Artificial intelligence in medical imaging of the liver
2019-02-21LiQiangZhouJiaYuWangSongYuanYuGeGeWuQiWeiYouBinDengXingLongWuXinWuCuiChristophDietrich
Li-Qiang Zhou, Jia-Yu Wang, Song-Yuan Yu, Ge-Ge Wu, Qi Wei, You-Bin Deng, Xing-Long Wu, Xin-Wu Cui,Christoph F Dietrich
Abstract Artificial intelligence (AI), particularly deep learning algorithms, is gaining extensive attention for its excellent performance in image-recognition tasks. They can automatically make a quantitative assessment of complex medical image characteristics and achieve an increased accuracy for diagnosis with higher efficiency. AI is widely used and getting increasingly popular in the medical imaging of the liver, including radiology, ultrasound, and nuclear medicine. AI can assist physicians to make more accurate and reproductive imaging diagnosis and also reduce the physicians’ workload. This article illustrates basic technical knowledge about AI, including traditional machine learning and deep learning algorithms, especially convolutional neural networks, and their clinical application in the medical imaging of liver diseases, such as detecting and evaluating focal liver lesions, facilitating treatment, and predicting liver treatment response. We conclude that machine-assisted medical services will be a promising solution for future liver medical care. Lastly, we discuss the challenges and future directions of clinical application of deep learning techniques.
Key words: Liver; Imaging; Ultrasound; Artificial intelligence; Machine learning; Deep learning
INTRODUCTION
In the past few decades, many medical imaging techniques have played a pivotal role in the early detection, diagnosis, and treatment of diseases, such as computed tomography (CT), magnetic resonance imaging (MRI), ultrasound, positron emission tomography ( PET), mammography, and X-ray[1]. In clinical work, the interpretation and analysis of medical images are mainly done by human experts. Recently, medical doctors have begun to benefit from the help of computer-aided diagnosis. Artificial intelligence (AI) is intelligence applied by machines, in contrast to the natural intelligence displayed by humans. In computer science, AI research is defined as the study of "intelligent agents": any device that perceives its environment and takes actions that maximize its chance of successfully achieving its goals[2], which provides the version that is used in this article. Note that they use the term "computational intelligence" as a synonym for artificial intelligence. Russell & Norvig who prefer the term "rational agent" write "The whole-agent view is now widely accepted in the field"[3]. AI has made significant progress which allows machines to automatically represent and explain complicated data[4]. It is widely applied in the medical field,especially some domains that need imaging data analysis, such as radiology[5],ultrasound[6], pathology[7], dermatology[8], and ophthalmology[9]. The emergence of AI can meet the desire of healthcare professionals for better efficacy and higher efficiency in clinical work.
In liver medical imaging, physicians usually detect, characterize, and monitor diseases by assessing liver medical images visually. Sometimes, such visual assessment, which is based on expertise and experience, may be personal and inaccurate. AI can make a quantitative assessment by recognizing imaging information automatically instead of such qualitative reasoning[10]. Therefore, AI can assist physicians to make more accurate and reproductive imaging diagnosis and greatly reduce the physicians’ workload. There are two kinds of AI methods widely used in medical imaging currently, one is traditional machine learning algorithms,and the other one is deep learning algorithms.
In the present paper, we discuss the basic principle of AI and current AI technologies about liver diseases in medical imaging domain for improved accurate diagnosis and evaluation (Table 1). In addition, we discuss the challenges and directions of clinical application of deep learning techniques in the future.
TRADITIONAL MACHINE LEARNING ALGORITHMS
Traditional machine learning algorithms rely mainly on the predefined engineered features that well describe the regular patterns inherent in data extracted from regions of interest (ROI) with explicit parameters on the basis of expert knowledge. The meaningful or task-related features are defined in line with mathematical equations so as to be quantified by computer programs[11]. These features can then be used to further quantify other medical imaging characteristics, such as different lesion density, shape, and echo. Statistical machine learning models, like support vectormachines (SVM) or random forests, are fit to the most typical features to identify relevant imaging-based biomarkers. Gatos et al[12]have attempted to employ traditional machine learning algorithms to support the liver fibrosis diagnosis by ultrasound image. However, the predefined features usually do not have the ability to adapt to the imaging modality changes and their associated signal-to-noise ratio.
