FUS IS ESSENTIAL FOR SOMATIC GROWTH AND SEXUAL SIZE DIMORPHISM IN ZEBRAFISH
2019-05-29DANHongRENFanXIONGShuTingCHENXinYANGTianYiandMEIJie430070
DAN Hong, REN Fan, XIONG Shu-Ting, CHEN Xin, YANG Tian-Yi and MEI Jie(,,430070,)
Abstract: The Fused in sarcoma gene (FUS) is a RNA-binding protein implicated in the regulation of transcription and pre-mRNA splicing. Mutations in FUS lead to neurodegenerative diseases including ALS(amyotrophic lateral sclerosis) in mammals. However, little is known about fus function in fish species. Here,we generated frame-shift alleles in the zebrafish fus gene using CRISPR/Cas9 mutagenesis. Homozygous fus-/-zebrafish developed normally and were fully fertile, except that their body size and body weight were smaller than that of wild-type zebrafish at both the larvae and adult stages. In addition, the female-biased sexual size dimorphism in adult zebrafish was also eliminated when fus was mutated. The expression levels of several growth-related genes including gh1, ghra, igf1, igf2a, stat5.1 and socs6 were significantly reduced in fus-/-zebrafish larvae compared with the wild-type. However, there was no effect on locomotor activity in fus-/-zebrafish. Therefore, different from its functions in mammals, fus is not associated with motoneuron development and has a divergent function in regulating somatic growth and sexual size dimorphism in fish species.
Key words: Fus; Zebrafish; CRISPR/Cas9; Somatic growth; Sexual size dimorphism
The RNA-binding protein FUS (also known as TLS) belongs to the TET (TAF15/EWS/TLS) protein family. As a member of heterogeneous nuclear Ribonucleoprotein (hnRNP) complex, FUS plays functions in RNA splicing, process and transport in cytoplasm, and regulates many developmental processes,such as growth, differentiation, and morphogenesis in vertebrates[1]. FUS has been widely known for its association with amyotrophic lateral sclerosis (ALS),and FUS mutantion leads to muscular weakness with degeneration of motor neurons[2—4]. In mouse, loss ofFusfunction resulted in male sterile with a defect in the normal development of premeiotic spermatocytes[5].In frog,fusis a critical regulator of gastrulation development and controls the splicing events during this process[6].
Chromosomal rearrangements result in the formation and activation of fusion oncogenes that are frequently observed in a number of human cancers. Usually, FUS and its fusion oncogenes can significantly promote cell proliferation. Overexpression of FUSDDIT3 activated the IGF-IR and PI3K/Akt/GSK-3β signaling, while siRNA-mediated knockdown of FUS-DDIT3 led to a reduction of IGF2 mRNA expression[7]. FUS/ERG fusion protein activated the IGF-1 promoter and induced IGF-1 expression in human mesenchymal stem cells[8]. Knockdown of FUS in cultured mammalian cells led to impaired cellular proliferation[9].
In zebrafish embryos, both knockdown offuswith antisense morpholino oligonucleotides and overexpression of ALS-related FUS mutation sequences led to defects in motoneuron degeneration and swimming[10]. On the contrary, no ALS-like phenotype was observed infus-mutated zebrafish generated by CRISPR-Cas9 system, as thefusmutants showed normal motoneuron and swimming ability. However, both the mRNA and protein levels of gene expression were mildly changed in the brain[11]. On the downside, thefusknockout zebrafish was generated from the outbred zebrafish (AB × TU) in the previous study[11], in which the genetic background is not clear. In this study, we generatedfus-mutated zebrafish in AB line zebrafish utilizing the CRISPR/CAS9 technology.The results showed thatfusplayed an essential role in regulating somatic growth, as loss offusfunction led to a significant reduction of body weight, body length and sexual size dimorphism compared with wild-type zebrafish.
1 Material and method
1.1 Zebrafish lines and maintenance
The wild-type zebrafish (AB line) were maintained at 28.5℃ on a 14h light / 10h dark cycle. All experiments procedures involving zebrafish were approved by the institution animal care and used committee of Huazhong Agricultural University. All embryos used were collected by natural fertilized and the age was determined according to a previous report[12].
