Map-based cloning of a recessive gene v1 for virescent leaf expression in cotton(Gossypium spp.)
2018-11-10ZHANGYoupingWANGQiaolianZUODongyunCHENGHailiangLIUKeASHRAFJavariaLISiminFENGXiaoxuYUJohnandSONGGuoli
ZHANG Youping,WANG Qiaolian,ZUO Dongyun,CHENG Hailiang,LIU Ke,ASHRAF Javaria,LI Simin,FENG Xiaoxu,YU John Z.and SONG Guoli*
Abstract Background:Virescence,as a recognizable phenotype in the early development stage of cotton,is not only available for research on chloroplast development and photosynthesis but also for heterosis exploitation in cotton.Methods:In current study,for fine mapping of virescent-1(v1)in cotton,three populations with a total of 5 678 individuals were constructed using T582 which has the virescent trait.Tobacco rattle virus,TRV1 and TRV2(pYL156),were used as vectors for the virus-induced gene silencing(VIGS)assay.Results:The v1gene was fine-mapped to a 20 kb interval on chromosome 20 of tetraploid cotton.We identified only one candidate gene with four single nucleotide polymorphisms between parents,among which the single nucleotide polymorphism at the position of 1 082 base pair caused the change of amino acid residue from Arg(3–79)to Lys(T582).The relative expression of the candidate gene in virescent plants was extensively lower than that in normal plants.Nullification of the gene by VIGS significantly turned the green leaf of normal cotton plants into yellow.We named this candidate gene as GhRVL.Conclusions:This study will facilitate the further research on virescent formation,and will be useful for breeding of hybrid cottons.
Keywords:Cotton,Virescence,T582,VIGS
Background
The phenotype of the virescent mutant is characterized by yellowish leaves at the early stage,which gradually become normal green leaves at maturity.Virescent mutants have been found in an extensive list of plant species including cotton(Killough and Horlacher 1933),tomato(Richard and Charles 1954),cucumber(Aalders 1959),barley(Jain 1966),peanut(Benedict and Ketring 1972),soybean(Palmer and Mascia 1980),maize(Hopkins and Elfman 1984),tobacco(Archer and Bonnett 1987),rice(Iba et al.1991),Arabidopsis(López-Juez et al.1998),rape(Zhao et al.2000)and sugi(Hirao et al.2009).Many previous researches explored the mechanism of chloroplast development and photosynthesis in virescent mutants(Iba et al.1991;López-Juez et al.1998;Benedict and Kohel 1968;Fambrini et al.2004;Wang et al.2016a,2016b),which helped to elucidate the mechanism of chlorophyll(Chl)synthesis and degradation,and to explore the interaction of genes in nucleus and chloroplast(Karaca et al.2004;Sugimoto et al.2004)forthe complex expression patternssuch as temperature-induced virescent mutant(Turcotte and Feaster 1978)and transient or long-lasting virescent mutant(Percival and Kohel 1974;Endrizzi et al.1984).Further,many other studies on chlorophyll biosynthetic process in Arabidopsis and rice provided a critical understanding for virescent variations(Koncz et al.1990;Falbel and Staehelin 1994;Nakayama et al.1995;Kruse et al.1997;Zhang et al.2006).
The whole pathway of Chl biosynthesis is consisted of 27 genes that encode 15 enzymes from glutamyl-tRNA to Chl b in Arabidopsis(Zhu et al.2014;Wang et al.2016a,2016b).In Chl biosynthesis,the initial step is the insertion of Mg2+into protoporphyrin IX.Same as in Arabidopsis,the mutant cs directly involved in Chl biosynthesis by motivating the ATP-dependent binding of Mg2+into protoporphyrin IX(Rissler et al.2002;Ikegami et al.2007),exhibiting the yellowish leaf phenotype(Koncz et al.1990;Kobayashi et al.2008).While in rice,an oxygenase mutant gene ygl1(osCAO)led to yellow-green leaves and delayed chloroplast development at seedling stage(Wu et al.2007).On the other hand,a negative correlation has been found between protoporphyrin and heme biosynthesis in tetrapyrrole biosynthesis,suggesting that excess of heme will cause inhibition of chlorophyll synthesis which resulted in leaf color mutant with lack of chlorophyll(Cao et al.2009).For instance,elevated accumulation of the OsHO2(Heme oxygenase)into Mg-protoporphyrin IX in rice caused the leaf color mutant(Li et al.2014b).
