Diagnosis of Laron syndrome using monoplex-polymerase chain reaction technology with a whole-genome amplification template:A case report
2019-04-22AdinaNeumannMiguelngelAlcntaraOrtigozaAriadnaGonzlezdelngelFelipeCamargoDiazEstherpezBayghen
Adina Neumann, Miguel Ángel Alcántara-Ortigoza, Ariadna González-del Ángel, Felipe Camargo-Diaz,Esther López-Bayghen
Adina Neumann, Felipe Camargo-Diaz, Laboratorio de Investigación y Diagnóstico Molecular,Instituto de Infertilidad y Genética México SC, INGENES, México City 05320, México
Miguel Ángel Alcántara-Ortigoza, Ariadna González-del Ángel, Instituto Nacional de Pediatría,Torre de Investigación, Mexico City 04530, México
Miguel Ángel Alcántara-Ortigoza, Ariadna González-del Ángel, Laboratorio de Biología Molecular, Departamento de Genética Humana, Instituto Nacional de Pediatría, México City 04530, México
Esther López-Bayghen, Departamento de Toxicología, Centro de Investigación y de Estudios Avanzados del Instituto Politécnico Nacional (CINVESTAV-IPN), México City 07360,México
Abstract
Key words: Growth hormone insensitivity; Growth hormone receptor mutations;Intragenic deletions; Molecular diagnosis; Embryo diagnosis; Laron syndrome; Case report
INTRODUCTION
The prevalence of Laron syndrome (LS), first described in 1966[1]as insensitivity to growth hormone (GH) due to mutations in the extracellular domain of the GH receptor (GHR)[2,3], is not commonly diagnosed in Latin America.Moreover, in Mexico, there are less than 10 documented cases of LS[4-7].LS is typically found among the Jewish community[8], but with a very low prevalence.For Latin Americans, most cases are found in Ecuador, most likely of Jewish descendants, due to a mass migration during the 15thcentury[4].Therefore, LS is typically not assessed in most Preimplantation Genetic Testing (PGT), neither in Mexico nor in the rest of the world.
LS is an autosomal recessive disorder[9], characterized by dwarfism, abnormal development of the reproductive organs (micropenis or enlarged breasts)[10-12].Most LS patients have an increased risk for seizures[13]but a decreased risk for cancer[14-16].LS subjects have elevated GH levels with usually low insulin-like growth factor 1[17].To date, the diagnosis of LS is performed as a post-clinical presentation.Here, we present a case of a couple that underwent PGT to identify potential LS-free embryos to achieve a normal pregnancy.
CASE PRESENTATION
Chief complaints
A 31-year-old Jewish, Mexican woman [body mass index (BMI) = 21.76 kg/m2] and her husband, a 32-year-old Jewish, Mexican man (BMI = 24.5 kg/m2) decided to attend the INGENES Institute in Mexico City for selection of a healthy embryo for a second pregnancy, as her first pregnancy produced a child with LS.
History of present illness
Both parents were presented as fully developed adults with healthy sexual development.Upon the diagnosis of LS, the borne-child mutation in the GHR descendant as deletion of exons 5 and 6 (del5-6) of GHR.Subsequent clinical and molecular analysis of the parents confirmed that they were carriers (heterozygotes) of the del5-6.We proposed anin vitrofertilization (IVF) protocol, complemented with pre-implantation genetic diagnosis (PGT) using monoplex polymerase chain reaction(PCR) targeting the del5-6 mutation present in GHR, as an intervention for healthy embryo selection.
History of past illness
The only relevant history is five years before attending INGENES, the mother had one previous pregnancy, which resulted in a child with LS.Afterward, both patients underwent genetic testing, not at our facility, and were diagnosed as carriers for mutations in GHR that could develop LS.
Personal and family history
The mother had no additional medical complications, nor was not taking any medications.For the father, no causes of male infertility were suspected; furthermore,he was healthy.Both parents are Jewish.
Physical examination upon admission
IVF and embryo isolation:IVF, embryo biopsy and PGT were performed according to the standard protocols of the INGENES Institute as previously described (Cedillo 2016; Schaeffer 2017).The mother underwent two standard courses of controlled ovarian stimulation (Depot GnRH antagonist, Cetrotide 0.25 mg daily dose, Merck,Darmstadt, Germany).Stimulation was prolonged until the diameter of leading follicles was > 18 mm.Afterward, recombinant human chorionic gonadotropin (hCG,Choragon 1000 IU, Ferring Laboratories, Saint-Prex, Switzerland) was administered;then, after 36 h, the oocytes were retrieved with ultrasound guidance.All 14-18 mm follicles were aspirated, and 35 ova were collected (summed total for both stimulations).It was decided to proceed with fertilization and culture.
