制药废水厂微生物群落和多种抗性基因相关性分析
2019-06-11袁立霞罗晓张文丽蒋永丰
袁立霞 罗晓 张文丽 蒋永丰
摘要:为了研究多种生物处理工艺制药废水中微生物群落结构和抗性基因的分布特征、扩增情况及其相关性,采用Miseq高通量测序分析技术和荧光定量PCR技术对制药废水厂中活性污泥进行检测。荧光定量结果表明:sul1,sul2,tetO,tetQ,tetW,OXA-1和可移动遗传元件int1在制药废水厂中各个阶段均能被广泛地检测到,总抗性基因浓度范围为3.09×108~2.26×109 copies/g(干重),基因总浓度上升了7.3倍。Miseq测序结果表明:制药废水中主要优势菌门为Proteobacteria,Bacteroidetes,Firmicutes,Thermus和Gemmatimonadetes等门,其平均总相对丰度比例占到81.05%;冗余分析显示,Aeromicrobium与sul2呈较高程度的正相关性,可能是sul2在微生物群落中存在的可能的主要菌群;Rhodovulum和Rhodospirillaceae菌属与OXA-1和tetQ呈较高程度的正相关性,这些菌属是这些ARGs分布的主要菌属;Acinetobacter与tetO和tetW呈现较高程度的正相关性,可能是tetO和tetW在微生物群落中存在的可能的主要菌属。因此,相关抗性基因的增值和分布与相关特定菌属有关,可以通过控制相关菌属的丰度来消减工业废水厂中的抗性基因。
关键词:微生物生态学;抗性基因;荧光定量PCR;Miseq;微生物群落结构
中图分类号:X703文献标志码:A
Abstract: In order to study the distribution, amplification and correlation of microbial community structure and resistance genes in pharmaceutical wastewater, a variety of biological treatment processes are used to treat pharmaceutical wastewater. Combining Miseq high throughput sequencing analysis technology and fluorescence quantitative PCR technology, activated sludge in pharmaceutical wastewater plants is detected. The results show that sul1, sul2, tetO, tetQ, tetW, OXA-1 and INT1 are widely detected at all stages of pharmaceutical wastewater treatment plant. The total resistance gene concentration ranges from 3.09×108 to 2.26×109 copies/g (dry weight), and the total gene concentration increases by 7.3 times. Miseq sequencing results show that the dominant bacteria in pharmaceutical wastewater are Proteobacteria, Bacteroidetes, Firmicutes, Thermus and Gemmatimonadetes, with an average relative abundance ratio of 81.05%. Redundancy analysis shows that Aeromicrobium is positively correlated with sul2 to a high degree, which might be sul2 existing in microbial communities. Among the possible major microflora, Rhodovulum and Rhodospirillaceae are highly positively correlated with OXA-1 and tetQ, which are the main microflora of ARGs distribution. Acinetobacter has a high degree of positive correlation with tetO and tetW, which may be the main possible microflora of tetO and tetW in microbial community. The results show that appreciation and distribution of antibiotic resistance gens are connected with concerned specific bacterial genus. The controlled abundance of concerned bacterial genus could subduce resistance gene in industrial wastewater factory.