1.2 Target gene disruption by CRISPR/Cas9
The target gene knockout by CRISPR/Cas9 was performed as previously described[13]. Genomic and mRNA sequence information of zebrafishfusgene were collected from Ensembl (www.ensembl.org).The online tool ZIFIT Targeter (http://zifit.partners.org/ZiFiT/CSquare9Nuclease.aspx) was used to design the guide RNA (gRNA) target sites, and the target sequence is GGGGGTTATGGAGGACAGTC. The gRNA was transcribed by Transcript Aid T7 High Yield Transcription Kit (Thermo Fisher Scientific,USA) according to the manufacturer's protocol.PCS2-Cas9 expression vector was linearized by Restriction enzymesXbaI and transcribed following the instructions of the Ambion Mmessage mMACHINE®Kit High Yield Capped RNA Transcription Kit.Zebrafish embryos at one-cell stage were co-injected with Cas9 mRNA (300 ng/μL) and gRNA (20 ng/μL).For rapidly genotyping and screening mutant zebrafish, PCR was performed to amplify genomic DNA containing the target site, and the primer sequences for PCR reaction were MF: 5′-GCCAAA CCTCAAGCCATGGGTAAG-3′ and MR: 5′-GTTG GAGCTGTATCCTCCCTG-3′.
1.3 Microscopic imaging and body length and body weight measurement
MS-222 (ethyl 3-aminobenzoate methanesulfonate, Sigma) was used to anesthetize different stages of zebrafish. Zebrafish embryos, larvae and adults were visualized and imaged by an inverted stereo fluorescence microscope (Leica DFC550) and Canon camera (EOS 750D), respectively. When imaging the embryos, a ruler with number marks was used for size comparison and length measurement. Body length and weight of adult zebrafish was measured by an electronic vernier caliper and analytical balances, respectively.
1.4 Probe synthesis and whole mount RNA in situ hybridization
Probe synthesis was performed according to a previous description[14]. Antisense probe ofgh1 was generated using a DIG RNA labeling kit (Roche,Mannheim, Germany), directly from PCR products that included a T7 RNA polymerase binding sequence at the 5′ end. The primer sequences were as following:gh1 forward, 5′-ATGGCTAGAGCATTG GTGCTG-3′,gh1 reverse, 5′-CAGGGTACAGTTG GAATCCAG-3′. Embryos/larvae were fixed with 4%paraformaldehyde (PFA) overnight at 4℃, and RNAin situhybridization was performed as previously described[15,16].
1.5 RNA isolation and quantitative real-time PCR(qRT-PCR)
Total RNA was isolated from each sample using TRIzol reagent (Promega, Madison, WI) and reversely transcribed into cDNA using the GoScript Reverse Transcription System (Promega) according to the manufacturer's instruction. qRT-PCR was performed on a Bio-Rad Real-Time PCR system by CFX96 Optics Module (Bio-Rad) to evaluate gene expression. All qRT-PCR reactions were performed in a volume of 20 μL as previously described[17]. 18S rRNA was used as an internal expression control.Each experiment was performed in triplicate and the data were analyzed using the 2-ΔΔCtmethod. The genespecific primer sequences were listed in Tab. 1.
1.6 Touch response test
The 2 days post fertilization (dpf) zebrafish larvae were placed in a 15 cm dish with embryonic medium and their tails were gently touched with a forcep. A ruler was placed at the bottom of the dish to calibrate the track lengths. Movements were imaged and video recorded using Canon camera (EOS 750D)with the speed of 40 frames per second. The swim distance in 1 second was calculated and used for statistical analysis.
1.7 Statistical analysis
All experiments data were performed at least in triplicate. Data were shown as mean±SD. Statistical analysis was conducted with SPSS 24.0 software(SPSS, USA) and graphs were plotted using Graph-Pad Prism 5 (La Jolla, CA). Two-tailed t-test was used for statistical analysis to determine statistical differences between different groups. Statistical significance was represented by asterisks (*P<0.05; **P<0.01; ***P<0.001).

