m6ARegulates Neurogenesis and Neuronal Development by Modulating H istone Methyltransferase Ezh2
2019-07-12JunchenChenYiChangZhangChunminHuangHuiShenBaofASunXuejunChengYuJieZhangYunGuiYangQiangShuYingYangXuekunLi2
Junchen Chen,Yi-Chang Zhang,Chunmin Huang,Hui Shen Baof ASun,Xuejun Cheng,Yu-Jie Zhang,Yun-Gui Yang,Qiang Shu*,Ying Yang*,Xuekun Li2,*,k
1 The Children’s Hospital,School of Medicine,Zhejiang University,Hangzhou 310052,China
2 The Institute of Translational Medicine,School of Medicine,Zhejiang University,Hangzhou 310029,China
3 CAS Key Laboratory of Genomic and Precision Medicine,Collaborative Innovation CenteRof Genetics and Development,College of Future Technology,Beijing Institute of GenoMics,Chinese Academy of Sciences,Beijing 100101,China
4 University of Chinese Academy of Sciences,Beijing 100049,China
5 Sino-Danish College,University of Chinese Academy of Sciences,Beijing 101408,China
6 Institute of SteMCell and Regeneration,Chinese Academy of Sciences,Beijing 100101,China
KEYWORDS N 6-methyladenosine(m6A);Mettl3;Neurogenesis;
Abstract N6-methyladenosine(m6A),catalyzed by The methyltransferase coMplex consisting of M ettl3 and Mettl14,is The mostabundant RNAmodification inMRNAsand participates in diverse biologicalprocesses.However,The rolesand precisemechanisMs of m6Amodification in regulating neuronal development and adult neurogenesis remain unclear.Here,we exaMined The function of
Introduction
In The adultmammalian brain,adultneuralsteMcells(aNSCs)exist in specific regions,namely,The subventriculaRzone in lateralventriclesand The subgranulaRzone in The dentategyrusof The hippocampus[1,2].aNSCs can self-renew,and exhibitmultipotent capabilities of generating neurons,astrocytes,and oligodendrocytes.The newborn neurons can integrate into existing neural circuits such as those involved in physiological functions including learning and memory[2-4].Recent studies have shown that epigenetic modifications,such as DNAmodifications,histonemodifications,and non-coding RNAs,p lay essential roles in regulating neurogenesis and neuronal development[5-11].
N6-methyladenosine(m6A)modification is The most abundant RNAmodification in The MRNAs of eukaryotic cells.It is involved in Avariety of biological processes including The translation efficiency,degradation,subcellulaRlocalization,alternative sp licing,and secondary structure of RNA[12-21].m6Ais deposited bymethyltransferase-like 3(Mettl3)and several o The Rcomponents of The methyltransferase complex.It is recognized by its YT521-B homology (YTH)domaincontaining proteins and hnRNPA2B1,and is erased by The fat mass and obesity-associated protein (Fto)and The α-ketoglutarate-dependent dioxygenase alkB homolog 5(Alkbh5).Many studies have revealed that The modulation of m6A level is involved in diverse processes including The regulation of fate deterMination,The proliferation and differentiation of steMcells,homeostasis,DNAdamage response,adipogenesis,spermatogenesis,and circadian clock processes[14,18,22-25].
Recently,it has been found that m6Ais prevalent in mRNAs of The mammalian nervous system,and disp lays dynaMic features during embryonic and postnatal neuronal development[16].m6A eraseRFto-deficient Mice disp lay impaired neuronal activity and altered behaviors related to dopaMine signaling[26-28].In addition,The specific knockdown of Fto in The mouse medial prefrontal cortex(MPFC)can promote cued feaRmemory[29].OuRprevious study has also found that The constitutive deletion of Fto inhibits adult neurogenesis in vivo,and iMpairs spatial learning and memory inMice[30].Recently,it hasbeen revealed thatm6Amodification regulates axon development[31,32],and The deletion of Mettl14 oRMettl3 dysregulates embryonic cortical neurogenesis[17,33],postnatal cerebellaRdevelopment[34],and stress responses in Mice[35].All of The se studies suggest iMportant functions of m6Amodification in The neuronal system.However, The mechanistic role of m6Ain regulating The proliferation and differentiation of aNSCs remains largely unknown.
In The present study,we found that both Mettl3 and m6Aexhibit dynaMic and conservative patterns during The differentiation of aNSCs in vitro.Mettl3 dep letion significantly reduced m6Alevel and altered The proliferation and cell cycle progression of aNSCs.Mettl3 depletion also skewed The lineage comMitment more toward glia,and inhibited morphological maturation of newborn neurons both in vitro and in vivo.m6Aimmunoprecipitation combined With deep sequencing(MeRIP-seq)has revealed that m6Atags are predoMinantly enriched in transcripts related to neurogenesis and neuronal development.Mettl3 dep letion specifically dysregulates The expression of genes related to The cellcycle and neuronaldevelopment.Finally,we shoWthat Mettl3 dep letion reduces The levels of histonemethyltransferase Ezh2 and H 3K 27me3.The overexpression of Ezh2 could rescue The defective neurogenesis and neuronal development caused by Mettl3 dep letion.OuRresults thus uncoveRAcrosstalk between RNAmethylation and histonemodifications and demonstrate The regulatory role of m6Amodification in aNSC proliferation and differentiation.