The ova were fertilized by intracytoplasmatic sperm injection, and 25 embryos were produced.Only morphologically optimal embryos were considered for this study, using the criteria established by the Istanbul consensus Workshop on Embryo Assessment[18].Eleven embryos presented as good quality (AB/BB) embryos by Embryo day 5 and were biopsied.Using micromanipulation, 10-15 trophectoderm cells per embryo were isolated and placed into a 0.2 μL PCR tube; afterward, the embryos were frozen.
Embryo transfer and pregnancy test:For day 5 biopsied embryos, the resulting blastocysts were cryopreserved using the vitrification technique.The endometrial preparation was carried out with the transdermal application of 17-β-estradiol (Evorel 50; 150 μg/subcutaneous/every 48 h) and the luteal phase support was carried with Utrogestan (300 mg/day/vaginal).Clinical decisions about which and how many embryos to transfer were determined by the Physician and Specialist in Reproductive Medicine with the patient’s approval.Embryo implantation was confirmed on day 14 by β-hCG serum levels >10 mUI/mL or the presence of a fetal heartbeat by ultrasound at 6.5 to 8 wk.All the patient´s demographics, IVF cycle, PGT results,implantation rate, and IVF outcomes (pregnancies and miscarriages) were recorded by the Specialist.
Laboratory examinations
Whole genome amplification and next-generation sequencing:Biopsies were taken from trophectoderm from at-risk LS embryos.For each sample, the whole genome was amplified (WGA) using the SurePlex amplification system (Illumina San Diego,CA, United States) according to the manufacturer’s instructions.WGA products were quantified using Qubit 3.0 Fluorometer (Life Technologies).The library preparation was carried out with the VeriSeq PGS Library Prep Kit (Illumina Inc.).DNA indexing was performed to simultaneously analyze samples from different embryos using the Nextera XT 96-Index Kit (Illumina, Inc.).For library preparation, 5µl (0.2 ng/µL) of each WGA product from each sample were tagmented (tagged and fragmented) by the VeriSeq PGS transposome using the manufacturer's protocol and neutralized by adding 5 µL of neutralization buffer.The tagmented DNA was amplified with Index 1 primers (N701 and N712) and Index 2 primers (S503 and S504) to become the nextgeneration sequencing (NGS) library via a limited cycle PCR program.Each sample’s NGS library was purified to remove short fragments and primers.Finally, NGS libraries were pooled, denatured with HT1, and loaded to the VeriSeq PGS (Illumina Inc.) sequencing cartridge following the manufacturer's protocol.NGS library was sequenced with a MiSeq apparatus using the MiSeq Reporter Software.Chromosome composition was determined as indicated above.
Determination of GHR mutations presence:Successful WGA was verified by agarose electrophoresis, which revealed an optimal DNA concentration to carry-out the PCR amplification of a 186-bp specific fragment of the deletion breakpoint of exons 5 and 6 of theGHRgene (NM_000163.4, 5p13-p12, MIM+600946) using the primers “5606” and “5662” and according to conditions previously published[19].These primers would amplify a 186-bp fragment in both homozygous and heterozygous del5-6 genotypes, but not in normal wild-type samples.To distinguish the homozygous (affected) from heterozygous (carrier) del5-6GHRgenotype, a second end-point PCR assay was performed simultaneously with the primers “4947” and“5077” (19)to obtain a 269-bp fragment that evaluates the integrity of exon 5, which is present both in heterozygous del5-6 and wild-typeGHRgenotypes.These PCR assays were previously validated in genomic DNA samples derived from peripheral blood leukocytes from the unaffected obligate del5-6 heterozygous parents, as well as in their affected homozygous child with LS.All PCR assays were done in duplicate using 60 ng of WGA-derived DNA per reaction from at-risk LS and normal control,embryos (n= 3), the two heterozygous parents and the homozygous LS-affected child(DNA samples were isolated by standard genomic DNA isolation from blood), and water as a control.Amplicons were resolved by agarose gel electrophoresis stained with ethidium bromide and visualized and photographed under ultraviolet light(Figure 1).
Confirmation of implantation and fetus GHR mutational status:Embryo implantation was confirmed on day 14 by β-hCG serum levels > 10 mUI/mL or the presence of a fetal heartbeat by ultrasound at 6.5 to 8 wk.The genomic composition of the fetus was verified and confirmed by applying the same techniques (WGA obtained from amniotic cells and PCR) using amniocytes extracted after 21 wk of the ongoing pregnancy (Figure 2).
FINAL DIAGNOSIS
Eleven embryos were collected from 2 rounds of IVF; 27.3% were the wild type for GHR, 45.5% were heterozygotes, and 18.2% homozygous mutants.One embryo yielded no results.Eight embryos were determined acceptable for transference.The patients agreed to have two embryos transferred.