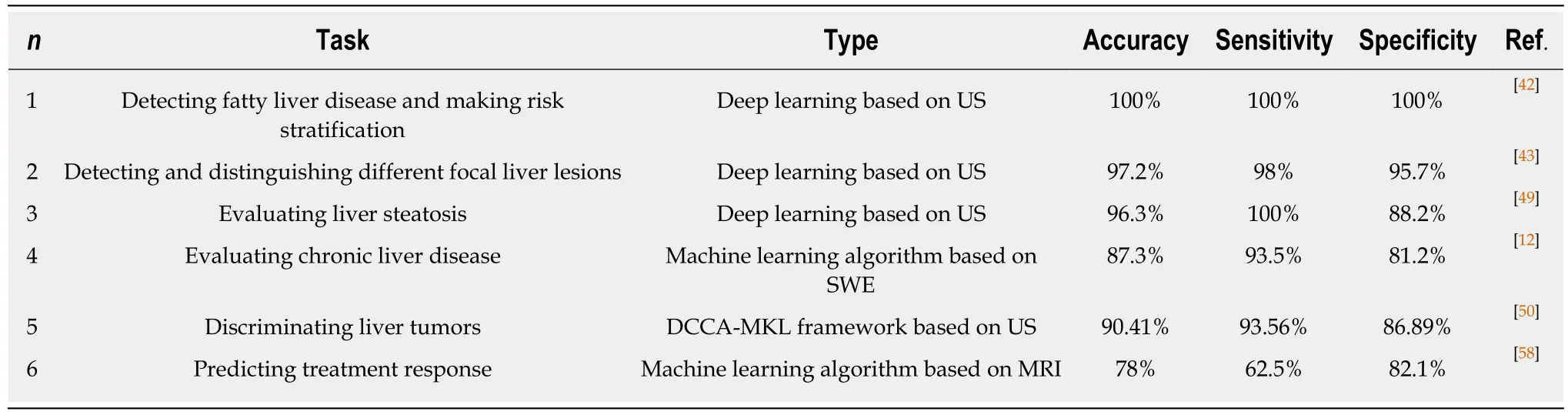
Table 1 Clinical application of artificial intelligence
DEEP LEARNING ALGORITHMS
As a subset of machine learning, deep learning is based on a neural network structure inspired by the human brain. In terms of feature selection and extraction, deep learning algorithms do not have to pre-define features[13,14]and do not necessarily require placing complexly shaped ROI on images. They can directly learn feature representations by navigating the data space, and carry out image classification and task procession. This data-driven mode makes it more informative and practical.Today, convolutional neural networks (CNNs) are the most popular type of deep learning architecture in the medical image analysis field[15]. CNNs usually perform end-to-end supervised learning through tagging data, while other architectures conduct unsupervised learning tasks through untagged data. CNNs consist of quite a few layers and the ‘hidden layer’ among them can complete feature extraction and aggregation by convolution and pooling operations. The fully connected layers can perform high-level reasoning before the final output outcomes. Some studies have found that deep learning methods have excellent performance on staging tasks in computed tomography (CT)[16], segmentation tasks in MRI[17], and detection tasks in ultrasound[18].
INPUT DATA AND TEACHING DATA
The input data and teaching date need to be prepared before the deep learning process. Collecting as many training data as possible can help reduce the risk of overfitting. For gray-scale ultrasound images and red-green-blue (RGB) color ultrasound images, such as color Doppler and shear wave elastography images(SWE), the channel of input data is one and three, respectively. Some researchers concatenated several types of images as one image and used the concatenated images as input data[12,19]. The data volume of input images is associated with the number of CNN parameters. More calculations and longer time are needed to train the large CNNs. Cropped images or resized images can be used to solve this problem. It is necessary for training data to perform image augmentation (such as mirrored images and rotated images) so as to reduce the risk of the overfitting problem, because a slight difference in position may lead to the inconsistency between examinations. For supervised learning, teaching data need to be prepared. The data which researchers want to predict from the input data can be used as teaching data, such as clinical diagnosis data and pathological diagnosis data. The form of output layer should be in the same form as the teaching data. The type of teaching data includes nominal variables, ordinal variables, continuous variables, and images.