Results
Mettl3 and m6Adisplay dynaMic expressions during aNSC differentiation
To investigate The role of Mettl3 in NSC differentiation and neural development,we first isolated aNSCs froMThe forebrains of adult(2-month-old)Wild-type(WT)Mice,as described in ouRprevious publications[36,37].The cultured aNSCs were positive foRThe neural steMcell markers Sox2 and Nestin(Figure S1A),and could incorporate thyMidine analog 5-bromo-2-deoxyuridine(BrdU)during The proliferation(Figure S1B and C).aNSCs generatedβ-III tubulin positive(Tuj1+)neurons and glial fibrillary acidic protein positive(GFAP+)astrocytes upon differentiation (Figure S1D).mRNAlevels of multiple pluripotency markers and lineagespecific markers underwent significant alterations during The processes froMproliferation to differentiation(Figure S1E). The se results indicate The homogeneity,self-renewalcapability,and multipotency of The cultured aNSCs.
We The n exaMined The expression of Mettl3 in aNSCs.We first performed immunof luorescence staining using Mettl3 and Mettl14 specific antibodies,and found that Mettl3 and Mettl14 resided in The nucleiof Nestin+/Sox2+aNSCs(Figure 1Aand B;Figure S2Aand B).Mettl3 and Mettl14 could also be detected in Tuj1+neuronal cellsand GFAP+glial cells differentiated froMaNSCs(Figure 1C and D;Figure S2C and D).Real-time PCR(RT-PCR)and Western blot results showed Asignificant increase in The expression of Mettl3 and Mettl14 during aNSC differentiation(Figure 1E-H;Figure S2E).During aNSC differentiation,The expression levels of Wtap,which encodes one component of The m6AWriteRcomp lex,and Fto,which encodes an m6Aeraser,were all increased,whereas The expression of Alkbh5,which encodes ano The Rm6Aeraser,did not shoWany significant change(Figure S2F).
G iven The dynaMic expression of Mettl3,we The n performed an RNAdot-blotassay to detectm6Alevel.Consistent with The expression pattern of Mettl3,we found that The global m6Alevel significantly increased froMThe proliferation to differentiation stages of aNSCs(Figure 1I and J).Immunostaining also showed that m6Aexisted in The mature neurons(NeuN+)of The hippocaMpus of The mouse brain(Figure S2G),which could be significantly dep leted by RNase treatment. The se resultssuggest The specificity of The m6Aantibody(Figure S2H).
Mettl3 regulates The proliferation and differentiation of aNSCs
We next aimed to study The regulatory roles of Mettl3 in The proliferation and differentiation of aNSCs.We adopted Alentivirus to deliveRAshort hairpin RNA(shRNA)to knock down Mettl3(Mettl3 KD)in mouse aNSCs(Figure 2Aand B).The dot-blot of m6Ashowed that Mettl3 dep letion significantly decreased globalm6Alevels compared to those of The controlgroup(Figure 2C and D).To assess The effect of Mettl3 deficiency on The proliferation of aNSCs,we first app lied ABrdU incorporation assay and found that The numbeRof BrdU+cellswas significantly decreased in Mettl3 KD aNSCs(Figure 2E-G),suggesting The inhibited proliferation of aNSCs.Itwas noticed that The percentage of Sox2+/Nestin+cells and The numbers of cell cycle markeRK i67+cells had not changed upon Mettl3 KD (Figure S3A-F).Taken toge The r, The se datAsuggest that Mettl3 deficiency inhibits The proliferation of aNSCs but does not affect The iRhomogeneity.
To fur The RexaMine The effects of Mettl3 on The proliferation of aNSCs,we analyzed The expression of multip le cyclins.We found that The MRNAlevels of Cyclin D 1 and D 2,which express throughout The whole cell cycle,did not shoWany obvious differences between control and Mettl3 deficient cells(Figure 2H).However,The mRNAlevels of Cyclin A1,A2,B1,B2,which are specifically expressed in The G 2/Mphase,were significantly decreased in Mettl3 deficient aNSCs compared to those of control cells(Figure 2H).Fur The rmore,we used floWcytometry to analyze The distribution of cells in each phase of The cell cycle.We found that The lack of Mettl3 increased The numbeRof cells in S phase,but decreased The numbeRof cells in G 2/Mphase(Figure 2I-K).The quantification of phosphor-histone H 3(p-H 3)immunof luorescence staining showed Aconsistently decreased numbeRof p-H 3+cells(Figure S3G). The se results indicate that Mettl3 deficiency dysregulates cell cycle progression.
We next studied The rolesof Mettl3 in regulating The differentiation of aNSCs.Immunof luorescence staining showed that Mettl3 dep letion decreased The numbeRof Tuj1+neurons(Figure 3A and B),but The overexpression of Mettl3 significantly increased The numbeRof Tuj1+neurons upon The differentiation of aNSCs foR2 days(Figure 3C-E).The results of qRT-PCRand Western blots consistently showed that The depletion of Mettl3 decreased The level of Tuj1 and increased The levelof GFAP(Figure 3F;Figure S4A),and that The overexpression of Mettl3 led to increased Tuj1 levels and decreased GFAP levels(Figure 3G;Figure S4B).
To fur The Rvalidate The effectsof Mettl3 in regulating aNSC differentiation,we app lied Adual luciferase reporteRassay to analyze The promoteRactivity of The neuronalcellmarker,NeuroD1,and glial cellmarker,Gfap,in aNSCs.Itwas found that The overexpression of Mettl3 could increase The promoteRactivity of NeuroD 1 and decrease The promoteRactivity of Gfap(Figures S4C and D).To fur The RdeterMine The function of Mettl3 in neuronal development,we also performed in utero electroporation.Itwas found that The percentage of neWborn non-neuronal cells(Tuj1-GFP+/GFP+)had significantly increased,meaning The significant decrease in Tuj1+GFP+cells,in The cortical p late of Mettl3 KD Mice(Figure S4E and F). The se resultsdemonstrate that Mettl3 regulates lineage comMitment during aNSC differentiation,With Apreference toward Aneuronal fate.
