RNA sequencing screening of differentially expressed genes after spinal cord injury
2019-07-17YiLiYingChenXiangLiJianWuJingYingPanRiXinCaiRiYunYangXiaoDongWang
Yi Li , Ying Chen , Xiang Li Jian Wu Jing-Ying Pan Ri-Xin Cai Ri-Yun Yang Xiao-Dong Wang
1 School of Biology & Basic Medical Sciences, Soochow University, Suzhou, Jiangsu Province, China
2 Department of Histology and Embryology, Medical College, Nantong University, Nantong, Jiangsu Province, China
3 Jiangsu Key Laboratory of Neuroregeneration, Co-innovation Center of Neuroregeneration, Nantong University, Nantong, Jiangsu Province,China
Abstract In the search for a therapeutic schedule for spinal cord injury, it is necessary to understand key genes and their corresponding regulatory networks involved in the spinal cord injury process. However, ad hoc selection and analysis of one or two genes cannot fully reveal the complex molecular biological mechanisms of spinal cord injury. The emergence of second-generation sequencing technology (RNA sequencing) has provided a better method. In this study, RNA sequencing technology was used to analyze differentially expressed genes at different time points after spinal cord injury in rat models established by contusion of the eighth thoracic segment. The numbers of genes that changed significantly were 944, 1362 and 1421 at 1, 4 and 7 days after spinal cord injury respectively. After gene ontology analysis and temporal expression analysis of the differentially expressed genes, C5ar1, Socs3 and CCL6 genes were then selected and identified by real-time polymerase chain reaction and western blot assay. The mRNA expression trends of C5ar1, Socs3 and CCL6 genes were consistent with the RNA sequencing results. Further verification and analysis of C5ar1 indicate that the level of protein expression of C5ar1 was consistent with its nucleic acid level after spinal cord injury. C5ar1 was mainly expressed in neurons and astrocytes. Finally, the gene Itgb2,which may be related to C5ar1, was found by Chilibot database and literature search. Immunofluorescence histochemical results showed that the expression of Itgb2 was highly consistent with that of C5ar1. Itgb2 was expressed in astrocytes. RNA sequencing technology can screen differentially expressed genes at different time points after spinal cord injury. Through analysis and verification, genes strongly associated with spinal cord injury can be screened. This can provide experimental data for further determining the molecular mechanism of spinal cord injury, and also provide possible targets for the treatment of spinal cord injury. This study was approved ethically by the Laboratory Animal Ethics Committee of Jiangsu Province, China (approval No. 2018-0306-001) on March 6, 2018.
Key Words: nerve regeneration; spinal cord injury; RNA sequencing; C5ar1; Itgb2; Socs3; CCL6; astrocytes; rats; neural regeneration
Graphical Abstract
RNA sequencing screening for differentially expressed genes after spinal cord injury
Introduction
Spinal cord injury (SCI) results in extremely high disability rate and serious consequences. Its pathophysiological changes are mainly divided into two stages: primary injury and secondary injury. The former refers to the damage caused by external force directly or indirectly to the spinal cord, including crush injury, contusion or transverse injury (Widrin,2018). Although the primary injury is transient, it triggers more complex secondary injuries, including neuronal apoptosis, glial cell proliferation and inflammatory responses,which further aggravate and extend the severity of injuries(Noller et al., 2017). Eventually, the primary injury can lead to the loss of sensory and/or motor function below the plane of damage, as well as cause dysfunction of various organs and cause chronic neuropathic pain (Park et al., 2018). SCI not only reduces the quality of life of patients, but also brings great emotional and economic burdens to families and society. However, current therapies are not totally satisfactory,and there is an urgent need for more effective treatments.
A growing number of researchers believe that an understanding of the mechanisms underlying SCI and its secondary damage is an important route to developing a treatment strategy for SCI (Dalbayrak et al., 2015; Garcia et al., 2016;Wang et al., 2018). Therefore, analysis starting at the molecular level for the gene expression and its corresponding regulatory pathways in the SCI process is required. However,research in the past often directly selected individual genes for detection and analysis of regulatory pathways (Park et al., 2016; Alizadeh et al., 2017; Tapia et al., 2017; Deng et al.,2018). The emergence and development of gene sequencing technologies, such as cDNA chips and gene chip technology, have largely addressed this problem (Uchida et al., 2010;Chamankhah et al., 2013). However, initial sequencing technology is limited in its use owing to its low resolution and accuracy (Marioni et al., 2008). The emergence of second-generation sequencing technologies such as RNA sequencing (RNA-Seq) technology, with its unprecedented sensitivity and specificity, in spite of its high cost (Narrandes and Xu, 2018), allow for better genome-wide sequencing.
RNA-Seq technology, also known as whole transcriptome shotgun sequencing, is a genome-wide sequencing technology that increases the coverage of DNA sequences(Ozsolak and Milos, 2011; Van Verk et al., 2013; Yi et al.,2018). Second-generation high-throughput sequencing is performed by extracting all transcribed RNA in a biological sample and then reversely transcribing them into cDNAs.On this basis, the overlapping assembly of the fragments is carried out, so that transcripts can be obtained to observe post-transcriptional changes, gene fusion, and gene expression changes (Maher et al., 2009). A global understanding of the up-and-down gene expression in the biological sample is thus formed (Ingolia et al., 2012). In recent years, if the procedures are tightly controlled, the results of RNA-Seq are generally more accurate and reliable so it is widely used in the molecular mechanism studies of various complex diseases, such as tumors, brain microangiopathy, depression,amyotrophic lateral sclerosis and SCI (Duan et al., 2015; Shi et al., 2017; Barham et al., 2018; Herman et al., 2018; Le et al., 2018; Yuan et al., 2018; Wang et al., 2019).
To this end, we applied RNA-Seq technology to efficiently analyze the differences in gene expression at different stages after SCI, and verified and functionally analyzed the selected genes to find possible determinants in the damage process(Chen et al., 2013). Our findings provide data for further study of the molecular mechanisms of SCI, and provide a reference for developing treatment strategies for SCI.