CNN
In 2006, Hinton et al[20]published a paper on "Science" that proposed an artificial neural network (ANN) with multiple hidden layers with excellent feature learning ability, which led to the study of deep learning. In 2012, Săftoiu et al[21]performed a study of the diagnosis of focal pancreatic lesions using ANN-assisted real-time endoscopic ultrasound (EUS) elastography and acquired ideal results. ANN is the main algorithm for driving deep learning and CNN is the most commonly used ANN for deep learning. In fact, as early as in the 1980s and 1990s, CNN performed with excellent results in several pattern recognition areas, especially handwritten digit recognition[22,23]. However, it is only suitable for the recognition of small pictures. Since the extended CNN achieved the best classification effect in ImageNet Large Scale Visual Recognition Challenge (LSVRC) in 2012, more researchers have begun to pay attention to it. CNN consists of input layer, hidden layer, and output layer. The hidden layer includes convolutional layers, pooling layers, and fully connected layers.Generally, a CNN model has many convolutional layers and pooling layers. The convolutional layer and the pooling layer are alternately set.
The convolution layer is composed of a plurality of feature maps, each feature map consists of many neurons, and each neuron is connected to a local region of the upper feature map through the convolution kernel which is a weight matrix[4]. The local weighted sum is then passed to a nonlinear function to obtain the output value of each neuron in the convolutional layer. The convolutional layers in CNN implement weight sharing in the same input and output feature map. This method can reduce the number of trainable parameters in the network and the complexity of the network model and make the network easier to train. The convolutional layer extracts various local features of its previous layer through the convolution operation. The first layer of convolution layer extracts low-level features and higher layers of convolutional layers extract more sophisticated features[24]. Increasing the depth of the network and the number of feature faces can improve the ability of deep learning, but it can easily lead to overfitting.
The pooled layer follows the convolutional layer and performs feature extraction again. Its role is mainly to semantically combine similar features and make the features robust to noise and deformation through pooling operations[4]. It is also composed of several feature maps. A feature map of the convolutional layer uniquely corresponds to a feature map of the pooled layer. The neurons in the pooled layer are connected to the local accepted domain of the convolutional layer, and the local accepted domains of different neurons do not overlap. The pooling layer obtains spatially invariant features by reducing the resolution of the feature map[25]. Common pooling methods include maximum pooling, mean pooling, and random pooling[26].Maximum pooling methods are commonly used in recent studies. When the classification layer adopts linear classifiers, the maximum pooling method can achieve a better classification performance than the mean pooling[27]. Random pooling has the advantage of maximum pooling, and it avoids overfitting due to randomness.
The fully connected layer follows the pooled layer and the convolutional layer.Each neuron in the fully connected layer is fully connected to all neurons in the previous layer. The fully connected layer can integrate local information with class discrimination from the convolutional layer or the pooled layer[28]. The activation function of each neuron generally uses the rectified linear unit (ReLU) function[29,30].The output value of the fully connected layer is passed to the output layer. The output layer performs regression tasks and multiple classification tasks by a softmax function. In order to reduce the risk of over-fitting of training, the dropout technique is often used in the fully connected layer[31]. Nodes within the CNNs which are dropped out with a certain probability at the training phase can prevent units from adopting too much. At present, the classification research on CNN mostly adopts ReLU function and dropout technique, and has obtained a good classification ability[28,31].
A prospective multicentre study aimed to evaluate liver fibrosis stages based on 2D-SWE images adopted a CNN model[32]. All the 2D-SWE images with the size of 250× 250 pixels were used as the input data and then the CNN model was triggered. This CNN model had four hidden layers and each convolutional layer followed with a max pooling layer. The first hidden layer contained 16 feature maps, and the remaining three hiden layers each contained 32 feature maps. These feature maps were obtained by applying 16 or 32 convolution filters (3 × 3 pixels) to the previous layer. A fully connected layer with 32 nodes was used to connect every neuron in the last fourth pooling layer so as to output the result of binary classification in the form of probabilities.