Mettl3 regulates neuronal development both in vitro and in vivo
To assess The effect of Mettl3 on neuronal development,we fur The Ranalyzed The morphology of neWly born neurons generated upon The differentiation of aNSCs.We observed that both The numbeRof dendritic branches and The total length of dendrites were significantly decreased in neWborn neurons afteRMettl3 KD(Figure 3H-K;Figure S4G),while The y were significantly increased upon Mettl3 overexpression(Figure 3H-K;Figure S4H).We also performed Mettl3 KD on cultured hippocampal neurons,and found that Mettl3 deficient neurons showed Adecreased numbeRof intersections and dendrites,Areduced total length of dendrites,and an increasedmean length of dendrites(Figure 3L-O;Figure S4I).Knocking down of Mettl3 by in utero electroporation consistently resulted in The numbeRof dendrites peRneuron also being significantly decreased and The mean length of dendrites being increased(Figure 3P-R). The se results suggest that Mettl3 promotes neuronal development both in vitro and in vivo.
m6Amodified transcripts are involved in neurogenesis and neuronal development

Figure 2 Mettl3 regulates The proliferation of aNSCsRT-PCRresults shoWing The MRNAlevelsof Mettl3 in controland Mettl3 KD aNSCs(A).Actin wasused asan internalcontrol(n=3).Western blot assays shoWing decreased Mettl3 in Mettl3 KD aNSCs coMpared to control saMp les(B).Gapdh was used as internal control.Dot-blot assay(C)and quantification(D)revealing The depletion of m6Ain Mettl3 KD aNSCs coMpared to The controlsaMples(n=3).Representative images of BrdU immunostaining in both control(E)and Mettl3 deficient(F)aNSCs.The quantification analysis of BrdU immunostaining in control and Mettl3 KD aNSCs(n=3)(G).The expression levels of multip le cyclin genes in control and Mettl3KD aNSCswere detected by qRT-PCR(H)(n=3).Actin wasused asan internalcontrol.FloWcytometry analysisof The cellcycle status of control(I)and Mettl3 KD aNSCs(J),and The percentage in each phase(K)(n=3).DatAare presented asmean±SEM,unpaired t-test,*,P<0.05;**,P<0.01;***,P<0.001.Scale bar,50μm.BrdU,5-bromo-2-deoxyuridine;KD,knockdown.
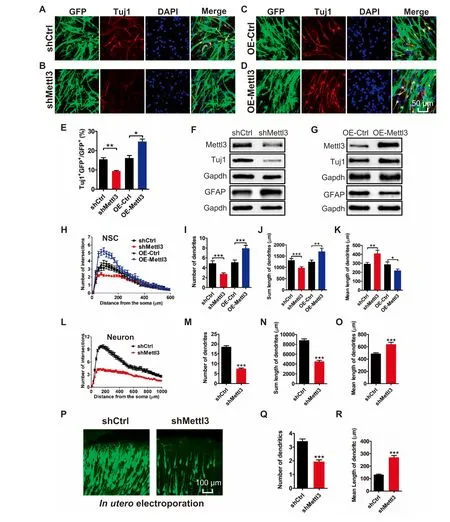
Figure 3 Mettl3 regulates The differentiation and neuronal development of aNSCsRepresentative immunof luorescence imagesof The differentiated aNSCsWith neuronal cellmarkeRTuj1 of control(A,C),Mettl3 KD(B),and Mettl3 OE(D)groups.Scale bar,50μm.Quantification of Tuj1+cells in differentiated aNSCs with Mettl3 KD and Mettl3 overexpression(n=3)(E).Western blot assays shoWing The protein levels of The neuronal cellmarker,Tuj1,and astrocyte marker,GFAP,in aNSCs(differentiation condition)With Mettl3 KD(F)and Mettl3 overexpression(G).Sholl analysis of newborn neurons generated upon The differentiation of aNSCs(H-K)(n=40).Sholl analysis of cultured hippocampal neurons(L-O)(control group:n=75;KD group:n=66).Representative images of in utero electroporation(P).The quantifications of The number(Q)and length(R)of dendrites.DatAare presented as mean±SEM,unpaired t-test,*,P<0.05;**,P<0.01;***,P<0.001.Scale bar,50μm.OE,overexpression.

Figure 4 Dynamicm6Amodification froMproliferation to differentiation of aNSCsTranscriptome-Wide distribution of m6Apeaks in aNSCs undeRThe conditions of proliferation and differentiation(A,B)(n=2).The most common sequencemotifamongm6Apeaks during proliferation and differentiation(C).Venn diagraMillustrating The m6Amodified genes in proliferating and differentiated samp les(D).GO analysisof The common m6Amodified genes shared between proliferating and differentiated saMples(E).GO analysis of The m6Amodified specific genes identified in proliferating saMp les(F)and differentiated saMples(G).Venn diagraMillustrating The totalm6A-tagged genes identified in proliferating and differentiated saMples,and differentially expressed genes identified in differentiated samples compared to proliferating samp les(H).GO analysis of The common genes shared between m6Atagged genes and differentially expressed genes(I).IGV tracks shoWing several genesWith differentialm6Amodification froMThe proliferation to differentiation of aNSCs(J).RNA-seq reads were used as input.TSS,transcription start site;GO,Gene Ontology;Proli,proliferation;D iffe,differentiation;IGV,Integrative GenoMics Viewer.