Keywords:microbial ecology; antibiotic resistance genes;fuorescence quantitative PCR;Miseq;microbial community structure
由于抗生素廣泛应用于人类和动物(畜禽/鱼)的疾病防治或动物的促进生长,大量残留抗生素进入环境,致使微生物在持续抗生素选择下产生了耐药性,形成了耐药菌[1-2],其风险比抗生素药物本身的污染风险更高[3],关于其行为特点和传播途径的研究日益增加。抗生素抗性细菌对抗生素的耐药性机理包括:细菌外膜不渗透性障碍、细菌外排泵系统、抗生素作用的靶位变化和抗生素的钝化失活等[4]。抗性基因是抗性菌具有抗性的主要原因之一,它可以通过多种形式的可移动遗传元件如质粒、整合子、转座子、插入序列等,突破细菌的种属关系广泛传播,加重抗性污染。FORSBERG等[5]证明人类病源菌能传播抗性到土著细菌中,并且由土著细菌携带的抗性基因能转导到病原菌的宿主细胞。抗性能潜在地影响环境中的微生物群落结构并对公共健康产生威胁[6],且碳青霉烯类抗生素耐药肠杆菌科细菌患病率上升迅速[7],这将对环境和人类健康构成严重的威胁[6,8]。
分析荧光定量PCR技术为研究环境中抗性基因和抗性微生物分布特征提供了有力的手段[8]。已有研究表明制药废水处理系统的进水中往往含有高浓度的抗生素及生产原料和中间体[9],对生物处理单元的微生物群落形成高强度选择性压力,为ARGs在污水中的持久存在和传播扩散提供了有利条件,并且制药废水处理系统出水中ARGs的浓度高于市政污水处理系统出水[10-11]。22种类型的ARGs能对5种类型的抗生素产生抗性,包括sul 1,sul 2和sul 3能对磺胺类抗生素产生抗性;tetO,tetQ,tetW,tetM和tetT对四环素类抗生素产生抗性;OXA-1和OXA-2能对β-内酰胺类抗生素产生抗性。可移动遗传元件(例如1类整合子)有利于形成多重耐药性并且加速地表水、土壤[12-13]和污水处理厂[14]中抗性基因的传播,并且可移动遗传元件(1类整合子)是细胞间转移抗性的指标[15]。本文主要研究多种抗性基因在制药废水系统生物处理单元中增值和传播以及微生物群落的动态变化及两者的相关性。
选取河北省某抗生素生产废水为研究对象,通过现场采样分析,重点考察污泥中β-内酰胺类、四环素类和磺胺类抗性基因及微生物群落结构在废水处理过程中的分布和相关性特征,揭示废水厂活性污泥中土著抗性基因的分布规律以及与微生物群落结构之间的互相作用,以期为评估废水处理厂中抗性基因环境风险提供依据,并为制定有效的ARGs(antibiotic resistance genes)环境污染控制方法提供理论基础。
1材料与方法
1.1样品采集
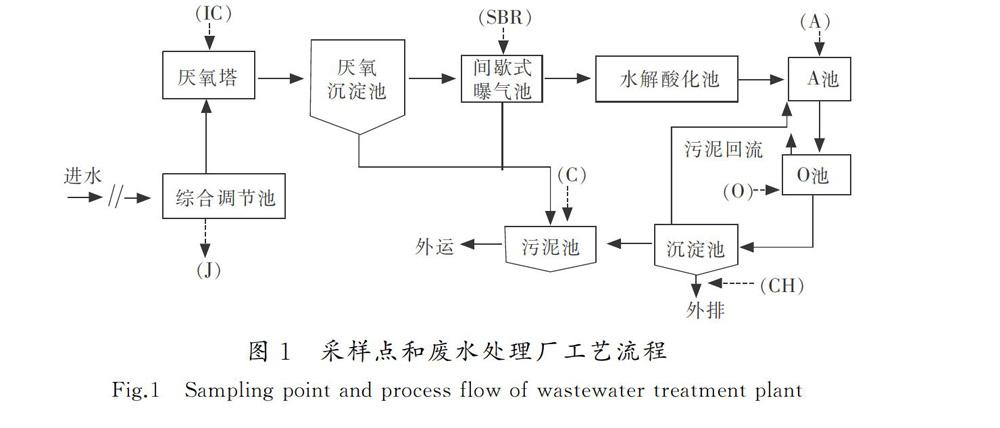
河北省某制药废水处理厂,该厂设计处理水量300 m3/d,主要采用生物处理法处理生产废水,采样点和废水处理厂工艺流程如图1所示。样品采用500 mL聚乙烯瓶,于2018年1月,采集不同污水处理单元活性污泥样品,分别标记为J(综合调节池)、IC(厌氧塔)、SBR(间歇式曝气池)、A(A池)、O(O池)、C(污泥池)、CH(出水)取样结束后,于冰盒中运送回实验室,离心图1采样点和废水处理厂工艺流程
Fig.1Sampling point and process flow of wastewater treatment plant(5 min,11 000 r/min)后称取5 g冷冻于-80 ℃冰箱中,以备DNA提取。