TRAINING AND TESTING WITH CNN
During the training phase, output data from CNNs and teaching data are fed to an error function. The errors are backpropagated to CNNs and force CNNs to adjust inner parameters to make the errors smaller. For multiple classification tasks, softmax cross entropy is commonly used as the error function. Different kinds of optimizers are used to adjust parameters within CNNs, such as stochastic gradient descent,AdaGrad[33], and Adam[34]. The learning processes are iterated with units of single input data, groups of input data, or all the input data. At present, minibatch learning is more popular than batch learning for the reason that the amount of calculations for batch learning process is very large. With minibatch learning, data are usually shuffled and assigned to different groups for each epoch. Repeating epochs result in decreased errors for the training phase and the testing phase. Sometimes, repeat of epochs would not necessarily result in a decrease of errors, due to the overfitting problem. In such a situation, early stopping technique might be useful to mitigate this problem. During the testing phase, output values from the trained CNN are compared with teaching data. Methods for evaluating the performance of model include sensitivity, specificity, area under the receiver operating characteristic curve(AUC), and other parameters.
CLINICAL APPLICATIONS
Focal liver lesion detection
Deep learning algorithms combined with multiple image modalities have been widely used in the detection of focal liver lesions (Table 2). The combination of deep learning methods with CNNs and CT for liver disease diagnosis has gained wide attention[35].Compared with the visual assessment, this strategy may capture more detailed lesion features and make more accurate diagnosis. According to Vivantil et al by using deep learning models based on longitudinal liver CT studies, new liver tumors could be detected automatically with a true positive rate of 86%, while the stand-alone detection rate was only 72% and this method achieved a precision of 87% and an improvement of 39% over the traditional SVM mode[36]. Some studies[37-39]have also used CNNs based on CT to detect liver tumors automatically, but these machine learning methods may not reliably detect new tumors because of the insufficient representativeness of small new tumors in the training data. Ben-Cohen et al developed a CNN model predicting the primary origin of liver metastasis among four sites (melanoma, colorectal cancer, pancreatic cancer, and breast cancer) with CT images[40]. In the task of automatic multiclass categorization of liver metastatic lesions,the automated system was able to achieve a 56% accuracy for the primary sites. If the prediction was made as top-2 and top-3 classification tasks, the accuracy could be up to 0.83 and 0.99, respectively. These automated systems may provide favorable decision support for physicians to achieve more efficient treatment.
CNN models which use ultrasound images to detect liver lesions were also developed. According to Liu et al by using a CNN model based on liver ultrasound images, the proposed method can effectively extract the liver capsules and accurately diagnose liver cirrhosis, with the diagnostic AUC being able to reach 0.968. Compared with two kinds of low level feature extraction method histogram of oriented gradients(HOG) and local binary pattern (LBP), whose mean accuracy rates were 83.6% and 81.4%, respectively, the deep learning method achieved a better classification accuracy of 86.9%[41]. It was reported that deep learning system using CNN showed a superior performance for fatty liver disease detection and risk stratification compared to conventional machine learning systems with the detection and risk stratification accuracy of 100%[42]. Hassan et al used the sparse auto encoder to access the representation of the liver ultrasound image and utilized the softmax layer to detect and distinguish different focal liver diseases. They found that the deep learning method achieved an overall accuracy of 97.2% compared with the accuracy rates of multi-SVM, KNN(K-Nearest Neighbor), and naive Bayes, which were 96.5, 93.6, and 95.2%, respectively[43].
An ANN based on18F-FDG PET/CT scan, demographic, and laboratory data showed a high sensitivity and specificity to detect liver malignancy and had a highly significant correlation with MR imaging findings which served as the reference standard[44]. The AUCs of lesion-dependent network and lesion-independent network were 0.905 (standard error, 0.0370) and 0.896 (standard error, 0.0386), respectively.The automated neural network could help identify nonvisually apparent focal FDG uptake in the liver, which was possibly positive for liver malignancy, and serve as a clinical adjunct to aid in interpretation of PET images of the liver.