Materials and Methods
Experimental animals
A total of 49 specific-pathogen-free 8-week-old adult female Sprague-Dawley rats weighing 200-220 g were purchased from the Laboratory Animal Center of Nantong University in China (license No. SYXK (Su) 2017-0046). All the surgical interventions and postoperative animal care were performed according to the National Institutes of Health (USA) guidelines for care and use of laboratory animals (NIH Publication No. 85-23, revised 1996). This study was approved ethically by the Laboratory Animal Ethics Committee of Jiangsu Province, China (approval No. 2018-0306-001) on March 6, 2018.
SCI model establishment
Forty-nine adult female Sprague-Dawley rats were randomly divided into a sham group (n = 10) and an SCI group (n =39). The rats in the SCI group were randomly and equally subdivided into three given time groups, 1, 4 and 7 days after SCI. Animal models were prepared in accordance with a previous method (Donnelly et al., 2012). Under anesthesia, a 1-cm incision was made at the eighth thoracic segment (T8)to open the lamina and expose the spinal cord layer by layer.In the sham group, no damage was inflicted on the spinal cord and the incision was sutured layer by layer. In the SCI group, after fixation of the spine, a spinal cord hitter (IH-0400, PSI, Fairfax, VA, USA) inflicted a 1.5 N impact to the exposed spinal cord, followed by layer by layer suturing. The model rats were postoperatively injected intraperitoneally with 5-mL lactated Ringer's solution (Anhui BBCA Pharmaceutical Co., Hefei, China) to supplement the blood volume,and then penicillin (100,000 units) (Shandong Lukang Pharmaceutical Co., Jining, China) was given daily for 1 week,and the bladder was massaged twice daily for artificial urination until it was able to urinate spontaneously.
Postoperative rat behavior testing
Postoperative rat behavioral testing was performed using the Basso, Beattie & Bresnahan (BBB) locomotor rating scale(Basso et al., 1995). Preoperative training was performed as follows: the rats were placed in a round, open basin to walk freely, and kept in the central area as much as possible. The test time was 4 minutes. Its movement range, trunk movement and coordination of the knee, hip and ankle during walking were observed. At least two testers performed each test and the mean of scores taken.
RNA-Seq technology and bioinformatics analysis
After anesthesia, a 5-mm spinal cord segment, including the injured portion, was taken from each rat, and total RNA was extracted according to the conventional method. Agilent 2200 (Agilent Technologies, Santa Clara, CA, USA) was used to test the integrity of samples. A sample was used for the establishment of a transcriptome library if the RNA-integrity number was more than 8. Qualified samples were sent to RiboBio Corporation (Guangzhou, China) for RNA-Seq analysis. Gene ontology (GO) analysis and temporal expression analysis were performed. Genes related to SCI and post-injury repair were subsequently screened for biological properties. Genes related to SCI were found by Chilibot database (http://www.chilibot.net/) and literature searches.
Real-time polymerase chain reaction
The qualified RNA samples were used as templates to reverse-transcribe cDNA in real-time polymerase chain reaction (PCR) assay. Then cDNA (250 mg/L) 2 μL, Fast Start Universal SYBR Green Master 10 μL, Primer (sense) 0.6 μL, Primer (antisense) 0.6 μL, and quantified to 20 μL with ddH2O. Primer sequences are shown in Table 1. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was the internal control, and the untemplated wells were used as negative controls. The relative quantitative analysis used was the 2-ΔΔCtmethod.
Western blot assay
Spinal cord tissues were added to the lysate at a ratio of 10 mL/g for protein sample preparation. The protein concentration was determined by the bicinchoninic acid method.The samples were separated on a 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis gel at 60-110 V for 2 hours, and electrotransferred onto polyvinylidene difluoride membranes at 300 mA for 90 minutes. The membranes were blocked with Tris-buffered saline with Tween 20 containing 5% skim milk powder for 2 hours, then incubated with the primary antibodies (rabbit-anti-C5ar1, 1:800, Proteintech Group Inc., Wuhan, China; rabbit-anti-GAPDH, 1:1000,Abcam, Cambridge, MA, USA) at 4°C overnight. The next day, they were rinsed, then incubated with the secondary antibody (horseradish peroxidase-labeled goat anti-rabbit IgG, 1:1000, Abcam) for 2 hours at room temperature. The protein bands werevisualized with enhanced chemiluminescence (Bio-Rad laboratories Inc., Hercules, CA, USA), and analyzed using a Chemi DocTMXRS+ gel imaging system(Bio-Rad laboratories Inc.), and Image Lab 5.1 software(Bio-Rad laboratories Inc.).
Immunofluorescence histochemistry
Under anesthesia, the animals were perfused with normal saline and 4% paraformaldehyde. The 5-mm spinal cord segments, including the injured portion, were excised. Frozen sections were then made, blocked with 0.5% Triton-X 100 and 3% bovine serum albumin, incubated with primary antibodies (rabbit anti-C5ar1, 1:400, Proteintech Group Inc.;rabbit anti-Itgb2, 1:400, Abcam; mouse anti-glial fibrillary acidic protein (GFAP), 1:500, Abcam; mouse anti-NF200,1:500, Abcam; goat anti-GFAP, 1:500, Abcam), overnight at 4°C. After rinsing with phosphate buffer saline, the samples were incubated with secondary antibodies (Alexa Fluor®488-labeled donkey anti-rabbit IgG antibody, 1:1000, Thermo Fisher, Waltham, MA, USA; Alexa Fluor®568-labeled donkey anti-mouse IgG, 1:1000, Thermo Fisher; Alexa Fluor®647-labeled donkey anti-goat IgG antibody, 1:1000, Thermo Fisher)at room temperature for 2 hours in the dark. The samples were rinsed with phosphate buffer saline and mounted with DAPI-containing sealant. They were observed using laser confocal microscope (SP7, Leica, Wetzlar, Germany) and recorded. No primary antibodies were used in controls.