To systematically illuMinate The function of m6Ain neurogenesis and neuronal development,we performed m6Aimmunoprecipitation combined With deep sequencing(MeRIP-seq)to detect The m6Apeaksand exp lore The iRdistribution in The transcriptome of aNSCs undeRproliferation and differentiation conditions(Figure S5A).We observed in both proliferating and differentiated aNSC samp les that The m6Apeakswere predoMinantly located in The coding sequence(CDS)and that The 3′untranslated regions(3′UTR)were especially enriched neaRstop codon regions(Figure 4Aand B). The m6Adistribution pattern disp layed Ahigh degree of siMilarity between proliferation and differentiation conditions,and was enriched in The m6Aconsensus motif,which was consistent With previous reports[15,18](Figure 4C).Bioinformatic analysis revealed that 9309 and 7411 m6Amethylated mRNAs were detected in proliferation and differentiation conditions,respectively(Table S1),whereas6569m6AmethylatedmRNAsoverlapped between The two conditions(Figure 4D).Gene ontology(GO)analysis showed The functional enrichment of The se 6569 overlapped genes was related to The transcription,neurogenesis,neuronal differentiation and cell cycle related pathways(Figure 4E).Fur The rmore,2740 unique m6Amethylated RNAs in proliferating samp les were enriched in terms related to DNAreplication and The cell cycle,while 842 unique m6Amethylated genes in differentiated samp les were enriched in terms related to neurogenesis,neuronal development and differentiation(Figure 4fand G).
To deterMine The relationship between m6Amodification and gene expression,we next performed RNA-seq in The se two conditions to uncoveRany global transcriptomealterations(Figure S5B).It was found that 8404 genes displayed altered expression froMproliferation to differentiation conditions of aNSCs:4378(52.09%)MRNAs were up-regulated while 4026(47.91%)geneswere down-regulated(Table S2).Among The se 8404 genes,5526(65.75%)genesweremodified bym6A,including 3065 (55.47%)up-regulated genes and 2461(44.53%)down-regulated genes(Figure 4H;Table S3).GO analysis showed that The se 5526 geneswere enriched in pathways related to The cell cycle,nervous systeMdevelopment and neuronal differentiation (Figure 4I).Key exaMp les include,Sox11 and Cdk12,involved in proliferation,and Notch2 and Neurog2,related to neuronal differentiation(Figure 4J;Figure S5C).Taken toge The r, The se datAsuggest thatm6Amodification p lays Akey role in regulating The gene expressions of aNSCs.
Mettl3 regulatesm6Amodification of neurogenesis-related genes
G iven that Mettl3 regulates neurogenesis and neuronal development,we next sought to investigate whe The RMettl3 KD affected gene expression related to those two areas.We first exaMined m6Adistribution patterns in control and Mettl3 KD aNSCs(in Aproliferation condition)(Figure S5A),and observed that Mettl3 KD had not altered The overallm6Adistribution patterns in transcripts coMpared to those of The control saMp les(Figure 5A).Fur The rmore,m6Awas enriched in The samem6Aconsensusmotifs of both Mettl3 KD and controlsaMp les,even though The globalm6Alevelhad been significantly decreased(Figure2C;Figure 4C;Figure 5A).
To exaMine The effects of Mettl3 dep letion on gene expression,we performed RNA-seq in control and Mettl3 KD samp les(Aproliferating condition)(Figure S5B).RNA-seq datAanalysis showed that Atotal of 1226 genes exhibited altered expressions in Mettl3 KD saMples coMpared With those of control saMp les(Fold change>1.5),including 844(68.84%)up-and 382(31.16%)down-regulated genes(Figure 5B;Table S4).Among The up-regulated genes,371 genes(43.96%)were methylated by m6A,while 176 downregulated genes(46.07%)weremethylated bym6A(Figure 5B and C).GO analysis showed that The m6Atagged up-regulated genes were enriched in areas of neuronal differentiation,neurogenesis and nervous systeMdevelopment(Figure 5D and E).The representative IGV images showed that m6Atagged down-regulated genes were enriched in terms related to The neuronaldevelopment,such as VegfAand Syt4,and cell cycle,cell proliferation,such as Tet1 and E2f7(Figure 5F).The analysis using STRING database showed The interaction between proteinscoded bym6Atagged transcripts(Figure5G).Meanwhile,we also performed GO analysis of differentially expressed genes Without m6Amodification,and found that The se genes showed loWcorrelations With neuronal development and The cell cycle(Figure S5E and F).Toge The r, The se results suggest that altered m6Amodification induced by Mettl3 dep letion impacts The expression of genes related to cell cycle progression and neuronal development.
Ezh2 rescues The deficits of neuronal development and neurogenesis induced by Mettl3 depletion
One recent study hasshown thatm6Aregulates specific histone modifications[38].Through analyzing m6Asequencing data,we observed that The transcripts of histonemethyltransferase Ezh2,which plays iMportant roles in neurogenesis and neuronal development[37,39,40],were tagged withm6Amodification(Figure S6A).The region With The largestm6Apeak was on exon 10 of Ezh2 and this was validated by m6ARIP followed by qPCR(Figure S6B).m6Aenrichment at Ezh2 was significantly decreased upon Mettl3 dep letion,as confirmed by m6A-IP-qPCR(Figure S6C).Western blot results showed that The protein level of Ezh2 was significantly decreased upon Mettl3 dep letion,whereas itsmRNAlevel did not change(Figure 6Aand B;Figure S6D).Consistently,The protein level of H 3K 27me3 was also decreased afteRMettl3 dep letion while no observable changeswere noted in The levels of H 3K 4me3(Figure 6C).We fur The Rconstructed Amutant Ezh2 plasMid(The site Within The biggestm6Apeak on exon 10 was denoted in Figure S6Aand wasmutated).The overexpression of WT ormutant Ezh2 showed siMilaRtranscription efficiencies(Figure S6E and G),and nei The Rof The Mwere observed to affect any protein and MRNAlevels of Mettl3 in ei The RaNSCs oRN 2Acells(Figure S6F;Figure S6H and I).Consistentwith The effectsof Mettl3 KD on The expression of Ezh2,Mettl3 overexpression notonly significantly increased The global levelof m6A,butalso upregulated The levelsof Ezh2 and H 3K 27me3(Figure S6J-L). The se results suggest that Mettl3 regulates Ezh2,but not vice versa.