Diffuse liver disease staging
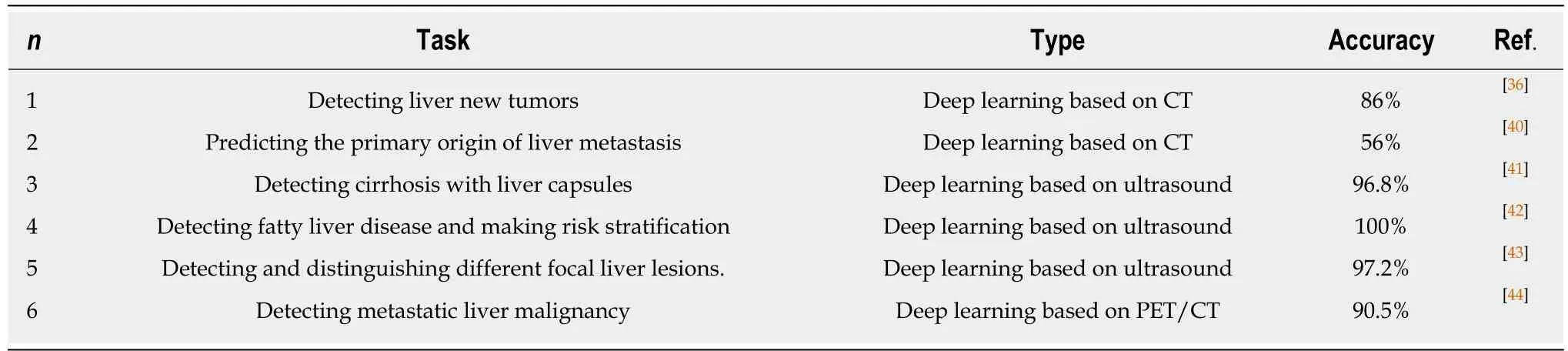
Table 2 Liver leision detection
There are many medical imaging methods combined with deep learning for staging of liver fibrosis diseases (Table 3). Yasaka et al[45]performed a retrospective study to investigate the performance of a deep CNN (DCNN) model with gadoxetic acidenhanced hepatobiliary phase MR images in the staging of liver fibrosis and found that the fibrosis score obtained through deep learning (FDLscore) was correlated significantly with pathologically evaluated fibrosis stage (Spearman's correlation coefficient = 0.63, P < 0.001). The AUCs for diagnosing fibrosis stages cirrhosis (F4),advanced fibrosis (≥ F3), and significant fibrosis (≥ F2) were 0.84, 0.84, and 0.85,respectively. They made a similar study to predict liver fibrosis stage by using a deep learning model based on dynamic contrast-enhanced portal phase CT images and found that the fibrosis score acquired from deep learning based on CT images (FDLCTscore) had a strong correlation with pathologically evaluated liver fibrosis stage(Spearman's correlation coefficient = 0.48, P < 0.001). The prediction of F4, ≥ F3, and ≥F2 could be possible by using the FDLCTscore with AUCs of 0.73 (0.62-0.84), 0.76 (0.66-0.85), and 0.74 (0.64-0.85), respectively[46]. Comparing the two models, the performance of the DCNN model based on CT images was not high, and the reason may be the difference in imaging modality’s ability to capture the features of liver parenchyma. However, CT is more readily available than MRI in clinical settings and the performance is expected to be improved by applying new technologies or using high-performance computers in the future. Wang et al[32]conducted a prospective multicenter study to evaluate the performance of the innovatively developed deep learning radiomics of elastography (DLRE), which could achieve quantitative analysis of the heterogeneity in two-dimensional shear wave elastography images for assessing liver fibrosis stages in chronic hepatitis B. In the training cohort, AUCs of DLRE for F4, ≥ F3, and ≥ F2 were 1.00 (0.99-1.00), 0.99 (0.97-1.00), and 0.99 (0.97-1.00),respectively, which were 0.13, 0.18, and 0.25 higher than those of 2D-SWE. This strategy showed an excellent diagnostic performance in predicting liver fibrosis stages compared with 2D-SWE. It is valuable and practical that the noninvasive techniques can provide an alternative to invasive liver biopsy and make accurate diagnosis of liver fibrosis stages.