Statistical analysis
All data are expressed as the mean ± SD. Both data analysis and image generation were performed by Graph Pad Prism 6 (GraphPad Software Co., San Diego, CA, USA). The BBB score data were analyzed by repeated-measure analysis of variance for homogeneity of analysis, and then analysis of variance was used for comparison between groups. PCR and western blot data were analyzed by one-way analysis of variance for homogeneity of variance, and then analysis of variance was used for comparison between groups. A value of P < 0.05 was considered statistically significant.
Results
Changes in locomotor function of SCI rats
The general state of SCI rats was confirmed based on their BBB score to ensure that the SCI animal model had been successfully prepared. Subsequent experimental procedures were then performed. Compared with the sham group, the BBB score was significantly lower at each time point after SCI. The hindlimb movement was completely lost at 1 day after injury, and the BBB score was 0. The score on the hindlimb function was still low at 4 days after injury, and slightly restored at 7 days after injury (Figure 1).This indicates that the hindlimb function of SCI rats had some but limited self-repair within 1 week after SCI. These results indicated that the animal model of SCI had been prepared successfully.
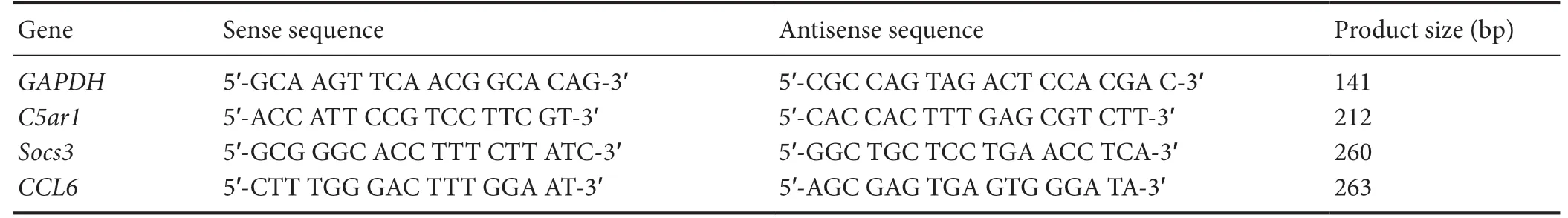
Table 1 Primer sequences for polymerase chain reaction
Screening of differentially expressed genes after SCI
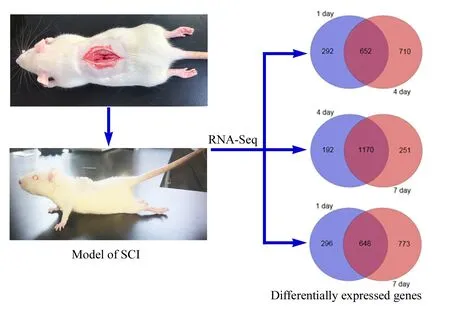
A large number of differentially expressed genes were screened by high-throughput sequencing at the genome-wide level using RNA-Seq technology. To further understand the specific distribution of differential transcripts,comparisons between the sham group and experiment groups were performed using Cuffdiff. The results of the Venn plot showed that compared with the sham group, the number of genes that changed significantly at 1, 4 and 7 days after SCI were 944, 1362 and 1421, respectively, of which 652 genes were differentially expressed at both 1 and 4 days, 648 genes at both 1 and 7 days, and 1170 genes at both 4 and 7 days (Figure 2).
GO functional analysis of differentially expressed genes was performed to further explore the function of the differentially expressed genes. The results showed that among the top 20 groups of GO enrichments, multiple biological processes related to damage and repair were involved, such as GO: 0006954 (inflammatory response), GO: 0050729 (positive regulation of inflammatory response), GO: 0009612 (response to mechanical stimulus); and GO: 0045766 (positive regulation of angiogenesis) (Figure 3).
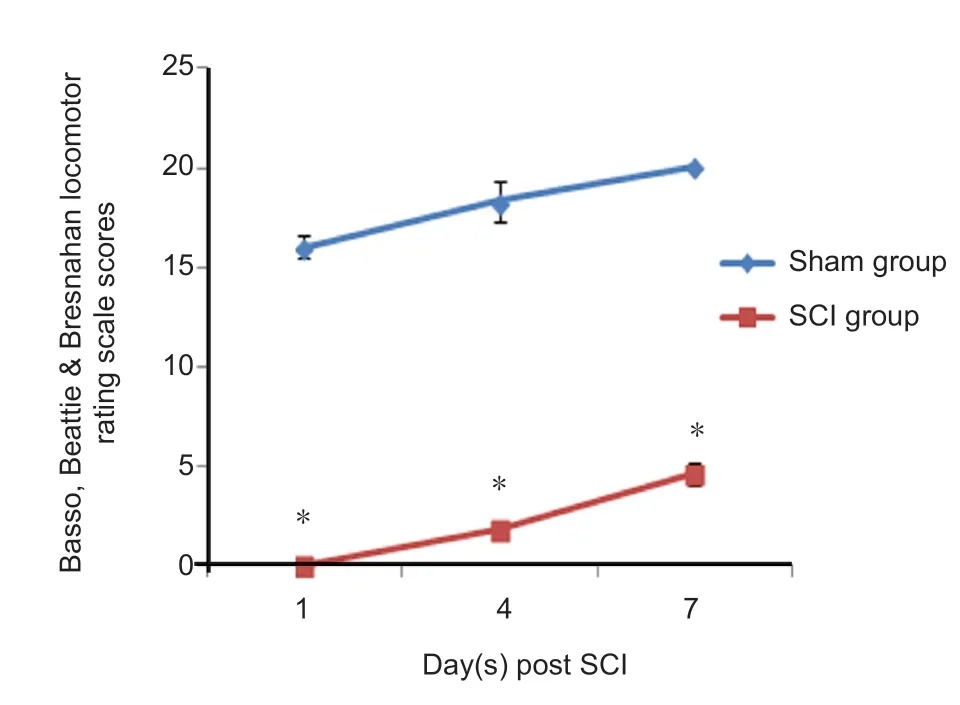
Figure 1 Basso, Beattie & Bresnahan locomotor rating scale for thehindlimb function in rats.Data are expressed as the mean ± SD (repeated-measure analysis of variance). *P < 0.05, vs, sham group. All experiments were conducted five times. SCI: Spinal cord injury.