Next,we analyzed The effect of knocking down Mettl3 on The expression of exogenous Ezh2.We found that The protein levels of both endogenous and exogenousWT Ezh2 were significantly decreased upon Mettl3 KD(Figure 6D and E),while mRNAlevels remained unaltered(Figure S6D).However,Mettl3 knockdown exhibited less effect on The protein level of mutated Ezh2(Figure 6D and E).Toge The r, The se results suggested Mettl3 deposited m6Amodification regulates Ezh2 expression at The translational level.
We finally exaMined whe The REzh2 could rescue The deficits of aNSCs induced by Mettl3 depletion.The results showed that The overexpression of Ezh2 could increase proliferation(indicated by The numbeRof BrdU+cells)(Figure 6fand G)and promote neuronal differentiation (indicated by The numbeRof Tuj1+cells)of aNSCs induced by Mettl3 KD(Figure 6H and I).Fur The rmore,The dendritic numbeRand length,and The numbeRof intersections of neWborn neurons,were also increased upon Ezh2 overexpression(Figure 6J-M).Taken toge The r, The se results indicate that overexpression of Ezh2 could rescue The deficits of neurogenesis and neuronal development induced by Mettl3 dep letion.

Figure 5 Mettl3-mediated m6Aregulates gene expression in proliferating aNSCsTranscriptome-wide distribution of m6Apeaks in Mettl3 KD samples(n=2)(A).Venn diagraMillustrating The up-regulated and downregulated genes and m6A-modified transcripts in Mettl3 KD saMples(B).Volcano plot shoWing differentially expressed genes between control and Mettl3 KD saMples(C).GO analysis of m6A-tagged up-regulated(D)and down-regulated(E)transcripts in Mettl3 KD samples.FouRexamplesof IGV tracks showing that The enrichment of m6Ain several transcripts,which are related to neurogenesisand neuronal development,were significantly decreased in Mettl3 KD saMp les(F).RNA-seq reads were used as input.The interaction network shoWing differentially expressed transcriptsWith m6Amodification.Genes related to cell cycle and neuronal differentiation are marked in red and gray,respectively,whereas genes related to both processes aremarked in green(G).

Figure 6 Ezh2 is regulated by Mettl3 and can rescue The deficits of neuronal development and neurogenesis induced by Mettl3 knockdownWestern blot assay shoWing Mettl3 KD led to Asignificant decrease in Ezh2(A,B),and H 3K 27me3(C)at The protein level(n=3).Western blot assay(D)and quantification(E)shoWing that Mettl3 KD significantly decreased expression of WT Ezh2,but had lesseffect on The expression of mutant Ezh2(n=3).Representative images(F)and quantification(G)shoWing that The overexpression of Ezh2 rescued The reduced proliferation induced by Mettl3 KD(n=3).Representative images(H)and quantification(I)shoWing that The overexpression of Ezh2 rescued The inhibited neuronal differentiation induced by Mettl3 KD(n=3).Ezh2 rescued The iMpaired morphologicalmaturation of newborn neurons induced by Mettl3 KD(n=30)(J-M).DatAare presented asmean±SEM,unpaired t-test,*,P<0.05;**,P<0.01;***,P<0.001.Scale bar,50μm.
Discussion
Adult neurogenesis is amulti-step event involving The maintenance of The steMcell pool,lineage comMitment,maturation,and The establishment of neural circuits.All of The se processes are precisely and intensively regulated by genetic and epigeneticmechanisMs.Here,we have demonstrated The iMportant roles of Mettl3-mediated m6A methylation in regulating The neurogenesis and neuronal development through modulating The expression of histonemethyltransferase Ezh2(Figure S6M).
m6Ahasbeen shown tomodulate The self-renewal,differentiation,and lineage deterMination of multip le steMcell types by regulating gene expression,especially The expression of key transcription regulators[18,23,25,41,42].The distribution pattern of m6Ais highly conservative With an abundance in 3′UTRsand neaRThe stop codonsobserved acrossdifferent cell types[16,18,19].OuRresults have shown that,in The transcriptomes of aNSCs,m6Aexhibits siMilaRdistribution features,and is enriched at siMilaRmotifs. The se highly conservative features of m6Amodification indicate its important function in different cell types.
DynaMicm6Amodification hasnotonly been found during neuronal development,but also can be induced by neuronal activity and learning,pointing to The function of m6Ain The neuronal system[16,26,27,29,43].OuRprevious study showed that The deletion of Fto inhibits adult neurogenesis aswell as learning and memory through its regulation of BDNF[30].The deletion of ei The RMettl3,Mettl14,oRYthdf2 could disturb embryonic cortical neurogenesis in Mice[17,33].Ectopic expression of Mettl3 induced neuronal defects in The cerebellum[34].OuRpresent results indicate that Mettl3 depletion not only inhibits The proliferation and cell cycle progression of aNSCs,also skews The iRlineage comMitmentmore toward gliAduring The differentiation in vitro.Moreover,ouRstudy also showed thatm6Amodification regulates The morphologicaldevelopmentof neurons.We found that,besides The RNAs tagged bym6Athat are highly overlapping between proliferation and differentiation conditions,m6Aalso specially tags unique MRNAs undeRei The Rproliferation oRdifferentiation conditions.It is plausible to speculate that The dynaMic and specific distribution patterns of m6Atagging on transcripts are highly correlated to its function in aNSCs.In this way, The se results fur The Rhighlight The function of m6Amodification in neuronal development and adult neurogenesis.