Focal liver lesion evaluation
The CNN is also greatly useful in evaluation of liver lesions. By using CNN models based on dynamic contrast-enhanced CT images in unenhanced, arterial phase, and delayed phase, a clinical retrospective study[47]investigated the diagnostic performance for the differentiation of liver masses. Masses were diagnosed according to five categories [category A, classic hepatocellular carcinomas (HCCs); category B,malignant liver tumors other than classic and early HCCs; category C, indeterminate masses or mass-like lesions (including early HCCs and dysplastic nodules) and rare benign liver masses other than hemangiomas and cysts; category D, hemangiomas;and category E, cysts] with a sensitivity of 0.71, 0.33, 0.94, 0.90, and 1.00, respectively.Median accuracy of the CNN model with dynamic CT for categorizing liver masses was 0.84. Median AUC for differentiating categories A-B from C-E was 0.92.
A new method[36]to automatically evaluate tumor burden in longitudinal liver CT studies by using a CNN model was developed and the tumor burden volume overlap error was 16%. This work is of great importance with the reason that the tumor burden can be used to evaluate the progression of disease and the response to therapy. The authors also performed liver tumor volumetric measurements to evaluate disease progression and response to treatment by tumor delineation with global and patient specific CNNs trained on a small annotated database of delineated images in longitudinal CT follow-up[48]. This method can automatically select the most appropriate CNN model for the unseen input CT scan and obviously improve therobustness from 67% for stand-alone global CNN segmentation to 100% in liver tumor delineation.
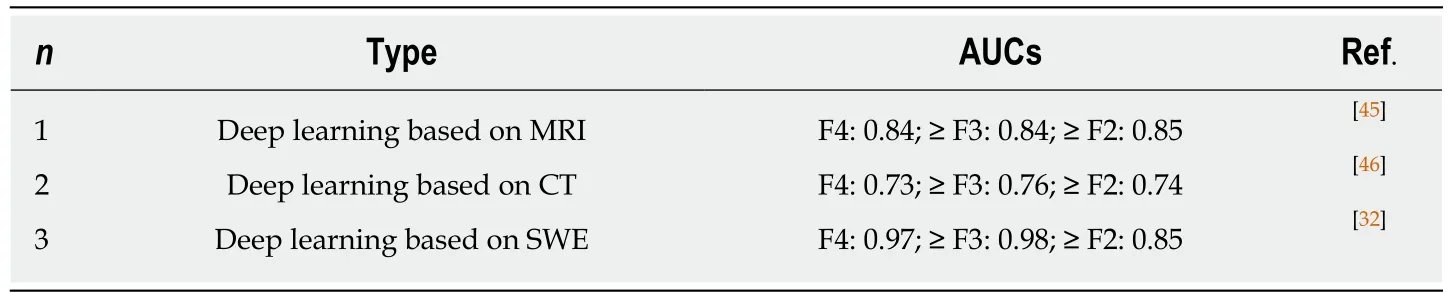
Table 3 Diffuse liver disease staging
Byra et al proposed a deep CNN model with transfer learning for liver steatosis assessment in B-mode ultrasound images[49]. The pre-trained deep CNN on the ImageNet dataset extracted high-level features first, and then the SVM algorithm classified images. The steatosis level was evaluated by the features and the Lasso regression method. Compared with the hepatorenal index and the gray-level cooccurrence matrix algorithm, whose accuracy rates were 90.9% and 85.4%, the CNN-based approach achieved significantly better results, with an AUC of 0.977, sensitivity of 100%, specificity of 88.2%, and accuracy of 96.3%.