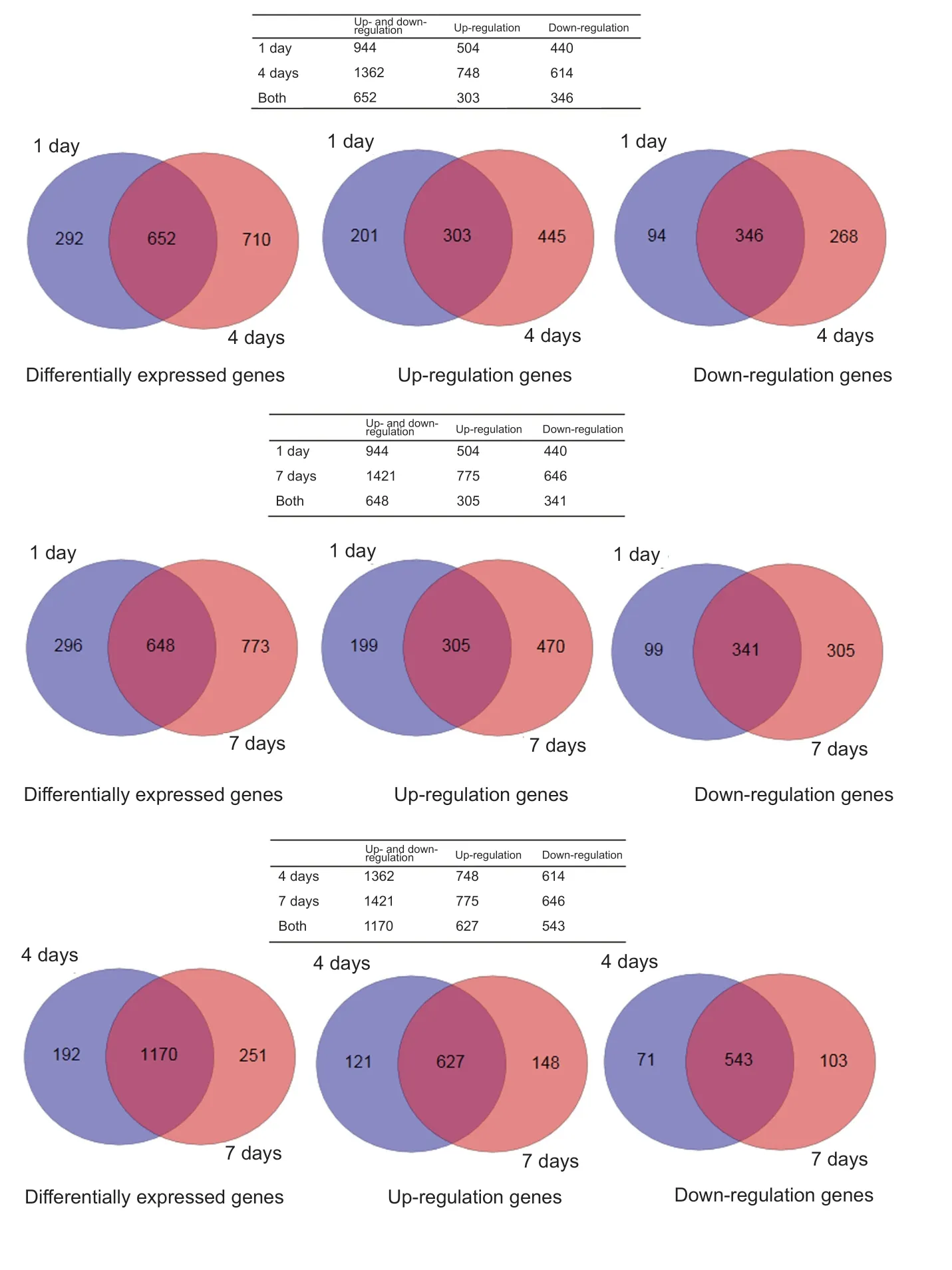
Figure 2 Venn plots of differentially expressed genes in the tissues from the sham group at 1, 4, and 7 days after spinal cord injury.Differentially expressed genes were screened by high-throughput sequencing at the genome-wide level using RNA-Seq technology.