The virescent trait with a photoperiod-sensitive genetic male sterility in cotton can reduce the cost of hybrid breeding and promote the exploitation of heterosis to overcome the limitations of seed production in the 3-line system(Zhao et al.2000;Duncan and Pate 1967;Ma et al.2013).To date,out of twenty virescent genes,seven single recessive nuclear genes from more than 30 mutants of the tetraploid cotton have been mapped in genetic linkage groups(Endrizzi et al.1984).Previously,Killough and Horlache found that a virescent mutant plant in Mebane varieties of upland cotton controlled by a recessive single nucleus gene v1which has been transferred into T582(Kohel et al.1965)and mapped into XVII linkage goup(Kohel 1983).Subsequent genetic mapping ofF2population (T582×Hai7124)showed that the v1gene resided on chromosome 20 of D subgenome with the closest genetic distance of 10.3 cM to CIR094(Hu and Zhou 2006).Therefore,current study was conducted to clone the v1gene with molecular markers which developed from the cotton genomesequenceusingmap-based techniques(Wang et al.2012;Li et al.2014a;Li et al.2015;Lu et al.2015).
Methods
Plant materials and phenotypic data collection
T582 is a multi-recessive marker stocking with cu,fg,cl1,gl1,and v1genes,while 3–79 and Pima 90 are sea island cottons with non-virescent trait.Chi-square tests were performed on three populations,including(i)population I of T582×(T582×3–79)BC1F1,(ii)population II consisting of(3–79×T582)×T582 BC1F1,and(iii)population III having BC1F1of(Pima 90×T582)×T582.A total of 1 200 plants from the population I,2 193 plants from the population II,and 2 285 plants from the population III were screened for virescent to non-virescent ratio of 1:1,respectively.
DNA extraction and polymerase chain reaction(PCR)analysis
The genomic DNA of fresh leaf tissues from all populations and their parents was extracted following cetyl trimethyl ammonium bromide(CTAB)method(Paterson et al.1993).PCR was conducted with a total volume of 10 μL consisting of 1 μL 10 × PCR buffer(TransGen Biotech Co.,Ltd.,Beijing),0.5 μL 2.5 mmol·L-1dNTPs(TransGen Biotech Co.,Ltd.,Beijing),0.2 μL each 10 μmol·L-1primer(GenScript Co.,Ltd.,Nanjing),30 ng template DNA and 0.1 μL 5 U·μL-1Taq DNA polymerase(TransGen Biotech Co.,Ltd.,Beijing).The PCR amplification was conducted as follows:3 min at 94°C;followed by 27 cycles of 30 s at 94°C,30 s at 55 °C,and 1 min at 72 °C.The final extension was done at 72°C for 5 min.The amplified PCR products were then separated by the 8.0%polyacrylamide gels.
Development of molecular markers for fine mapping
Based on the closest marker(CIR094)to v1(Hu and Zhou 2006),we used the SciRoKo34 software to get further SSR(Simple sequence repeats)primers(Kofler et al.2007).When no polymorphic SSR was found in a fine mapping interval,we explored arbitrary sequence from every 500 base pairs(bp)of the interval through the Primer 5.0 software.To analyze the polymorphism and linkage map of all markers,we used parental plants and six of each virescent and non-virescent individuals from the population I.