The roles exactly p layed by m6Amodification are undeRintensive study.Previous studies have reported thatm6Aregulatesgene expression throughmodulating sp licing and The efficiency of translation[19,44-46].Although Mettl14 itselfdoes nothave catalytic activity,The deletion of Mettl14 could significantly reduce globalm6Alevels,disturb embryonic cortical neurogenesis,iMpaiRstriatal-mediated behavioRand function through affecting global transcriptomes[17,47].All The se studies indicate The rolesof RNAmethyltransferase-mediatedm6Ain regulating gene expression during development and during The operation of various physiological functions such as that of neuronal function.One recent study has indicated that m6A modification affects histone modification.Mettl14 hap loinsufficiency led to genome-Wide changes in histonemodification including increased H 3K 27me3,H 3K 27ac,and H 3K 4me3[38].However,we observed that The level of The methyltransferase Ezh2 was significantly decreased in Mettl3 deficient cells,while its overexpression could rescue Mettl3 deficiency induced phenotypes.Whe The Rthis discrepancy was due to The specific cell types undeRstudy requires fur The RexaMination.
In summary,ouRstudy has revealed The critical roles of Mettl3-mediatedm6Amodification in regulating cell cycle progression,lineage comMitment and neuronal development of aNSCs through modulating histone methyltransferase Ezh2. The se findings highlight The function of epitranscriptoMic mechanisMs in neuronal development and adult neurogenesis.
Materials andmethods
Animals
Mice were housed in standard conditions in The Laboratory Animal CenteRof Zhejiang University on A12 h light/dark scheduleWith freeaccess to food and water.The pregnantMice were purchased froMShanghai SLAC Laboratory Animal CoMpany,China.All animal experiments were conducted according to protocols approved by The Zhejiang University Animal Care and Use ComMittee.
Isolation and culture of aNSCs
The isolation of NSCs froMThe forebrain of adultMice was performed as described previously[36,37].aNSCs were cultured in DMEM/F-12 mediuMcontaining 20 ng/Ml FGF-2(Catalog No.100-18B-B;PeproTech,Rocky Hill,NJ),20 ng/Ml epidermal groWth factor(Catalog No.100-15;PeproTech),2%B27 supplement(Catalog No.12587-010; The rmo FisheRScientific,Grand Island,NY),1% antibiotic-antimycotic(Catalog No.15140-122; The rmo FisheRScientific),and 2mML-glutaMine(Catalog No.25030-149; The rmo FisheRScientific)in A5%CO2incubatoRat 37°C.
In vitro proliferation and differentiation assay
FoRThe in vitro proliferation assay,aNSCs were cultured on coverslips With mediuMsupplied With 5μMBrdU foR8 h.FoRThe in vitro differentiation assay,aNSCs were cultured on coverslipsWith proliferation medium,and The n transferred into differentiation mediuMcontaining 1μMretinoic acid(Catalog No.R-2625;Sigma,Saint Louis,MO)and 5μMforskolin(Catalog No.F-6886;Sigma)foR48 h.
Immunof luorescence staining
Afterwashing With PBS foR30Min,cell samp les on coverslips oRbrain sections were blocked With 3%normal goat seruMand 0.1%triton X-100 in PBS foR1 h at rooMteMperature.Samples were incubated with primary antibodies oveRnight at 4°C.The second day,cells oRsections were washed With PBS foR30Min,and The n incubated With Fluorophoreconjugated secondary antibodies foR1 h at rooMteMperature.FoRBrdU immunostaining,saMp leswere pretreated With 1MHCl at 37°C foR30Min before The app lication of block solution.The folloWing primary antibodieswere used:Mettl3(Catalog No.21207-1-AP;Proteintech,Rosemont,IL),Mettl14(Catalog No.HPA038002;ATLAS,Bromma,Sweden),m6A(Catalog No.202003;Synaptic SysteMs,Goettingen,Germany),NeuN(Catalog No.AB2237;Millipore,Burlington,MA),Nestin(Catalog No.556309;BD PharMingen,San Jose,CA),SOX 2(Catalog No.sc-365823X;SantACruz Biotechnology,Dallas,TX,USA),Tuj1(Catalog No.G 712A;Promega,Madison,WI),GFAP(Catalog No.Z0334;DAKO,SantAClara,CA),BrdU(Catalog No.ab6326;Abcam,Cambridge,MA),and K i67(Catalog No.AB9260;Millipore).
Electroporation and luciferase assays
Electroporation was performed With an electroporator(Catalog No.AAB-1001;AmaxALonza,Germany)as described previously[37].Briefly,The cultured aNSCs were collected and resuspended With 100μl nucleof ection solution and electroporated using The manufacturer’s protocol.The electroporated cells were cultured With fresh proliferation mediuMand The mediuMwas rep laced With differentiation medium(5μMforskolin and 1μMretinoic acid)on The second day.48-60 h later,The cellswere collected foRAluciferase assay With AluMinometeRaccording to The manufacturer’s protocol(Promega).0.1μg Renilla-luciferase p lasMids and 2μg NeuroD 1-/G fapluciferase p lasMidswere used foReach electroporation,respectively.To knock down Mettl3,short hairpin RNA(shRNA)targeting Mettl3(5′-taagcacactgatgaatcttt-3′,Qiagen,H ilden,Germany)was cloned into to lentivirus-U 6 vectors.