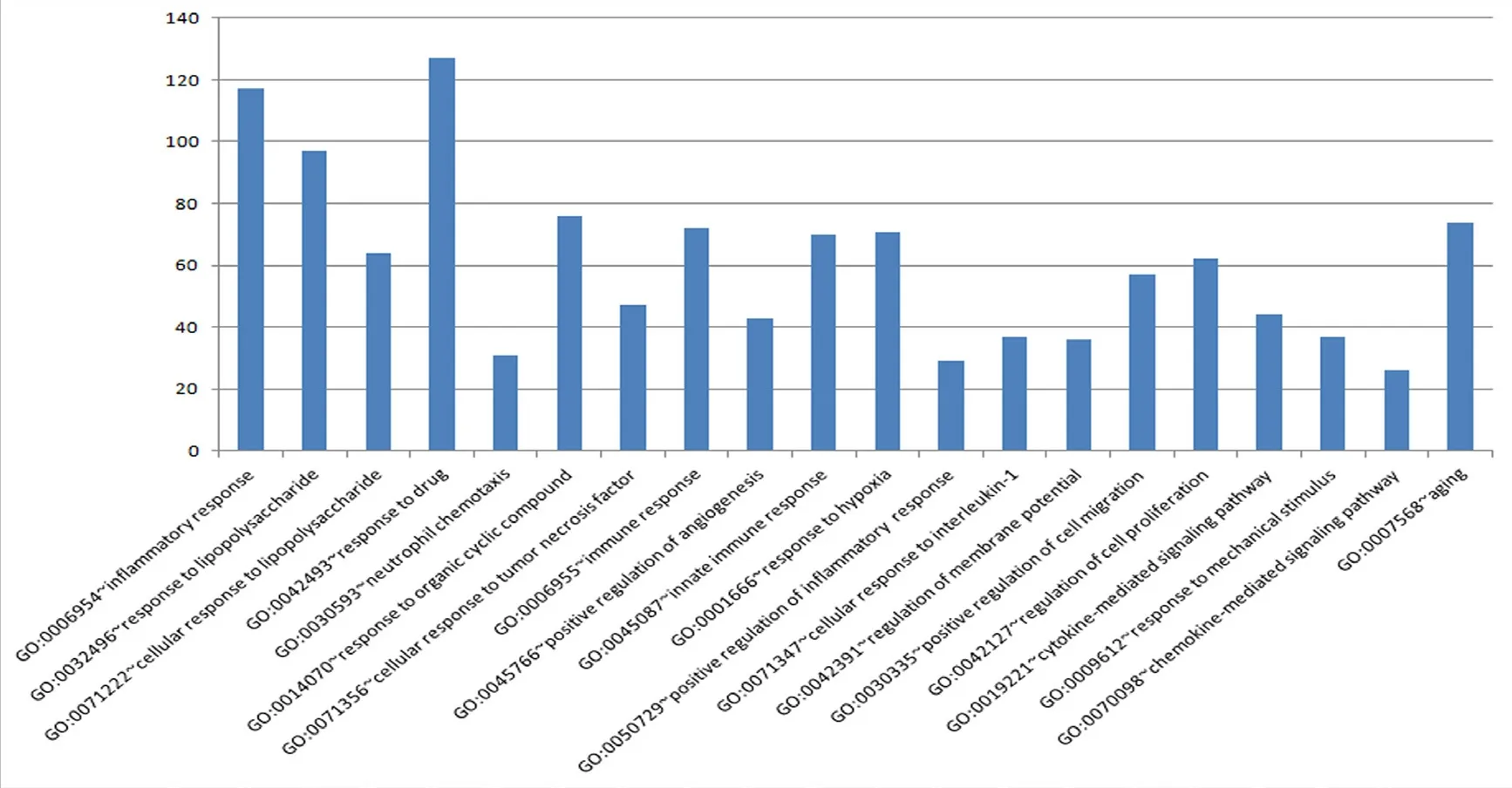
Figure 3 GO functional analysis of differentially expressed genes.The abscissa represents various terms, and the ordinate represents numbers of genes.
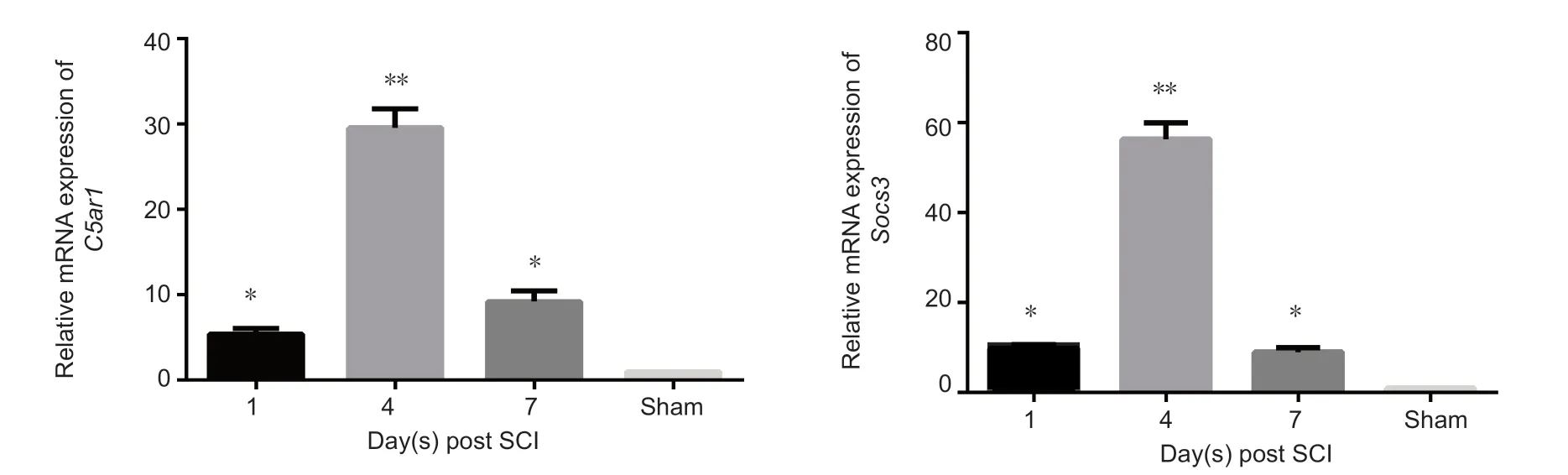
Figure 4 Relative gene expression of C5ar1, Socs3 and CCL6 in the spinal cord after SCI.Data (2-ΔΔCt ratio to GAPDH mRNA) are expressed as the mean ± SD (n = 10 in sham group, n = 13 in SCI groups at 1, 4, and 7 days) relative to sham group(mean = 1). *P < 0.05, **P < 0.01, vs. sham group (one-way analysis of variance). All experiments were conducted five times. Sham: Sham group; 1 d, 4 d and 7 d: 1, 4, 7 days after SCI; SCI: spinal cord injury.