Fine mapping of the v1gene
We used 1 200 plants(581 virescentand 619 non-virescent)of the population I for the initial mapping by SSR markers.Joinmap 3.0 software was used to analyze linkage between v1gene and molecular markers(Van Ooijen and Voorrips 2001).Subsequently,we also used 1 095 virescent plants and 1 098 non-virescent plants from the population II and 1 106 virescent plants and 1 179 non-virescent plants from the population III for the fine mapping of v1.
Identification and sequence analysis of the v1candidate genes
We obtained the sequence of the predicted gene from the Cotton GenomeProjectdatabase(http://cgp.genomics.org.cn/page/species/index.jsp)and the G.hirsutum genome database(http://mascotton.njau.edu.cn/info/1054/1118.htm),and further confirmed by BLAST(Basic local alignment search tool)searches using the EST(Expressed sequence tag)database.The full-length v1candidate gene in virescent and non-virescent plants was amplified by the 5′-ATGGCTTCC GTGCTTGGAACCTCAA-3′,5′-TCAGCTGAAAACCT CATAGAATTTC-3′primer pair.PCR assays were conducted to amplify the region which was subsequently cloned by One Step Cloning Kit(Vazyme,Nanjing)into PBI121 vector(TaKaRa,Dalian),and sequenced by the Genewiz(Beijing,China).Sequence alignments were performed with NCBI-BLAST and sequences were analyzed using ClustalW2(https://www.ebi.ac.uk/Tools/msa/clustalw2/).
Quantitative reverse transcription-PCR(qRT-PCR)analysis
Total RNA was isolated using EASY spin Plant RNA Kit(Aidlab,Beijing)from fresh leaves of the 3–79 and T582,respectively.Afterwards,PrimeScript™II 1st strand cDNA Synthesis Kit(TaKaRa,Dalian)was used to synthesize the first-strand cDNA.qRT-PCR was carried out in a total volume of 20 μL containing 10 μL SYBR®Premix Ex Taq II(2×),2 μL cDNA,0.4 μL ROX Reference Dye II(50×),0.8 μL of 10 μmol·L-1forward and reverse primers each and 6 μL ddH2O.PCR wasconducted in ABI PRISM@7500-Fast Real-Time PCR system under the following conditions:30 s at 95 °C;40 cycles of 5 s at 95 °C and 30 s at 60 °C;followed by 15 s at 95 °C,1 min at 60 °C,15 s at 95°C(Cheng et al.2016).qRT-PCR was carried outbythegene-specificprimers(5′-ATTGCCACT GTCATCCCCAACTGCT-3′,5′-TCAGCTGAAAACCT CATAGAATTTC-3′)and actin(Genbank ID:AY305733)(5′-ATCCTCCGTCTTGACCTTG-3′,5′-TGTCCGTCA GGCAACTCAT-3′)was employed as an internal control.Last,relative gene expression was quantified using the 2–ΔΔCTmethod.
VIGS(virus-induced gene silencing)assay
Tobacco rattle virus,TRV1 and TRV2(pYL156),were used as vectors for the VIGS(Virus-induced gene silencing)assay.The cotton phytoene desaturase(PDS)was used to check the efficiency of VIGS(Tuttle et al.2008;Pang et al.2013),since the silencing of PDS gene caused the loss of carotenoids and chlorophyll which resulting in white leaves.The pYL156 was employed as the negative control.A 250 bp fragment of the GhRVL gene was amplified from cDNA library of CCRI(China Cotton Research Institute,former name of ICR,CAAS)12-Dgl leaf tissues by PCR using the primer pair 5′-CTCA CATCCTGCTCGGTTTATTCTC-3′,5′-AGAAGAAAG AGAACTCCTAGCTGAA-3′,and inserted into the vector pYL156 by BamHI and KpnI double digestion to construct pYL156-RVL.Four vectors were transformed into GV3101 strain of Agrobacterium tumefaciens by freeze-thaw method.The Agrobacterium cultures were grown overnight in LB medium having 25 μg·mL-1rifampicin,50 μg·mL-1kanamycin and gentamycin at 28 °C.The bacteria were harvested at 4 000 r·min-1for 5 min and re-suspended in infiltration solution(LB medium having10mmol·L-1MgCl2,10mmol·L-1MESand 200 μmol·L-1acetosyringone).The three different bacterial cultures of pYL156-RVL,pYL156-PDS and pYL156,respectively,mixed with pTRV1 at 1:1 ratio after staying at 25°C for 4 h.The seedling stage of CCRI 12-Dgl was used for VIGS and the procedures for the infiltration infection were carried out according to the previous description(Gao and Shan 2013).The phenotype of the infiltrated plants was examined 1 week later.Total RNA was isolated from the true leaves of silenced and controlled plants,respectively.qRT-PCR was performed to confirm the silencing of the GhRVL gene in theVIGS plants.