Total RNAisolation,reverse transcription,and quantitative real-time PCR
Total RNAwas extracted froMaNSCs using TRIzol reagent(Catalog No.15596018; The rmo FisheRScientific).Total RNAwas isolated and The concentration was quantified using ANanoD rop spectrophotometeR2000( The rmo FisheRScientific).0.5μg total RNAwas used foRreverse transcription(RT)using ART reagent kit(Catalog No.R223-01;Vazyme,Nanjing,China).Quantitative real-time PCR(qRT-PCR)was performed using SYBRG reen(Catalog No.Q71502;Vazyme).All real-time PCRreactionswere performed in triplicate,and The resultswere analyzed using TheΔΔCtmethod.
Western blot
The collected cells were washed With PBS and resuspended With RIPAbuffer(Catalog No.ab156034;Abcam,Cambridge,MA,USA)containing 1×protease inhibitoRcocktail(Catalog No.04693124001;Sigma,Saint Louis,MO,USA). The samples were centrifuged at 4°C for 20Min at 14,000 rpMand The supernatantswere collected.Samp leswere The n denatured foR5Min at 95°C and The n subjected to SDSpolyacrylaMide gel electrophoresis.The folloWing primary antibodies were used:anti-Mettl3(Catalog No.21207-1-AP;Proteintech), anti-Mettl14 (Catalog No. HPA038002;
ATLAS),anti-Tuj1(Catalog No.G 712A;Promega),anti-GFAP(Catalog No.3670;Cell Signaling,Danvers,MA),anti-tubulin(Catalog No.ab15246;Abcam),anti-Ezh2(Catalog No.3147;Cell Signaling),anti-H 3K 27me3(Catalog No.07-449;Millipore),anti-H 3K 4me3(Catalog No.ab8580;Abcam),anti-H istone3(Catalog No.ab1791;Abcam),and anti-Gapdh(Catalog No.AM4300; The rmo FisheRScientific).Secondary HRP conjugated antibodieswere app lied foR1 h at rooMteMperature.The signal was detected by Tanon Detection system(Tanon 5200,Shanghai,China)and The intensity of immuno-blot bands was normalized to those of Gapdh oRTubulin.
m6Adot-blot assay
FoRm6Adot-blot,total RNAsaMp les were denatured at 65°C,and The n spotted onto Hybond N+membranes(Catalog No.NP1096;GE Healthcare,Buckinghamshire,UK).Membraneswere blocked With 5%Milk foR1 h at rooMtemperature,and The n incubated with primary antibodiesovernight at 4°C.On The next day,membranes were incubated With HRP-conjugated secondary antibodies foR30Min at room temperature.The signal was detected by Tanon Detection system,and The signal density was quantified using Photoshop sof tware.
Cell cycle analysis
To analyze The cell cycle aNSCs,propidiuMiodide(PI)staining was performed according to The manufacturer’s instructions(Multi Sciences,Hangzhou,China).In brief,afteRsubculture foR24 h,aNSCs were harvested and fixed With absolute ethanol.The pellet was The n dislodged in PBS and stained With propidiuMiodide solution at rooMteMperature foR30Min.The cells were The n analyzed using Cytof LEX(Beckman Coulter,Boulevard Brea,CA)and datAwere analyzed using FloWJo sof tware.
In utero electroporation(IUE)
In utero electroporation wasperformed asdescribed previously[48].Briefly,The timed pregnant C57 Mice(E13.5)were anes The tized using isof lurane.The uterine hornswere The n exposed and ba The d in warMPBS.2μl recombinant p lasMid(final concentration 1.5mg/Ml)Mixed With fastgreen(0.01%)wasmanually Microinjected into The lateral ventricle With Aglass Micropipette(H irschmann DE-M16).FoRelectroporation,five 100-Microsecond pulses of 35 V With A900-Microsecond intervalwere delivered across The uterus using an electroporator(Catalog No.101438;BEX,Tokyo,Japan).AfteRelectroporation,The uterine horns were p laced back into The abdoMinal cavity to alloWThe embryos to continue normal development.The pregnantMice were sacrificed at scheduled time points as indicated and The embryos were harvested foRfur The Ranalysis.
mRNApurification,m6AMeRIP-seq and m6AMeRIP-qPCR
m6AMeRIP-seq was carried out as previously described With somemodifications[16,25,49].In brief,mRNAswere purified froMtotal RNAsusing ADynabeads®mRNApurification kit(Catalog No.61006; The rmo FisheRScientific),and digested With DNase I to remove any potential DNAcontaMination.mRNAs were fragmented to around 100 nt using an RNAfragmentation reagent(Catalog No.AM8740; The rmo FisheRScientific)through incubation at 90°C foR1Min,and were The n precipitated With ethanol. The m6Apolyclonal antibody(Catalog No.202003;Synaptic SysteMs)was incubated With 40μl DynabeadsTMProtein A(Catalog No.10001D; The rmo FisheRScientific)in 500μl IPP buffer(150mMNaCl,0.1%NP-40,10mMTris-HCl,pH 7.4)foR1 h at rooMteMperature.The recovered mRNAswere denatured at 75°C foR5Min and put on ice immediately.5μg of fragmented mRNAs were added to The antibody-bead Mixture followed by The incubation at 4°C foR4 h.AfteRextensivewashing by IPP buffeRfive times, The m6A-containing RNAs were eluted foRtWice With 300μl 0.5mg/Ml N6-methyladenosine(Catalog No.P3732;Berry&Associates,Dexter,MI)at rooMteMperature foR1 h using gentle rotation.The eluted samp les were combined toge The Rand extracted With Acid Phenol(pH 4.3-4.7),followed by standard ethanol precipitation.The recovered RNAwassubjected to The cDNAlibrary construction by using KAPAStrandedMRNA-Seq K it IlluMina®p latform(Catalog No.KK 8401;KAPA,Boston,MA)and sequenced on The HiSeq 3000 platform.