TREATMENT
Three 2-embryos transfers were performed (2 normal homozygous and 4 heterozygous carriers) were selected for transfer.The first 2 transfers were unsuccessful, whereas the final transfer, with 2 heterozygous embryos, resulted in a clinical pregnancy (β-hCG serum levels = 252.28 mUI/mL and the presence of one fetal heartbeat sac).The genomic composition of the fetus was verified, applying the same techniques but using DNA from amniocytes, extracted after 21 wk of the ongoing pregnancy.The fetus was confirmed as a heterozygous healthy carrier.
OUTCOME AND FOLLOW-UP
At 36 wk, the mother delivered a healthy baby.No physical deformities were present,suggesting the absence of LS.Moreover, at age 11 mo, the child was clinically confirmed as normal by the family pediatrician.
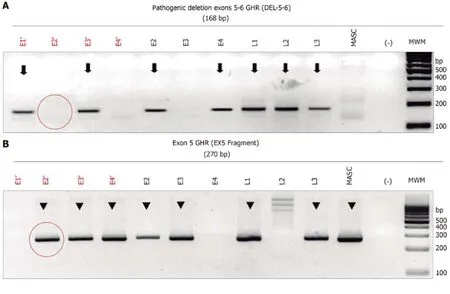
Figure 1 Electrophoresis of the monoplex polymerase chain reaction assay to identify the deletion of exons 5 and 6 (del5-6) of the growth hormone receptor gene for in vitro fertilization embryos.
DISCUSSION
In the review by Zvi Laron in 2015, the author indicates that the use of PGT can aid LS patients to have healthy children; however, a search of the literature yielded no studies supporting this claim[8].Here, we present a case study where a couple underwent IVF to select embryos that were wild-type or heterozygous carriers of the GHR mutations previously characterized in the parents.The procedure was successful, as a fetus amnion-analysis result excludes in the fetus in gestation the clinical and molecular diagnosis of LS attributable to a homozygous genotype for the pathogenic deletion that eliminates exons 5 and 6 ("del5-6") of theGHRgene previously characterized in the parents.Likewise, the absence of the "del5-6" fragment excludes the healthy carrier/heterozygous character in the fetus for LS due to the pathogenic deletion that eliminates exons 5 and 6 of theGHRgene.Eventually, the child was born and was physiologically normal.
One key concern for the procedure was the DNA source.With most LS patients,sample DNA comes from blood or buccal samples.With embryos, the DNA is more limited as for actual day 5-trophectoderm biopsies from blastocyst produce 10 to 15 cells for extraction of genomic DNA.To improve the signal, WGA was performed.With our method and using control samples (the parents, their affected child, and normal embryos), we can show the specificity of the monoplex PCR to determine the del5-6 of GHR.Moreover, when we applied the method to the embryo cohort, we determined that a minority of embryos would be unsuitable for implantation.When the selected embryos were implanted, we were able to confirm the genotype of the fetus, demonstrating the applicability of the method.
There are over 70 documented mutations associated with the development of LS[20].For Latin Americans, the E180 polymorphism is the most common due to a mass migration to Ecuador[21]; nevertheless, the E180 polymorphism has been found in Brazil, Argentina, and Mexican-descendants in the United States[4].Recently, in Monterrey, Mexico, three patients were identified with LS, and the key mutations that were identified as possible causes were not of GHR[5-7].Here, we demonstrate that the LS in the parents was associated with the del5-6 of GHR.This is the first report to demonstrate the presence of this mutation in Mexico.

Figure 2 Electrophoresis of the monoplex polymerase chain reaction assay to determine the growth hormone receptor mutational status in the fetus of the pregnant mother.
Interestingly, the connection with this family and other locations where the mutation is present is difficult to discern.Other than the family being Jewish, there is not another connection.This suggests that, for the Jewish population, members may want to undergo genetic analysis of common diseases.
CONCLUSION
In conclusion, to detect the del5-6 of GHR using WGA embryonic DNA and monoplex-PCR can identify LS at-risk embryos during PGT.Moreover, we provide evidence that WGA is useful and can serve as a template to diagnose other monogenic diseases pre-implantation in patients undergoing fertility treatments.
ACKNOWLEDGEMENTS
We would like to express our gratitude to the participants of the study, to Tania Rojas for their technical assistance in PGT, to the members of the INGENES-IVF Laboratory,and to Dr.Leonardo M.Porchia for his contributions in preparing the manuscript.
杂志排行
World Journal of Clinical Cases的其它文章
- Overview of organic anion transporters and organic anion transporter polypeptides and their roles in the liver
- Value of early diagnosis of sepsis complicated with acute kidney injury by renal contrast-enhanced ultrasound
- Value of elastography point quantification in improving the diagnostic accuracy of early diabetic kidney disease
- Resection of recurrent third branchial cleft fistulas assisted by flexible pharyngotomy
- Therapeutic efficacy of acupuncture combined with neuromuscular joint facilitation in treatment of hemiplegic shoulder pain
- Comparison of intra-articular injection of parecoxib vs oral administration of celecoxib for the clinical efficacy in the treatment of early knee osteoarthritis