Results
Genetic analysis of virescent traits
The leaves of T582 are virescent at early stage(Fig.1a),while 3–79 and Pima 90 have green leaves during the entire growth season(Fig.1b).Among three backcross populations constructing for the fine mapping of the v1gene,581 virescent plants and 619 non-virescent plants derived from the population I(T582×(T582×3–79)BC1F1)fit at 1:1 ratio(χ2=1.2033,P > 0.05).Similarly,1:1 ratio(χ2=0.0041,P > 0.05)has been found between the 1 095 virescent plants and 1 098 non-virescent plants of population II(BC1F1segregating plants of(3–79×T582)×T582).And,the ratio of 1 106 virescent plants to 1 179 non-virescent plants from population III which derived from the BC1F1of(Pima90×T582)×T582 was 1:1(χ2=2.3322,P > 0.05).These results showed that a recessive nuclear gene v1controlled the virescent phenotype in T582,which is also consistent with previous studies(Killough and Horlacher 1933;Kohel et al.1965).

Fig.1 Comparison of the virescent and non-virescent leaves.a Virescent leaf.b Non-virescent leaf
Primary mapping of the v1gene
Previous study showed that CIR094 was identified near the v1gene with 10.3 cM distance on chromosome 20 of tetraploid cotton(Hu and Zhou 2006).A linkage marker Dt_chr11–5923(VS12)was obtained by genome-wide molecular markers(Lu et al.2015)which is closer to the v1gene than BNL2570 and MUSS143 analyzed by 1 200 individuals of population I.There was approximately 5 Mb distance between CIR094 and VS12 markers,which was confirmed by BLAST search in the whole genome sequence of G.hirsutum and G.raimondii(Wang et al.2012;Li et al.2015).Fifteen primer pairs of polymorphic SSR markers which designed by the SciR-oKo 34 software were screened to the 5 Mb intervals from G.hirsutum genome database.With the linkage analysis by JoinMap 3.0,v1was initially mapped to a 275 kb region between VS2 and VS3(Fig.2a),which was further lessened to a 100 kb region between VS13 and VS14(Fig.2b)with the assistance of virescent and non-virescent plants of the population I.
Fine mapping of the v1gene
As no polymorphic SSR is available for the 100 kb interval,the polymorphism of 96 SSR markers from every 500 bp of the 100 kb interval was screened by T582 and 3–79,and obtained five polymorphic arbitrary markers named VS17,VS18,VS19,VS20,and VS21.Then,1 095 virescent plants and 1 098 non-virescent plants from the population II were used to further refine the range of the v1gene,which mapped it approximately 20 kb interval between VS18 and VS19(Fig.2c).Meantime,five polymorphic arbitrary markers were verified between the virescent and non-virescent plants of population III,which were consistent with the fine mapping results of population II(Fig.2c).As no polymorphic SSR and recombinant individuals were found in all three mapping populations,it was concluded that v1gene was located in the 20 kb interval between VS18 and VS19.Information about all markers was listed in Table 1.
