS 0804-A, AOCS 0804-B, AOCS 0804-C, AOCS 0804-D. Champaign, IL: AOCS, 2004.
[13] Miller A, Rennie D. Report of the certification process for A2704–12 soybean certified reference materials tenth batch. Urbana, IL: AOCS, 2017.
[14] Wu LQ, Wang J, Yang B, et al. Current status and trend of study on protein content certified reference materials. Acta Metrol Sin, 2010, 31: 125–127 (in Chinese). 武利庆, 王晶, 杨彬, 等. 蛋白质含量标准物质研究现状及趋势. 计量学报, 2010, 31: 125–127.
[15] Pritchard C, Quaglia M, Mussell C, et al. Fully traceable absolute protein quantification of somatropin that allows independent comparison of somatropin standards. Clin Chem, 2009, 55(11): 1984–1990.
[16] Arsene CG, Ohlendorf R, Burkitt W, et al. Protein quantification by isotope dilution mass spectrometry of proteolytic fragments: cleavage rate and accuracy. Anal Chem, 2008, 80(11): 4154–4160.
[17] Phinney KW, Bunk DM, Zhang NF. Certificate of analysis: Bovine serum albumin (7% solution) (SRM 927e). USA: NIST, 2016.
[18] Margolis S, Paule R, Christensen RG, et al. Certificate of analysis: Angiotensin I (human) (SRM 998). USA: NIST, 2016.
[19] Bunk DM, Welch MJ, Hagwood CR. Certificate of analysis: Human cardiac troponin complex (SRM 2921). USA: NIST, 2016.
[20] Kato M, Kinumi T, Yoshioka M, et al. Reference material certificate: C-reactive protein solution (NMIJ CRM 6201-c). JAP: AIST, 2017.
[21] Song DW. Reference material certificate: Troponin Ⅰ (GBW09229). CHN: NIM, 2016.
[22] Xue J, Liang GM, Crickmore N, et al. Cloning and characterization of a novel Cry1A toxin fromwith high toxicity to the Asian corn borer and other lepidopteran insects. FEMS Microbiol Lett, 2008, 280(1): 95–101.
[23] Zhou ZS, Liu YX, Liang GM, et al. Insecticidal specificity of Cry1Ah tois determined by binding of APN1 via domain II Loops 2 and 3. Appl Environ Microbiol, 2017, 83(4): e02864–16.
[24] Li XY, Li SY, Lang ZH, et al. Chloroplast-targeted expression of the codon-optimized truncatedgene in transgenic tobacco confers a high level of protection against insects. Plant Cell Rep, 2013, 32(8): 1299–1308.
[25] Dai PL, Zhou W, Zhang J, et al. The effects of Bt Cry1Ah toxin on worker honeybees (and). Apidologie, 2012, 43(4): 384–391.
[26] Dai PL, Zhou W, Zhang J, et al. Field assessment of Btcorn pollen on the survival, development and behavior of. Ecotoxicol Environ Safe, 2012, 79: 232–237.
[27] Geng LL, Cui HJ, Dai PL, et al. The influence of Bt-transgenic maize pollen on the bacterial diversity in the midgut of. Apidologie, 2013, 44(2): 198–208.
[28] Jiang WY, Geng LL, Dai PL, et al. The Influence of Bt-transgenic maize pollen on the bacterial diversity in the midgut of Chinese honeybees,. J Integr Agric, 2013, 12(3): 474–482.
[29] Zhou ZS, Yang SJ, Shu CL, et al. Comparison and optimization of the method for Cry1Ac protoxin preparation in HD73 strain. J Integr Agric, 2015, 14(8): 1598–1603.
[30] Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 1970, 227(5259): 680–685.
[31] Edman P. Method for determination of the amino acid sequence in peptides. Acta Chem Scand, 1950, 4: 283–293.
[32] Tabashnik BE, Finson N, Chilcutt CF, et al. Increasing efficiency of bioassays: evaluating resistance toin diamondback moth (Lepidoptera:). J Econ Entomol, 1993, 86(3): 635–644.
[33] Finney DJ. Probit Analysis. Cambridge: Cambridge University Press, 1971.
[34] Russell RM, Robertson JL, Savin NE. POLO: a new computer program for probit analysis. Bull Entomol Soc Amer, 1977, 23(3): 209–213.
[35] Brun V, Masselon C, Garin J, et al. Isotope dilution strategies for absolute quantitative proteomics. J Proteomics, 2009, 72(5): 740–749.
[36] Vogl J, Pritzkow W. Isotope dilution mass spectrometry — A primary method of measurement and its role for RM certification. MAPAN, 2010, 25(3): 135–164.
[37] Wu LQ, Yang B, Bi JM, et al. Development of bovine serum albumin certified reference material. Anal Bioanal Chem, 2011, 400(10): 3443–3449.
[38] Xue J, Zhou ZS, Song FP, et al. Identification of the minimal active fragment of the Cry1Ah toxin. Biotechnol Lett, 2011, 33(3): 531–537.
Preparation and purification of Cry1Ah protein candidate reference material
Lin Guo1,2, Lili Geng2, Xiaoxiao Sun2, Meiling Wang2, Changlong Shu2, and Jie Zhang2
1 Faculty of Agronomy, Jilin Agricultural University, Changchun 130118, Jilin, China 2 State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing 100193, China
With the rapid development of transgenic technology, the safety of genetically modified products has received extensive attention. Certified reference materials for the detection of genetically modified organisms play important roles in ensuring comparability and traceability of the qualitative and quantitative detection of genetically modified products. However, the development of protein reference materials is relatively slow, and one of the difficulties is the preparation of protein candidates with high purity. Thegene ofhas been used for the development of transgenic insect-resistant crops because of its excellent insecticidal activity against lepidopteran pests such as Asian corn borer, and has obtained transgenic lines with good insect resistance traits. In order to develop Cry1Ah protein certified reference material, it is urgent to establish a preparation and purification system. In this study, a system for preparing Cry1Ah protein by Bt expression system was optimized, and a high-purity Cry1Ah protein (size exclusion chromatography purity: 99.6%) was obtained by ion-exchange chromatography and size exclusion chromatography stepwise purification. The results of biological activity assay showed that there was no significant difference in the insecticidal activity of purified Cry1Ah protein and protoxin against diamondback moths (). Finally, the amino acid sequence of the activated Cry1Ah protein was determined using Edman degradation and mass spectrometry. In summary, the obtained Cry1Ah pure protein can be used for the development of protein reference materials.
genetically modified organisms, certified reference material,, Cry1Ah protein
January 29, 2019;
March 29, 2019
Supported by:National Major Special Project for the Development of Transgenic Organisms (No. 2014ZX08012-003), National Natural Science Foundation of China (No. 31501711).
Jie Zhang. Tel: +86-10-62815921; E-mail: zhangjie05@caas.cn
国家转基因生物新品种培育重大专项 (No. 2014ZX08012-003),国家自然科学基金 (No. 31501711) 资助。
2019-05-13
http://kns.cnki.net/kcms/detail/11.1998.q.20190510.1018.001.html

郭林, 耿丽丽, 孙晓晓, 等. Cry1Ah蛋白标准物质候选物的制备与纯化. 生物工程学报, 2019, 35(8): 1511–1519.Guo L, GengLL, SunXX, et al.Preparation and purification of Cry1Ah protein candidate reference material. Chin J Biotech, 2019, 35(8): 1511–1519.
(本文责编 陈宏宇)