Tab. 1 The primers for qRT-PCR
2 Result
2.1 Generation of fus-mutated zebrafish line
Quantitative RT-PCR (qRT-PCR) was performed to examine the expression levels offusin adult tissues and different embryonic stages (Fig. 1).Thefustranscript was maternally deposited and ubiquitously expressed at all embryonic stages, with the highest expression at 8 hours post fertilization (hpf)(Fig. 1A). In additon,fuswas predominantly expressed in the ovary, brain and kidney (Fig. 1B).
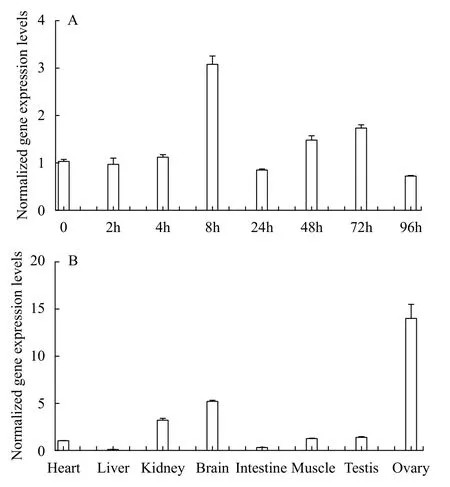
Fig. 1 Expression levels of fus mRNA during the embryo development and in adult tissues
To study the function offusduring zebrafish development, we used CRISPR/Cas9 system to generatefus-mutated zebrafish. The sgRNA targeting site was chosen in the third exon. Finally, 2 bp-deletion(namedfus-/-) zebrafish were identified and chosen to establish mutant lines (Fig. 2A). Zebrafish Fus protein consists of two founctional domains including RRM superfamily and zf-RanBP. The mutation infus-/-zebrafish resulted in the open reading frame (ORF)shift and caused the deletion of RRM superfamily and zf-RanBP domains (Fig. 2B). Sanger sequencing for wide-type (WT) and homozygous mutant further confirmed the type of mutation (Fig. 2C). To examine the stability offusmRNA in mutant zebrafish, qRT-PCR was executed to check the expression offusmRNA infus-/-mutants (Fig. 2D). The expression offusmRNA in mutant zebrafish decreased by 72.3%, 88.1% and 66.8% compared with wide-type at 48, 72 hpf and 7 dpf, respectively.
2.2 fus regulates somatic growth and sexual size dimorphism in adult zebrafish
All adultfus-/-zebrafish were fertile. Andfus-/-zebrafish embryos/larvae developed normally, except that their body sizes were smaller than that of wildtype embryos at 6 dpf (Fig. 3A). In addition, at 3 months post fertilization (mpf), we found that both male and femalefus-/-zebrafish displayed smaller body morphology in comparison with that of wildtype zebrafish (Fig. 3B), but no obvious body morphology difference was observed between heterozygous and wild-type zebrafish. To verify the defects in somatic growth, we measured the body length offusmutants and wild-type zebrafish at 6 dpf. A significant reduction of body length was detected infusmutant larvae (Fig. 3C), suggesting thatfusis required for normal somatic growth in zebrafish embryos. Further, the body length and body weight offusmutant and wild-type zebrafish were measured at 3 mpf (Fig. 3D—E). The results indicated that the length and weigth offusmutants were significant lower than that of wild-type in both male and female adult zebrafish. Moreover, for wild-type zebrafish, the length and weigth of females were significantly higher than that of males at 3 mpf (*P< 0.05; ***P<0.001).Noticeable, no significant difference of body length and weigth were observed between females and males at 3 mpf when loss offusfunction. Our data revealed thatfusis not only required for normal somatic growth, but also plays an important role in sexual size dimorphism in adult zebrafish.