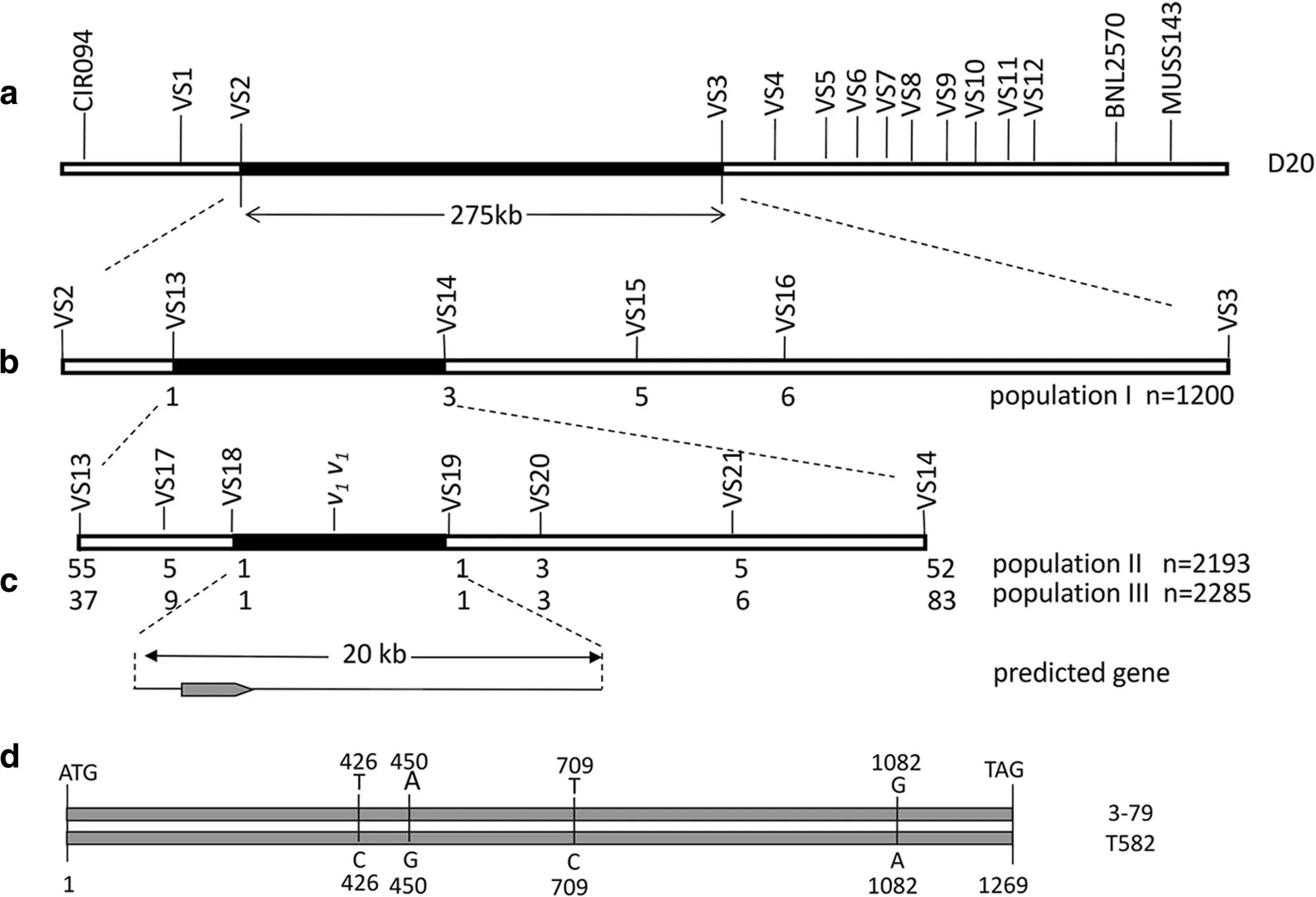
Fig.2 Genetic and physical maps of the v1gene and sequence analysis of the candidate gene on chromosome 20 of the D subgenome in cotton.a Linkage map of the scaffold assembled using 1 200 selected individuals from population I.v1was mapped between VS2 and VS3 markers at around 275 kb.b Additional mapping of v1.v1was further mapped to a 100 kb interval by VS13 and VS14 with the recombinants in population II.c Candidate region for v1and predicted gene.Candidate region for v1was identified between VS18 and VS19 at about 20 kb.One gene was found in that region.Numbers of recombinants between v1and markers were presented under the linkage map.d Sequence comparisons of the candidate gene in 3–79 and T582.SNPs at the sequence positions of 426,450,709 and 1 082 were shared by 3–79 and T582
Identification and sequence analysis of the v1candidate gene
One candidate gene Gh_D10G0283 which was identified in the 20 kb mapping interval based on genome annotation databases(http://cgp.genomics.org.cn/page/species/index.jsp and http://mascotton.njau.edu.cn//info/1054/1118.htm)and BLAST search result with EST and Unigene databases in NCBI.The length of open reading frame(ORF)was 1 269 bp encoding 422 amino acids(Fig.3).The candidate gene was homologous to the magnesium chelatase I gene(ChlI,AAM98163)of Arabidopsis,which derived the ATP-dependent insertion of Mg2+into protoporphyrin IX with yellowish leaf phenotype(Rissler et al.2002;Ikegami et al.2007).Sequence alignment between 3-79 and T582 showed 4 single nucleotide polymorphisms(SNPs)differences