1.2水样分析
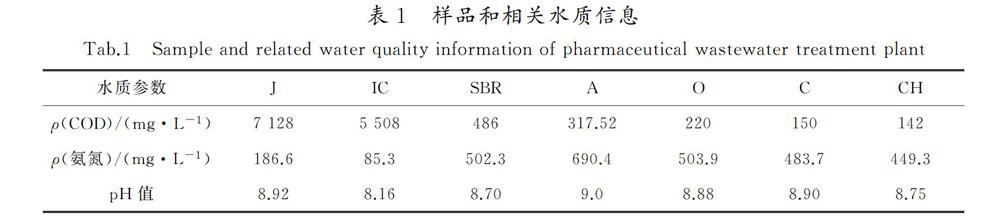
按照《水和废水监测分析方法》[16]国家标准方法分析常规化学指标,测定水中CODCr和氨氮,测定结果见表1。
1.5抗性基因的定量检测
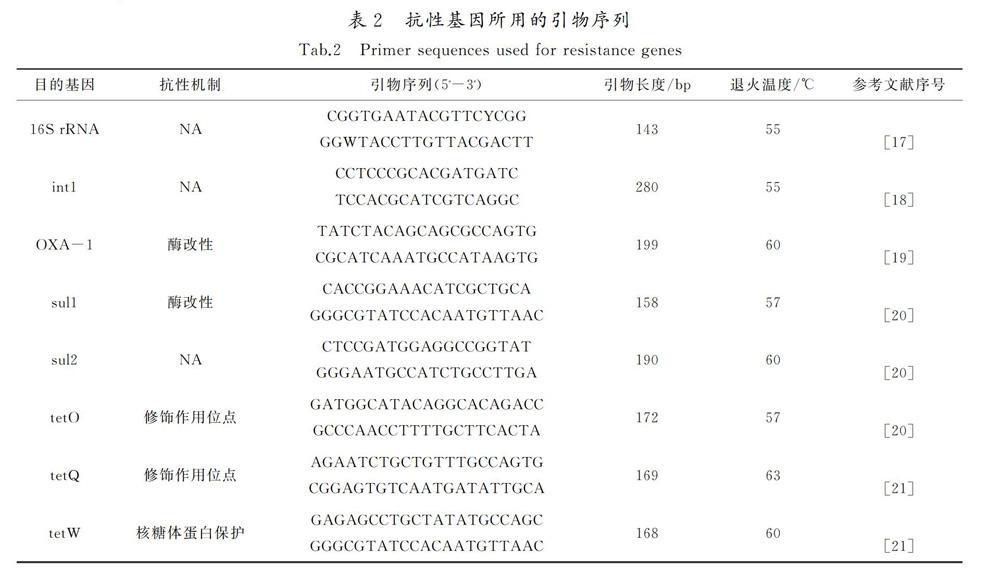
采用SYBR-Green 实时定量PCR方法对各基因进行定量分析,检测仪器为StepOne型荧光定量PCR仪(ABI,美国)。PCR产物经过克隆测序确认后,使用生工质粒提取试剂盒SK1131从阳性克隆子中提取质粒,用作标准曲线。使用NanoDrop微量分光光度计(Thermo Scientific,美国)测定质粒浓度。制作标准曲线时按照10倍梯度浓度稀释构建好的各质粒,于90 μL稀释液中加入10 μL质粒,做4~6个点,通过预实验选取合适标准品用于制备标准曲线。标准质粒、环境样品、阴性对照均做3个平行,取平均值进行计算。
质粒拷贝数换算公式为Q=[C×10-9×6.02×1023]/[N×660],式中:Q为质粒拷贝数,copies/μL;C为质粒浓度,ng/μL;N为克隆产物碱基数,(g/mol);M1为载体碱基数,2 692 bp(pMD 18-T);M2为载体碱基数,3 023 bp(pGEMX-T Easy)。
荧光定量PCR反应体系为12.5 μL 2×SYBR,1.0 μL DNA(10 ng/μL),0.5 μL引物F(10 mol/L),0.5 μL引物R(10 mol/L),10.5 μL水,总体积25.0 μL。荧光定量PCR反应程序为1)50 ℃,2 min;2)95 ℃,5 min;3)95 ℃,20 s;4)退火,30 s;5)72 ℃,31 s;6)Plate read,重复1)~5),39次重复;7)Melt-curve分析60 ℃ to 95 ℃。每隔0.2℃采集一次荧光以生成溶解曲线,根据溶解曲线的变化检测扩增结果的特异性。退火温度和反应时间根据引物不同进行调整。
1.6统计分析
抗性基因浓度结果和群落结构使用OriginPro 8.6软件(Origin Lab Corporation,USA)进行分析;使用MOTHUR软件计算各个样本Alpha多样性指标,以反映本次测序深度、物种均匀性等;使用R软件对样本绘制热图并分析;ARGs与群落结构相关性使用CANOCO 5.0(Microcomputer Power ,Ithaca,NY)软件分析,基因相关性使用SPSS 19.0软件进行相关性分析。
2结果讨论与分析
2.1微生物菌群耐药基因定量检测
利用荧光定量PCR的方法检测抗性基因的分布特征、变化情况和相对表达量,水处理阶段ARGs分布结果如图2所示。
2.2微生物群落多样性分析
2.3微生物群落和抗性基因相关性分析
微生物群落结构会影响ARGs的产生和丰度[32],但是微生物群落结构在环境样本中对ARGs扩增影響的研究还是有限的。通过高通量测序和荧光定量方法来分析污水处理系统中微生物群落和抗性基因分布及其相关性。
首先对制药废水厂的5个样本进行主成分分析,如图4 a)所示,然后选取抗性基因int1,OXA-1,sul1,sul2,tetO,tetQ和tetW作为环境因子,结合各样本微生物群落结构,选取制药废水厂中15种相对丰度较高的菌属作为样本,利用冗余分析(RDA)方法研究微生物与环境因子的相关性,结果见图4 b)。