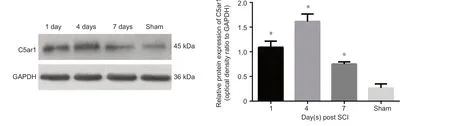
Figure 5 Relative protein expression of C5ar1 in the spinal cord after SCI.Data are expressed as the mean ± SD (n = 10 in sham group, n = 13 in SCI groups at 1, 4, and 7 days), showing C5ar1 expression by the optical density ratio of C5ar1 to its GAPDH in each group, *P < 0.05, vs. sham group (one-way analysis of variance). All experiments were conducted five times. Sham: Sham group; 1 d, 4 d and 7 d: 1, 4, 7 days after SCI; SCI: spinal cord injury.

Table 2 Genes closely linked to spinal cord injury or spinal cord development, which showed significant changes to expression after spinal cord injury
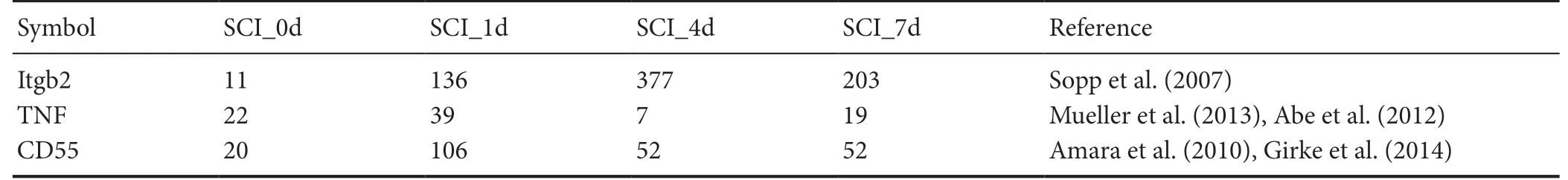
Table 3 Genes related to C5ar1
Temporal expression analysis of differentially expressed genes after SCI
Temporal expression analysis of differentially expressed genes at different time points was performed to screen for genes with a consistently significant change (fold change> 4) from 0 to 1 day, 1 to 4 days, and 4 to 7 days (Table 2).Searching the Chilibot database, the genes C5ar1, Socs3, and CCL6 found most strongly associated with SCI or spinal cord development were further screened.
Expression of some genes after SCI verified by real-time PCR
Real-time PCR results verified the changes of the above three genes at different stages after SCI. C5ar1 expression was upregulated 1 day after injury, peaked at 4 days and downregulated significantly at 7 days after SCI compared with the sham group (P < 0.05 or P < 0.01; Figure 4). The variations in Socs3 and CCL6 expression were the similar to that of C5ar1 between SCI and sham groups (P < 0.05 or P < 0.01; Figure 4).
Expression of C5ar1 protein in the spinal cord after SCI
We selected the gene C5ar1 from the verified differentially expressed genes because it is involved in the response to injury,inflammatory response and neuronal apoptosis. The expression trend of C5ar1 after SCI by western blot assay was observed.C5ar1 expression significantly increased after SCI compared
with that in the sham group (P < 0.05; Figure 5). C5ar1 expression peaked at 4 days after SCI. This finding was consistent with the RNA-Seq sequencing and real-time PCR results.
Bioinformatics analysis of C5ar1 in the spinal cord after SCI
Further bioinformatics analysis was performed on C5ar1.Itgb2, CD55 and TNF were most strongly associated with C5ar1 by searching the Chilibot database and query literature.These genes were also found to be differentially expressed temporally in the RNA-Seq results. Itgb2 expression was upregulated after 1 day of SCI, peaked at 4 days and downregulated at 7 days. TNF expression was upregulated at 1 day after SCI and downregulated at 4 days, but upregulated again at 7 days. CD55 expression was upregulated at 1 day after SCI and downregulated at 4 days until unchanged at 7 days. Therefore,only the expression trends of Itgb2 and C5ar1 were highly consistent (references are shown in Table 3).
Localization and expression of C5ar1 and Itgb2 in spinal cord tissue after SCI
The cellular localization and expression of C5ar1 in spinal cord tissue were observed by immunofluorescence histochemistry. The astrocyte and neuronal markers GFAP and NF200 were used for staining and localization (Trojanowski et al., 1986; Middeldorp and Hol, 2011; van Bodegraven et al., 2019). C5ar1 was mainly found in astrocytes and neurons, and its expression level was differentially expressed with time after SCI. C5ar1 was expressed in the cells at 1 day after SCI; both the immunofluorescence expression and the number of positive cells increased at 4 days. At 7 days, the immunofluorescence expression of cells decreased slightly(Figures 6 & 7). This result was highly consistent with the changing trends of results from RNA-Seq, real-time PCR and western blot assay.
The expression of Itgb2, which was highly consistent with the changes in C5ar1 expression, was mainly expressed in astrocytes of the spinal cord tissue after SCI (Figure 8).
Discussion
In recent years, researchers have focused on analyzing the complex pathological processes of SCI and its molecular mechanisms to determine the effects of changes to the microenvironment on SCI repair. This approach attempted to find techniques that might protect nerve tissue cells by improving the microenvironment around SCI and enhancing axonal regeneration, such as drug therapy, genetic modification, cell transplantation or tissue engineering. The RNA-Seq technology used in this study investigated the differences in transcriptome levels at different stages after SCI. The results of this study showed that compared with the sham group,the number of differentially expressed genes at 1, 4 and 7 days after SCI were 944, 1362 and 1421, respectively. Of the differentially expressed genes those genes of interest were confirmed after screening by GO functional analysis and temporal expression analysis. Multiple biological processes associated with injury repair were enriched, suggesting that these genes may play an important role in SCI and post-injury repair processes. The differentially expressed transcripts between samples were selected by the difference multiples(log2|fold_change|>2) and significant levels (q value < 0.05).Subsequent n searching of the Chilibot database, selected those genes with persistent and significant changes (fold change > 4) that were strongly associated with SCI, some of which were verified by real-time PCR. The results showed that the differential expression of C5ar1, Socs3 and CCL6 at different stages after SCI was consistent with the results of RNA-Seq sequencing.