at 426,450,709,and 1 082 positions,respectively(Fig.2d).According to protein sequence alignment,the SNP at position of 1 082 caused amino acid residue mutant from Arg(3–79)to Lys(T582),while the other SNPs had synonymous substitution.Furthermore,qRT-PCR showed that the relative expression of the candidate gene in T582 was significant lower than that of 3–79(Fig.4a),suggesting that the candidate gene may cause the formation of virescent leaf.
Silencing of the v1candidate gene leading to yellow leaf
Functional analysis of the v1gene was performed in CCRI 12-Dgl(Cheng et al.2016)using VIGS to validate its role in the formation of virescent leaf in cotton.The 250 bp interference fragment was inserted into a TRV2(tobacco rattle virus)vector to construct the VIGS vector.One week after the Agrobacterium-mediated infection,the mutant phenotypes of the VIGS-treated plants started to emerge.The plants injected with pYL156-PDS revealed a photo-bleaching of leaves(Fig.5c),while yellow-green leaves ofthe plants infiltrated with pYL156-RVL were observed(Fig.5b).Meanwhile,the plants infiltrated with pYL156 had no effecton non-virescent leaf(Fig.5a).To check the silencing efficiency,RNA was extracted from leaves of VIGS plants for qRT-PCR.Expression of the candidate gene in the plants infected by pYL156-RVL which was reduced largely as comparing with the plants infected by pYL156(Fig.4b).Therefore,we concluded that silencing of the candidate v1gene caused yellow leaves,and the candidate gene was subsequently named as GhRVL(Gossypium hirsutum regulator of virescent leaf)gene.