對7个样本进行主成分分析,结果(见图4 a))表明,PC1(主成分1)表示两组间差异中可以解释全面分析结果的56.67%,PC2(主成分2)表示两组间差异中可以解释全面分析结果的31.61%,两点之间的距离越近,表明2个样本之间的微生物群落结构相似度越高,差异越小。从图4 a)中可以看出,A,O和C这3个样本分布较近,菌群相似度较高;J与CH样本与其他样本单元分布较远,微生物群落结构相似度较低,有明显的差异性,说明制药废水厂中进水、出水和废水处理单元的微生物群落菌群有较大的不同。A,O和C这3个样本与IC和SBR样本分布相对较远,说明IC,SBR和A,O和C这3个样本菌群有较大的不同。且每组样本与样本之间的距离呈现一定的变化规律。
对A,O,C,IC和SBR单元中前15种相对丰度较高的菌属进行冗余分析,结果表明(见图4 b)),主轴1和主轴2共解释了微生物群落结构和水质、抗性基因参数的96.86%。从图4 b)来看,Aeromicrobium与sul2呈较高程度的正相关性,可能是sul2在微生物群落中存在的可能的主要菌群;Rhodovulum和Rhodospirillaceae菌属与OXA-1和tetQ呈较高程度的正相关性,这些菌属是这些ARGs分布的主要菌属;Acinetobacter与tetO和tetW呈现较高程度的正相关性,可能是tetO和tetW在微生物群落中存在的可能的主要菌属。
3结语
采用高通量测序技术和荧光定量PCR技术,通过检测废水厂活性污泥中抗生素抗性基因的绝对丰度和微生物菌群的相对丰度,进而综合分析制药废水处理厂中某些抗生素抗性基因和微生物群落结构的特征,结果显示:sul1,sul2,tetO,tetQ,tetW,OXA-1和可移动遗传元件int1在制药废水厂中各个阶段的检出频率均为100%,从IC池到C池,总抗性基因浓度范围为3.09×108~2.26×109 copies/g(干重),基因总浓度上升了7.3倍;在污泥样品中,变形菌门(Proteobacteria),拟杆菌门(Bacteroidetes),厚壁菌门(Firmicutes),栖热菌门菌门(Thermus),芽单胞菌门(Gemmatimonadetes)等为优势菌门,平均总相对丰度比例占到81.05%;Aeromicrobium与sul2呈较高程度的正相关性,可能是sul2在微生物群落中存在的可能的主要菌群;Rhodovulum和Rhodospirillaceae菌属与OXA-1和tetQ呈较高程度的正相关性,这些菌属是这些ARGs分布的主要菌属;Acinetobacter与tetO和tetW呈现较高程度的正相关性,可能是tetO和tetW在微生物群落中存在的可能的主要菌属。因此,相关抗性基因的增值和分布与相关特定菌属有关,可以通过控制相关菌属的丰度来消减工业废水厂中的抗性基因,研究结果为以后控制制药废水厂中抗生素抗性基因的传播提供了理论基础。
参考文献/References:
[1]HE L Y, YING G G, LIU Y S, et al. Discharge of swine wastes risks water quality and food safety: Antibiotics and antibiotic resistance genes from swine sources to the receiving environments [J]. Environment International, 2016, 92/93: 210.