A machine-learning algorithm that quantifies color information in terms of stiffness values from ultrasound shear wave elastography (SWE) images and discriminates chronic liver diseases (CLD) from healthy cases was introduced[12]. The highest accuracy of SVM model in the differentiation of healthy persons from CLD patients was 87.3%, and the sensitivity and specificity values were 93.5% and 81.2%,respectively. The present study provided novel objective parameters and criteria for CLD diagnosis through SWE images.
A novel two-stage multi-view artificial intelligence learning framework based on contrast-enhanced ultrasound (CEUS) for discriminating benign and malignant liver tumors achieved the best performance[50]. This method conducted deep canonical correlation analysis (DCCA) on three image pairs and generated total six-view features. A multiple kernel learning (MKL) classification algorithm then yielded the diagnosis result by these multi-view features. The mean classification accuracy,sensitivity, and specificity of DCCA-MKL framework were 90.41 ± 5.80%, 93.56 ±5.90%, and 86.89 ± 9.38%, respectively. DCCA-MKL achieved 17.31%, 10.45%, 24.00%,34.44%, 24.00%, and 10.45% improvements over A-P-SVM, on classification accuracy,sensitivity, specificity, Youden index, false positive rate, and false negative rate,respectively. The proposed DCCA-MKL framework based on liver CEUS has high evaluation and prediction performance for liver tumors. In future work, multi-modal deep neural network algorithm deserves to be investigated and this deep learning algorithm may more effectively fuse and learn feature representation of three-phase CEUS images.
Segmentation
Segmentation of the liver or liver vasculature with CT is of great importance in the diagnosis of vascular disease, radiotherapy planning, liver vascular surgeries, liver transplantation planning, tumor vascularization analysis, etc. Manual segmentation is time-consuming and prone to human errors. The application of deep learning models with the process to achieve automation was studied by some investigators. By using CNN, Bulat et al achieved accurate segmentation of the portal vein automatically from CT images with a Dice similarity coefficient of 0.83 for patients scheduled for liver stereotactic body radiation therapy[51]. Lu et al[52]reported that the liver could be located and segmented automatically via CNN from CT scans for patients planned for living donor liver transplant surgery or volume measurement with high accuracy and efficiency. Li et al[39]described a stand-alone liver tumor segmentation method based on a seven-layer CNN from CT images and achieved a 82.67% ± 1.43% precision. The CNN method has better performance than other machine learning algorithms. In addition, a novel, fully automatic approach to segment liver tumors from contrastenhanced CT images based on a multi-channelfully convolutional network (MC-FCN)was presented. The MC-FCN model provided greater accuracy and robustness than previous methods[53]. These automated segmentation solutions show the potentials of using deep learning to facilitate clinical therapy and achieve more precise medical care.
Liver image quality (IQ) evaluation
Automatical qualitative IQ evaluation based on a classification task (diagnostic vs nondiagnostic IQ) is greatly necessary and useful, because liver MRI as a powerful tool to evaluate chronic liver diseases and to detect focal liver lesions has many limitations, such as inconsistent image quality and decreased robustness related to long acquisition time, motion artifact, and multiple breath-holds, especially T2-weighted sequences(T2WI) are more easily affected by suboptimal image quality[54,55].Steven et al developed and tested a deep learning approach using CNN for automated task-based IQ evaluation of liver T2WI. They found that the CNN algorithm yielded a high negative predictive value when screening for nondiagnostic T2WI of the liver[56].The ability of real-time marking low-quality images allows the technologist to make timely adjustments and improve image quality by altering technical parameters, rerunning a sequence, or running additional sequences.
Treatment response prediction
Automatical prediction of an HCC patient’s possible response to transarterial chemoembolization before treatment by an accurate method is significant and worthwhile. It could minimize patient harm, reduce unnecessary interventions, lower health care costs and so on. Abajian et al reported that transarterial chemoembolization outcomes in HCC patients could be accurately predicted by combining clinical data and baseline MR imaging based on ML models. The overall accuracy of logistic regression (LR) and random forest (RF) models to predict treatment response was 78% (sensitivity 62.5%, specificity 82.1%, positive predictive value 50.0%, and negative predictive value 88.5%)[57]. This strategy can assist physicians to make optimal treatment selection in HCC patients.