Socs3 is a feedback inhibitor of the JAK/STAT pathway and plays a key role in regulating the immune response of JAK/STAT-mediated inflammatory cytokines (Croker et al.,2008). The structural basis for this effect is that Socs3 contains a kinase inhibitory region that in combination with the JAK/STAT/gp130 complex can inhibit downstream signaling and inflammatory cytokines (La Manna et al., 2018).In the central nervous system, Socs3 is expressed on almost all types of cells (Baker et al., 2009). A previous study has shown that the Socs3 protein is involved in the regulation and progression of microglial immune responses and, therefore, regulation of its expression may be a target for drug development in the control of inflammation (Cianciulli et al., 2017). After SCI, Socs3 was found to not only regulate the inflammatory response, but also to be associated with the formation of glial scars. For example, Socs3 may prevent the formation of glial scars (Li et al., 2010), and can regulate the proliferation and differentiation of NG2 cells in glial scars (Hackett et al., 2016). It had also been reported that increased Socs3 level was strongly associated with neuronal death (Park et al., 2014). The Socs3-dependent JAK/STAT pathway played a key role in neuronal loss and even acted in regulating axon regeneration during secondary injury of SCI (Liu et al., 2015). Multiple genes in zebrafish after SCI,equivalent to mammalian stat3, socs3, atf3, mmp9 and sox11 were upregulated (Hui et al., 2014). CCL6 is a member of the CC chemokine family and is normally expressed on microglia cells without stimulation. Under normal physiological environment or abnormal pathological conditions,CCL6 mediates the main immune effector cells of the central nervous system, or interaction between microglia and astrocytes, by activating the PI3-kinase/Akt pathway (Kanno et al., 2005). RNA-Seq screening indicated that both Socs3 and CCL6 were related to pathophysiological reactions after SCI,especially inflammatory reactions. They could be used as potential targets to reduce inflammatory responses as well as to improve the repair environment after SCI.
C5ar1 (CD88) is the most important receptor for the complement component C5a, which is structurally a typical G-protein coupled receptor that signals primarily through Gαi and Gα16 (Monk et al., 2007). C5ar1 is widely expressed in inflammatory cells, such as granulocytes, monocytes and macrophages, as well as in the central nervous system,especially in glial cells (Woodruff et al., 2010). Immunofluorescence histochemical staining of this study showed that C5ar1 was mainly present in astrocytes and neurons,which is consistent with previous findings. The widespread expression of C5ar1 means that it may be involved in various activities. First, damage to the central nervous system activates the complement system, triggering a cascade of biological effects after C5a binds to C5ar1, including recruitment of granulocytes, activation of glial cell and neuronal signal transduction pathways (Guo et al., 2013b). C5ar1 and C5a are also effectors of the neuropathic complement pathway and have multi-effect inflammatory effects, which can stimulate nuclear factor-kappa β, activator protein-1,and adenosine monophosphate to participate in protein signal transduction. Activation of C5ar1 is associated with the onset and severity of epileptic seizures and subsequent neuronal degeneration (Benson et al., 2015). Pathological changes in the hSOD1G93A mouse model of amyotrophic lateral sclerosis can be improved when antagonists inhibit C5a-C5ar1 receptor signaling (Lee et al., 2017). C5a-C5ar1 is also associated with permeability of the blood-brain barrier(Lee et al., 2017) and cognitive loss of Alzheimer's disease.After SCI, C5ar1 is highly expressed and specifically binds to C5a, inducing a chemotactic response that participates in neuronal apoptosis and subsequent inflammatory reactions.This process is inextricably linked to a series of secondary injuries (Kwon et al., 2004; Qiao et al., 2010; Thundyil et al., 2012; Guo et al., 2013a). It is generally believed that the expression level of C5ar1 in the central nervous system is simultaneously associated with the inflammatory state. Its expression produces a variety of inflammatory mediators during complement activation, such as interleukin-2, interleukin-6, and tumor necrosis factor. The GO analysis of this study showed that C5ar1 was involved in multiple biological processes, such as injury response, inflammatory reaction and neuronal apoptosis. C5ar1 was found to have temporal differential expression after SCI, as shown by the changes in immunofluorescence histochemical staining, real-time PCR and western blot assay. C5ar1 expression was upregulated at 1 day after SCI, peaked at 4 days, and downregulated at 7 days, but was still higher than that in the sham group. This trend was consistent with the RNA-Seq results. This suggests that C5ar1 begins to be expressed in the early stage after SCI and then increases its own protein level, providing a material basis for mediating secondary pathological changes following SCI.
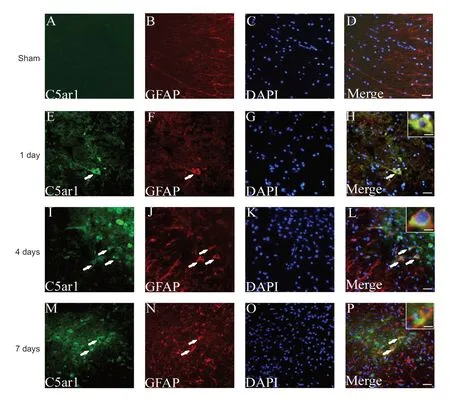
Figure 6 Immunofluorescence staining of C5ar1 and GFAP in spinal cord tissue of the SCI rats.Green fluorescence (DyLight 488) in A, E, I and M refers to C5ar1; red fluorescence (Dy-Light 568) in B, F, J and N refers to GFAP; blue fluorescence in C, G, K and O refers to DAPI labeled nuclei. D, H, L and P are merged figures. Arrows indicate double-positive cells. Sham:Sham group; 1 day, 4 days and 7 days: 1, 4, 7 days after SCI.Scale bars: 30 μm. Scale bars in the insets of H, L, and P: 10 μm.GFAP: Glial fibrillary acidic protein; SCI: spinal cord injury.