[2]RODRIGUEZ-MOZAZ S, CHAMORRO S, MARTI E, et al. Occurrence of antibiotics and antibiotic resistance genes in hospital and urban wastewaters and their impact on the receiving river [J]. Water Research, 2015, 69: 234-242.
[3]NAQUIN A, SHRESTHA A, SHERPA M, et al. Presence of antibiotic resistance genes in a sewage treatment plant in Thibodaux, Louisiana, USA [J]. Bioresource Technology, 2015, 188(11): 79-83.
[4]TONG Z, ZHANG X X, LIN Y. Plasmid metagenome reveals high levels of antibiotic resistance genes and mobile genetic elements in activated sludge [J]. Plos One, 2011, 6(10): e26041.
[5]FORSBERG K J, REYES A, WANG B, et al. The shared antibiotic resistome of soil bacteria and human pathogens [J]. Science, 2012, 337(6098): 1107-1111.
[6]PRUDEN A, PEI R, STORTEBOOM H, et al. Antibiotic resistance genes as emerging contaminants: Studies in Northern Colorado [J]. Environmental Science & Technology, 2006, 40(23): 7445.
[7]PEHRSSON E C, TSUKAYAMA P, PATEL S, et al. Interconnected microbiomes and resistomes in low-income human habitats [J]. Nature, 2016, 533(7602): 212-216.
[8]王玉倩, 薛秀花. 实时荧光定量PCR技术研究进展及其应用 [J]. 生物学通报, 2016, 51(2): 1-6.
[9]AYDIN S, INCE B, INCE O. Development of antibiotic resistance genes in microbial communities during long-term operation of anaerobic reactors in the treatment of pharmaceutical wastewater [J]. Water Research, 2015, 83(4): 337-344.
[10]LIU M, ZHANG Y, YANG M, et al. Abundance and distribution of tetracycline resistance genes and mobile elements in an oxytetracycline production wastewater treatment system [J]. Environmental Science & Technology, 2012, 46(14): 7551.
[11]GUO X, YAN Z, ZHANG Y, et al. Behavior of antibiotic resistance genes under extremely high-level antibiotic selection pressures in pharmaceutical wastewater treatment plants [J]. Science of the Total Environment, 2017, 612: 119-128.
[12]WANG F H, QIAO M, LYU Z E, et al. Impact of reclaimed water irrigation on antibiotic resistance in public parks, Beijing, China [J]. Environmental Pollution, 2014, 184(1): 247-253.
[13]XU Z, LI L, SHIRTLIFF M E, et al. Resistance class 1 integron in clinical methicillin‐resistant Staphylococcus aureus strains in southern China, 2001-2006 [J]. Clinical Microbiology & Infection, 2011, 17(5): 714-718.
[14]BEN-SHAHAR O, OBARA I, ARY A W, et al. Antimicrobial resistance of integron-harboring Escherichia coli isolates from clinical samples, wastewater treatment plant and river water [J]. Science of the Total Environment, 2012, 414(414): 680-685.
[15]LING A L, PACE N R, HERNANDEZ M T, et al. Tetracycline resistance and Class 1 integron genes associated with indoor and outdoor aerosols [J]. Environmental Science & Technology, 2013, 47(9): 4046-4052.
[16]國家环境保护总局. 水和废水监测分析方法[M].第4版. 北京:中国环境科学出版社, 2002.