In addition to predicting chemotherapy response, deep learning CNN models are also utilized for the prediction of radiotherapy toxicity. Ibragimov et al proposed a novel method to predict hepatobiliary toxicities after liver stereotactic body radiation therapies by using CNNs with transfer learning based on 3D CT. The CNNs were applied to find the consistent patterns in toxicity-related 3D dose plans and numerical pre-treatment features were inputted into the fully-connected neural network for more comprehensive prediction. The AUC of CNNs for 3D dose planned analysis to achieve hepatobiliary toxicity prediction was 0.79, and when combined with some pre-treatment features analysis, the AUC can reach 0.85[58]. This framework can implement accurate prediction of radiation toxicity and greatly helps in the progress of radiotherapy.
CONCLUSION
AI, especially deep learning, is rapidly becoming an extremely promising aid in liver image tasks, leading to improved performance in detecting and evaluating liver lesions, facilitating liver clinical therapy, and predicting liver treatment response. In the future, the development of AI is inseparable from physicians and the work of physicians will be closely linked with AI. Machine-assisted medical services will be the optimal solution for future liver medical care. We need to determine which specific radiology tasks are most likely to benefit from the deep learning algorithm,taking into account the strengths and limitations of these algorithms. In the context of the rapid development of AI technology, physicians must keep pace with the times and apply technology rigorously in order to become a technology driver and better serve patients.
CHALLENGES AND FUTURE PERSPECTIVES
There is considerable controversy about the time needed to implement fully automated clinical tasks by deep learning methods[59]. The debated time ranges from a few years to decades. The automated solutions based on deep learning aim to solve the most common clinical problems which demand a lot of long-term accumulation of expertise or are much too complicated for human readers, for example, lung screening CT, mammograms and so on. Next, researchers need to develop more advanced deep learning algorithms to solve more complex medical imaging problems, such as ultrasound or PET. At present, a common shortage of AI tools is that they cannot resolve multiple tasks. There is currently no comprehensive AI system capable of detecting multiple abnormalities throughout the human body.
A great amount of medical data which are electronically organized and amassed in a systematic style facilitate access and retrieval by researchers. However, the lack of curation of the training data is a major drawback in learning any AI model. To select relevant patient cohort for specific AI task or make segmentation within images is essential and helpful. Some segmentation algorithms using AI[60]are not perfect to curate data, as they always need human experts to verify accuracy. Unsupervised learning which includes generative adversarial networks[61]and variational autoencoders[62]may achieve automated data curation by learning discriminatory features without explicit labeling. Many studies have explored the possibilities of unsupervised learning application in brain MRI[63]and mammography[64]and more field applications of this state of the art method are needed.
It is of great significance to indicate that AI is different from human intelligence in numerous ways. Although various forms of AI have exceeded human performance,they lacked higher-level background knowledge and failed to establish associations like the human brain. In addition, AI is trained for one task only. The AI field of medical imaging is still in its infancy, especially in the ultrasound field. It is almost impossible for AI to replace radiologists in the coming decades, but radiologists using AI will inevitably replace radiologists who do not. With the advancement of AI technology, radiologists will achieve an increased accuracy with higher efficiency. We also need to call for advocacy for creating interconnected networks of identifying patient data from around the world and training AI on a large scale according to different patient demographics, geographic areas, diseases, etc. Only in this way can we create an AI that is socially responsible and benefits more people.
杂志排行
World Journal of Gastroenterology的其它文章
- Evolving role of magnetic resonance techniques in primary sclerosing cholangitis
- Cancer risk in primary sclerosing cholangitis: Epidemiology,prevention, and surveillance strategies
- Effect of Sheng-jiang powder on multiple-organ inflammatory injury in acute pancreatitis in rats fed a high-fat diet
- Preoperative rectosigmoid endoscopic ultrasonography predicts the need for bowel resection in endometriosis
- Short- and long-term outcomes of endoscopically treated superficial non-ampullary duodenal epithelial tumors
- Serum hepatitis B virus RNA is a predictor of HBeAg seroconversion and virological response with entecavir treatment in chronic hepatitis B patients