RNA-seq and MeRIP-seq datAanalysis
MeRIP-seq and RNA-seq was performed.The proliferation(proli)versus differentiation(differ)cells,and control versus mettl3 knockdown cellswere conducted in two biological rep licates using an IlluMinAHiSeq 3000 platform.RaWreads of each saMple(n=2 foReach group)were trimmed using The Trimmomatic sof tware foReach saMp le to remove adaptoRsequencesand baseswith loWquality[50].The processed reads With length largeRthan 35 nt were The n aligned to The mouse reference genome(version mm10,UCSC)using TopHat2[51]With default parameters.Only uniquemapped readsWith mapping quality no less than 20 were kept foRThe subsequent analysis.
FoRMeRIP-seq, The MACS2 sof twarewas used to identity m6A-enriched(version 2.0.10)[52],With The corresponding input saMple serving as control.MACS2 was run With default options except for‘-nomodel,-keepdup all’to turn of ffragment size estimation and to keep all uniquelymapping reads,respectively.To identify high confidential oRoverlapped m6Apeaks,peaks were intersected in ApairWise fashion among two replicates oRbetween two conditions using The BedTools package by setting‘-f0.5’[53].Fold changes foRm6Apeaks were obtained froMMACS2 output.
FoRRNA-seq, The numbeRof readsmapped to each Ensemble gene(release 68)were counted using The HTSeq python package[54],With The ‘union’overlap resolution mode and unstranded count feature by setting‘--mode=union’and‘--stranded=no’,respectively.The expression of transcripts wasquantified as reads peRkilobase of exonmodel perMillion mapped reads(RPKM). The se RNAs,which aremethylated by m6Ain both conditions,are defined as overlapped m6Amethylated RNAs.
Motifidentification withinm6Apeaks
Motifs enriched in m6ApeaksWithin allMRNAswere identified using HOMERsof tware(v4.7)[55].BEDTools’shuffleBed(version 2.16.2)was used to generate randoMpeaks Within totalMRNAs as background sequences[53]. The motiflength was restricted to 5-6 nucleotides.
Statistical analysis of differentially expressed genes
To identify differentially expressed genes,The R-package DEGseq was used With fold-change≥1.5,P≤1×10-3and The method MARS(MA-plot-based method With randoMsamp lingmodel)as The parameters[56].
Gene ontology analysis
Gene ontology(GO)analysis was performed using The DAVID database[57].Enrichment maps were generated by Cytoscape(version 3.5.0)With The Enrichment Map p lugin[58].Each enriched GO function terMis represented by Anode and The node size is proportional to The numbeRof genes in its corresponding function terMin The enrichment maps.The thicknessof each edge represents The numbeRof common genes between two linked nodes.SiMilaRGO functions are categorized as one cluster.The function terMand The numbeRof genes in each clusteRare labeled.Agene interaction network was generated using STRING[59].
Statistical analysis
All datAare expressed as mean±SEM.G raphPad Prism(G raphPad Sof tware Inc.)was used foRstatistical analysis.Unpaired student’s t-testwasused to deterMine The differences between two groups;Atwo-way ANOVAanalysis followed by Bonferronimultip le-comparison test was used to deterMine differences between multip le groups.P<0.05 was considered statistically significant.
DatAavailability
The raWsequence datAreported in thispapeRhavebeen deposited in The Genome Sequence Archive[60]in BIG DatACenter[61],Beijing Institute of GenoMics(BIG),Chinese Academy of Sciences,as GSA:CRA001248,which is publicly accessible at http://bigd.big.ac.cn/gsa.
Authors’contributions
XL and QS conceived and designed The project.JC,HS,XC,and Y-JZ performed aNSC culture,neurogenesis assay,m6Adot-blot,RT-PCR,and Western blot assays.JC,YZ,CH,and BS performed RNA-seq and MeRIP-seq datAanalysis.YY did RNA-seq and MeRIP-seq library construction.XL,JC,YZ,BS,YY,YGY,and QS analyzed The data.XL and YY Wrote The manuscript With input froMall o The Rauthors.All authors had read and approved The finalmanuscript.
Competing interests
The authors declared no competing financial interests.
AcknoWledgments
XL was supported in part by The International Collaboration PrograMof The Ministry of Science and Technology of China(G rant No.YS2017YFGH 001214),The National Natural Science Foundation of China(G rant Nos.31771395 and 31571518),and The National Key R&D PrograMof China(G rant No.2016YFC0900400).YY was supported by The National Natural Science Foundation of China(G rant No.31770872),The Youth Innovation Promotion Association(G rant No.CAS2018133)and The National Key R&D PrograMof China,SteMCell and Translational Research(G rant No.2018YFA0109700).QS was supported in part by The National Key R&D Program of China (G rant No.2017YFC1001703),The Key R&D PrograMof Zhejiang Province(G rant No.2017C03009),and Zhejiang Provincial PrograMfoRThe Cultivation of H igh-level Innovative Health Talents(2016-6),China.
Supplementary material
Supp lementary datAto this article can be found online at https://doi.org/10.1016/j.gpb.2018.12.007.
杂志排行
Genomics,Proteomics & Bioinformatics的其它文章
- Progress and Challenges foRLive-cell Imaging of Genomic Loci Using CRISPR-based Platforms
- Single-cell Analysis of CAR-T Cell Activation Reveals AMixed TH 1/TH 2 Response Independent of Diあerentiation
- Global Quantitative Mapping of Enhancers in Rice by STARR-seq
- Integrating Culture-based Antibiotic Resistance Prof ileswith Whole-genome Sequencing DatAfoR11,087 Clinical Isolates
- Chronic Food Antigen-specific IgG-mediated Hypersensitivity Reaction as ARisk FactoRfoRAdolescent Depressive Disorder
- Transcriptome and Regulatory Network Analyses of CD19-CAR-T Immuno The rapy foRB-ALL