[17]GAZE W H, ZHANG L, ABDOUSLAM N A, et al. Impacts of anthropogenic activity on the ecology of class 1 integrons and integron-associated genes in the environment [J]. Isme Journal, 2011, 5(8): 1253.
[18]CHEN H, ZHANG M. Occurrence and removal of antibiotic resistance genes in municipal wastewater and rural domestic sewage treatment systems in eastern China [J]. Environment International, 2013, 55(4): 9.
[19]翟文超. 抗生素抗性基因在抗生素制药废水处理过程中的分布特征及控制原理研究 [D]. 天津: 南开大学, 2014.
ZHAI Wenchao. The Fate and Control Principle of Antibiotic Resistance Genes in Pharmaceutical Wastewater Treatment Systems[D].Tianjin: Nankai University, 2014.
[20]LUO Y, MAO D, RYSZ M, et al. Trends in antibiotic resistance genes occurrence in the Haihe River, China [J]. Environmental Science & Technology, 2010, 44(19): 7220.
[21]AMINOV R I, GARRIGUESJEANJEAN N, MACKIE R I. Molecular ecology of tetracycline resistance: development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protection proteins [J]. Applied & Environmental Microbiology, 2001, 67(1): 22.
[22]WANG J, MAO D, MU Q, et al. Fate and proliferation of typical antibiotic resistance genes in five full-scale pharmaceutical wastewater treatment plants [J]. Science of the Total Environment, 2015, 526(4): 366-373.
[23]PEAK N, KNAPP C W, YANG R K, et al. Abundance of six tetracycline resistance genes in wastewater lagoons at cattle feedlots with different antibiotic use strategies [J]. Environmental Microbiology, 2007, 9(1): 143-151.
[24]GUO X, XIA R, HAN N, et al. Genetic diversity analyses of class 1 integrons and their associated antimicrobial resistance genes in Enterobacteriaceae strains recovered from aquatic habitats in China [J]. Letters in Applied Microbiology, 2011, 52(6): 667-675.
[25]LIU Y J, WANG X C, YUAN H L. Characterization of microbial communities in a fluidized-pellet-bed bioreactor for wastewater treatment [J]. Desalination, 2009, 249(1): 445-452.
[26]HOA P T P, NONAKA L, VIET P H, et al. Detection of the sul1, sul2, and sul3 genes in sulfonamide-resistant bacteria from wastewater and shrimp ponds of north Vietnam [J]. Science of the Total Environment, 2008, 405(1/2/3): 377-384.
[27]MOURA A, PEREIRA C, HENRIQUES I, et al. Novel gene cassettes and integrons in antibiotic-resistant bacteria isolated from urban wastewaters [J]. Research in Microbiology, 2012, 163(2): 92-100.
[28]TENNSTEDT T, SZCZEPANOWSKI R, BRAUN S, et al. Occurrence of integron-associated resistance gene cassettes located on antibiotic resistance plasmids isolated from a wastewater treatment plant [J]. Fems Microbiology Ecology, 2010, 45(3): 239-252.
[29]DU J, REN H, GENG J, et al. Occurrence and abundance of tetracycline, sulfonamide resistance genes, and class 1 integron in five wastewater treatment plants [J]. Environmental Science & Pollution Research, 2014, 21(12): 7276-7284.
[30]王麗梅, 罗义, 毛大庆, 等. 抗生素抗性基因在环境中的传播扩散及抗性研究方法 [J]. 应用生态学报, 2010, 21(4): 1063-1069.
WANG Limei, LUO Yi, MAO Daqing, et al. Transport of antibiotic resistance genes in environment and detection methods of antibiotic resistance, 2010, 21(4): 1063-1069.
[31]康晓荣. 超声联合碱促进剩余污泥水解酸化及产物研究 [D]. 哈尔滨: 哈尔滨工业大学, 2013.
KANG Xiaorong. Study on Hydrolysis and Acidification of Activated Sludge Enhanced by Ultrasound Combined with Alkaline [D]. Harbin:Harbin Institute of Technology, 2013.
[32]YANG Y, LI B, ZOU S, et al. Fate of antibiotic resistance genes in sewage treatment plant revealed by metagenomic approach [J]. Water Research, 2014, 62(7): 97-106.第40卷第2期河北科技大学学报Vol.40,No.2