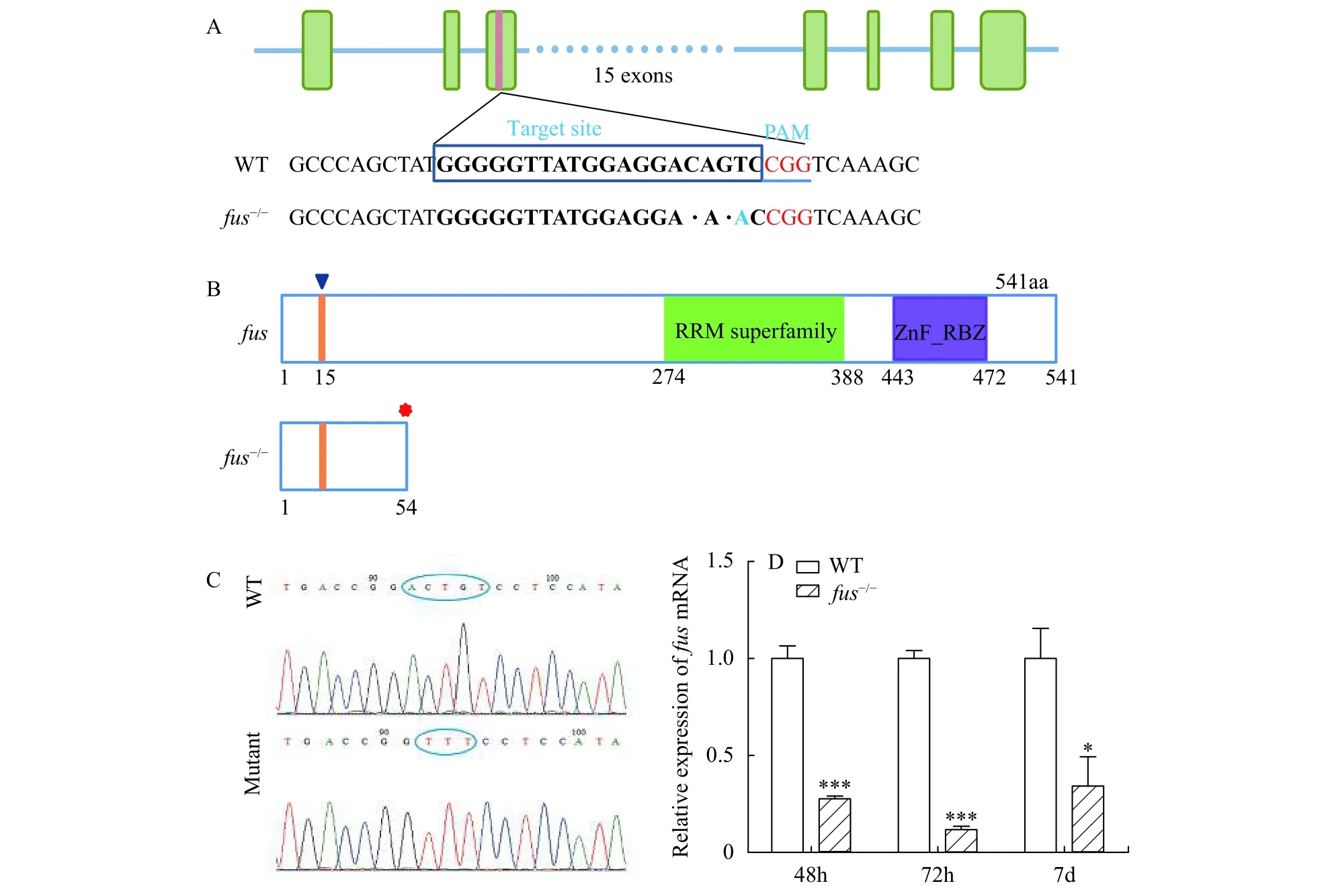
Fig. 2 Generation of fus-mutated zebrafish
2.3 fus mutation causes dysregulated expression of growth-related genes and has no effect on locomotor activity in zebrafish
Growth hormone (GH)/Insulin-like growth factor (IGF) signal is the key signal pathway for regulating fish body growth[18,19].stat5.1/stat5bhas been reported to play an important role in regulating somatic growth and sexual size dimorphism in adult zebrafish[17]. Loss of the function of suppressor of cytokine signaling 6 (socs6) leads to growth retardation[20].Therefore, qRT-PCR was performed to compare the expression of growth-related genes includinggh1,ghra,igf1,igf2a,stat5.1 andsocs6 between wild-type andfus-/-zebrafish larvae. The results indicated that expression levels of all selected growth-related genes were significantly reduced infusmutants compared with wild-type zebrafish at 7 dpf (Fig. 4A). Wholemountin situhybridization (WISH) on embryos/larvae at 48, 72, 96 hpf and 6 dpf further demonstrated a dramatic reduction ofgh1 mRNA in pituitary offus-/-zebrafish (Fig. 4B).