Figure 7 Immunofluorescence staining of C5ar1 and NF200in spinal cord tissue of the SCI rats.Green fluorescence (DyLight 488)in A, E, I and M refers to C5ar1;red fluorescence (DyLight 568) in B, F, J and N refers to NF200; blue fluorescence in C, G, K and O refers to DAPI labeled nuclei; and D, H, L and P are merged figures.Sham: Sham group; 1 day, 4 days and 7 days: 1, 4, 7 days after SCI.Arrows indicate double-positive cells. Scale bar: 30 μm. Scale bars in the insets of H, L, and P: 10 μm. NF200: Neurofilament-200;SCI: spinal cord injury.
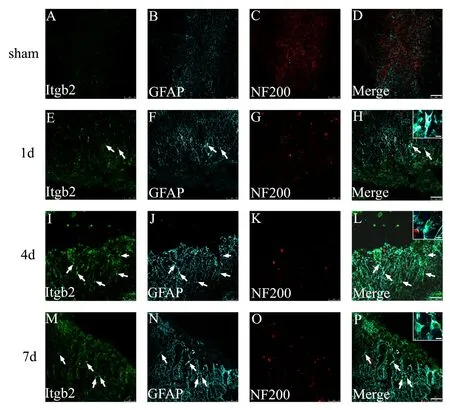
Figure 8 Immunofluorescence staining of Itgb2, GFAP and NF200 in spinal cord tissue of the SCI rats.Green fluorescence (DyLight 488)in A, E, I and M refers to Itgb2;cyan fluorescence (DyLight 647)in B, F, J and N refers to GFAP;red fluorescence (DyLight 568)in C, G, K and O refers to NF200;and D, H, L and P are merged figures. Sham: Sham group; 1 day, 4 days and 7 days: 1, 4, 7 days after SCI. Arrows indicate double-positive cells. Scale bars: 75 μm. Scale bars in the insets of H, L, and P: 10 μm. GFAP: Glial fibrillary acidic protein; NF200: neurofilament-200; SCI: spinal cord injury.
Among the differential genes identified above, we were most interested in C5ar1 (CD88). Therefore, we performed further analysis on C5ar1. By searching the Chilibot database and searching relevant literature, we found that many genes,such as Itgb2, CD55 and TNF, may be associated with C5ar1,and the expression trends of Itgb2 and C5ar1 are highly consistent. Itgb2 expression was upregulated at 1 day after SCI,reached the highest peak at 4 days then decreased at 7 days.Using immunofluorescence histochemistry, Itgb2, localized to astrocytes in the spinal cord sections, also exhibited temporal differential expression after SCI. Itgb2 (CD18) belongs to a family of β2 integrins. It is a transmembrane glycoprotein distributed on the cell surface that plays an important role in cell adhesion, cell surface signaling transduction and immune response. In the models of cerebral hemorrhage injuries, the symptoms of acute brain injury and cerebral edema can be reduced and even lower mortality rates in CD18-deficient mice compared with wild mice (Titova et al.,2008a, 2008b). As reported by Gris et al. (2004), the use of monoclonal antibodies to block Itgb2 significantly reduced secondary injury after SCI and promoted the recovery of motor and sensory functions in SCI rats. These findings indicate that, after SCI, an increase in Itgb2 level will aggravate secondary damage. However, further investigations on the relationship between C5ar1 and Itgb2, as well as the expression of CD55 and TNF and their roles and relationship with C5ar1, are required after SCI.
In conclusion, the differentially expressed genes, which play important roles in a series of pathological reactions after SCI and post-injury repair process, could be screened by RNA-Seq technology, and subjected to bioinformatic analysis. C5ar1 and Itgb2, two prominent genes of particular interest, were selected from all the differential genes, and subjected to various experimental techniques to verify changes in their expression and analyze their functions. On the one hand, it provides a basis for further elucidation of the molecular mechanism of SCI; on the other hand, it suggests targets for the treatment of SCI. Although several differentially expressed genes were found, only a few were studied and confirmed as participants in SCI. A more comprehensive study may be conducted in the future.
Author contributions:Study design and experimental implementation:YL, YC, XDW; experimental implementation: XL, JW, JYP, RXC, RYY;fundraising: XDW. All authors approved the final version of the paper.
Conflicts of interest: There are no conflicts of interest.
Financial support:This study was supported by the National Natural Science Foundation of China, No. 31570983 (to XDW); the Priority Academic Program Development of Jiangsu Higher Education Institutes of China. The funders did not participate in data collection and analysis,article writing or submission.
Institutional review board statement:The studies were approved by the Lab Animal Ethical Committee of Jiangsu Province, China (approval No.2018-0306-001) on March 6, 2018.
Copyright license agreement:The Copyright License Agreement has been signed by all authors before publication.
Data sharing statement:Datasets analyzed during the current study are available from the corresponding author on reasonable request.
Plagiarism check: Checked twice by iThenticate.
Peer review: Externally peer reviewed.
Open access statement:This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-Non-Commercial-ShareAlike 4.0 License, which allows others to remix, tweak,and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
Open peer reviewer: Li Yao, Wichita State University, USA.
Additional file:Open peer review report 1.
杂志排行
中国神经再生研究(英文版)的其它文章
- Novel miRNA, miR-sc14, promotes Schwann cell proliferation and migration
- Knowledge domain and emerging trends in Alzheimer's disease: a scientometric review based on CiteSpace analysis
- Lesions of mediodorsal thalamic nucleus reverse abnormal firing of the medial prefrontal cortex neurons in parkinsonian rats
- Mesenchymal stem cell-derived exosomes promote neurogenesis and cognitive function recovery in a mouse model of Alzheimer's disease
- Identification of microRNAs and messenger RNAs involved in human umbilical cord mesenchymal stem cell treatment of ischemic cerebral infarction using integrated bioinformatics analysis
- miR-15b-5p targeting amyloid precursor protein is involved in the anti-amyloid eflect of curcumin in swAPP695-HEK293 cells