It has been previously reported that knockdown offusin zebrafish resulted in impaired motoneurons[2].To explore whether thefus-/-zebrafish has similar phenotype, we evaluated motor activity offus-/-embryos by performing the touch-evoked escape response test[11]. In the test, 2 dpf embryos were placed in the middle of a 15 cm diameter dish with embryonic medium and a 15 cm ruler was included for scale.If the motoneurons of embryos were damaged, the response of embryos would be slower. After slightly touching the tail with a forcep, we measured the length of the swim tracks. No significant difference of swim distance was observed between wild-type andfus-/-larvae (Fig. 5). Therefore,fusregulates the somatic growth but is not associated with motoneuron development in zebrafish.
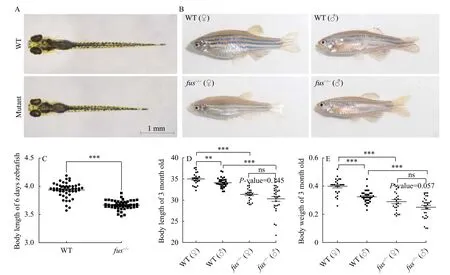
Fig. 3 Loss of fus function affects body growth and sexual size dimorphism in zebrafish
3 Discussion
The RNA-binding protein FUS has been shown to regulate many important biological processes, including the gene transcription, DNA repair and alternative splicing of neuronal genes during neuronal differentiation[21—24]. In rat, mouse and zebrafish model,overexpression of human wild-type FUS or ALS-related FUS mutation sequences developed the phenotypes of ALS. FUS mutation resulted in phenotypes of ALS[2,3,25]. However, loss of FUS function in mice produces other phenotypes different from ALS, such as male sterility and defective B-lymphocyte development[5,26,27]. In the present study, we generatedfusmutant zebrafish by CRISPR-Cas9 system to analyze its roles in fish development. Thefus-/-zebrafish displayed a significant reduction in body length and body weight, suggesting an important role offusin body growth and development.
Although knockdown offuswith morpholino resulted in phenotypes of ALS in zebrafish embryos[10],non-specific effects of morpholinos were frequently observed in zebrafish studies. Moreover, the phenotypes caused by genetic mutations and gene knockdowns are frequently different in zebrafish, due to the genetic compensation induced by deleterious mutations[28]. In addition, knockdown offusin frog led to defects in gastrulation development[6]. Accordingly,gene editing technology such as CRISPR-Cas9 could be utilized to create a genetic knockout model to studyfusfunction in zebrafish. In our study, the mRNA offusgene was instable in thefusmutant zebrafish. And this phenomenon was also reported in some other gene mutants[19,29,30]. In comparison with the recent report thatfusmutants produced by CRISPR-Cas9 system showed no ALS-like phenotype and normal development[11], our studies indicated that the body size and weight offusmutant zebrafish were smaller than that of wild-type (Fig. 3). The phenotypic difference might be the reflection of different genetic backgrounds between the previous outbred zebrafish (AB × TU) strain and the AB strain, because the genetic background of outbred zebrafish(AB × TU) is not so clear. Although loss offusfunction in the mouse caused male sterile[5], thefusmutant zebrafish were fully fertilized. These data suggest thatfushas divergent functions in different organisms.

Fig. 4 The expression of growth-related genes were dysregulated in zebrafish embryo/larvae
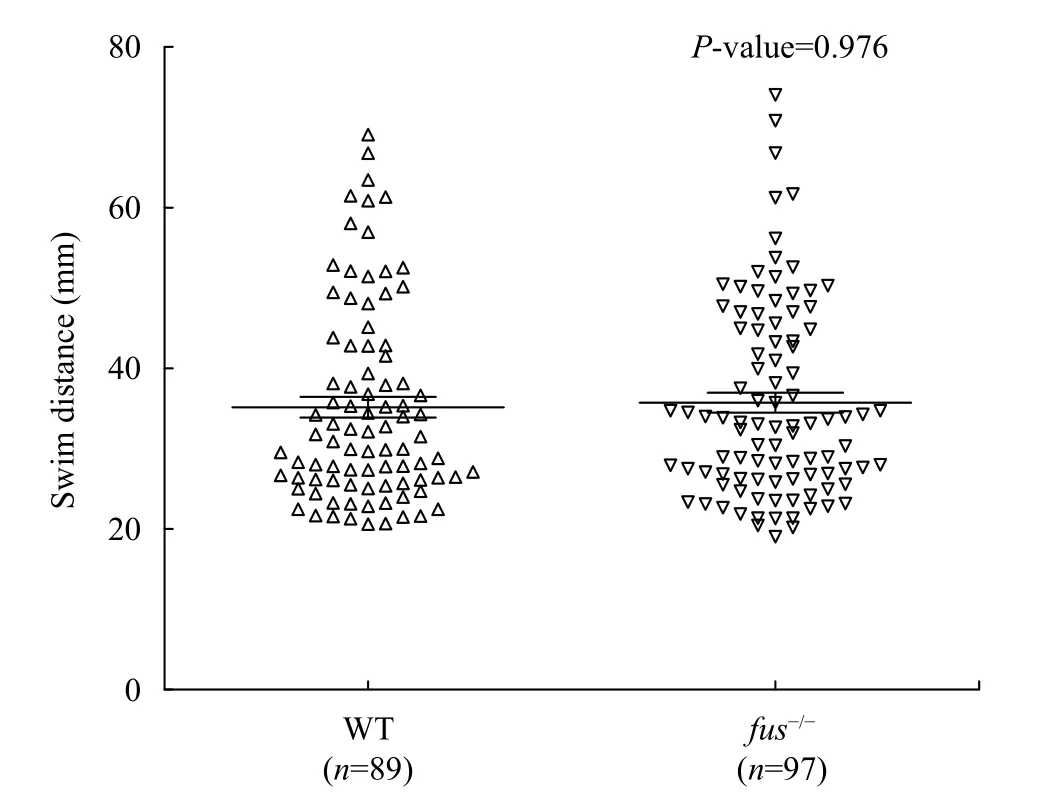
Fig. 5 Touch evoked escape response of wild-type and mutant larvae
Sexual size dimorphism, a phenomenon describing the different growth rate between male and fe-male individuals, has been widely observed in cultivated fish species, such as tilapia (Oreochromis niloticus) and yellow catfish (Pelteobagrus fulvidraco)[19].GH/IGF signal is an important signal pathway to regulate body growth. Previous studies demonstrated that sexual dimorphic expression of GH/IGF signaling genes,ghrelin/GHSR,leptinand its receptor might contribute to the sexual size dimorphism in fish species[31—34]. GH stimulates the synthesis and secretion of IGF-I through the ERK, PI3K/Akt and JAK-STAT signaling pathways[35]. FUS-DDIT3 and FUS/ERG fusion protein activated the IGF-IR and PI3K/Akt/GSK-3β signaling and regulated IGF-1 and IGF-2 mRNA expression in mammals[8,36]. In zebrafish, the expression of growth-related genes includinggh1,ghra,igf1,igf2a,stat5.1 andsocs6 were inhibited infus-/-zebrafish compared with that in wild-type (Fig. 4).Moreover, the body length and weight were also significantly reduced infus-/-zebrafish (Fig. 3). Sexual size dimorphism has been observed in adult zebrafish.stat5.1/stat5bplays a critical role in regulating somatic growth and sexual size dimorphism in adult zebrafish[17]. The significant reduction ofstat5bexpression infus-/-zebrafish may contribute to the inhibi-tion of somatic growth and sexual size dimorphism in adult zebrafish. As a splicing factor,Fusknock-down in frog caused intron retention ofFgf8 andFgfr2.However, we found thatgh1andstat5.1 were normally spliced in thefusmutants (data not shown). Sofusmay regulate the mRNA stability of genes involved in axis growth in thefusmutants[6]. Our results provided a novel view thatfusmediates zebrafish body growth and sexual size dimorphism via regulating the expression of a number of growth-related genes including GH/IGF axis genes andstat5b. And functional divergence offusgene is existed in the vertebrate evolution. As a RNA-binding protein, the mechanism how Fus regulates gene expression needs to be further clarified.