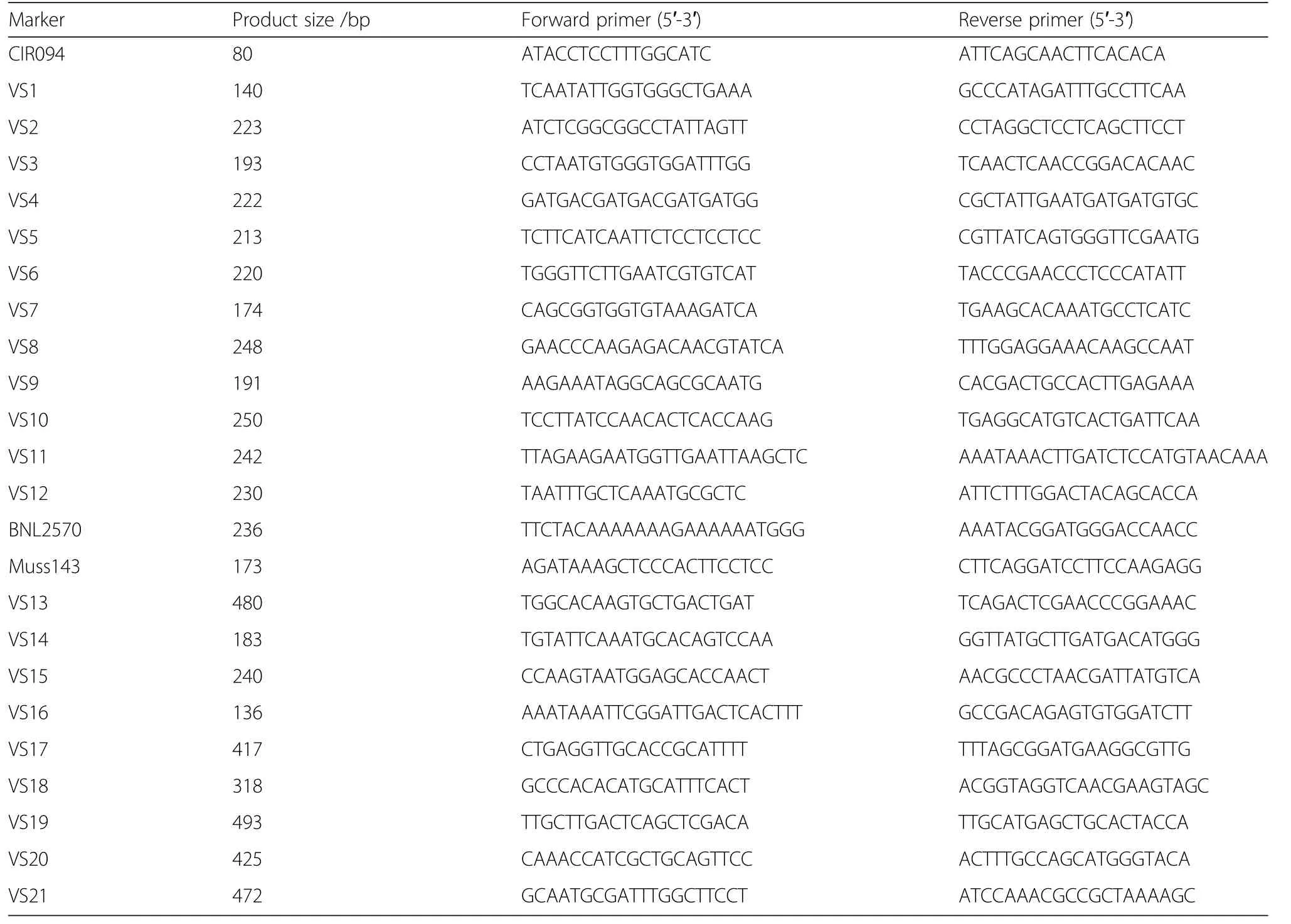
Table 1 List of polymorphic molecular markers for mapping of the recessive v1gene

Fig.3 The sequence of the v1candidate gene
Discussion
T582 serves as a basic tool for scientific study into the mechanism of cotton metabolism,inheritance and development due to its multiple recessive marker stocking with cu,fg,cl1,gl1,and v1(Kohel et al.1965).The candidate gene of the pigment glands which related gene gl1would provide the prospects of fabricating gossypol-free cotton seeds(Cheng et al.2016).In addition,many important genes were subdivided into different linkage groups through T582(Percival and Kohel 1974;Endrizzi et al.1984).Especially,seven of the 20 virescent genes were reported at one recessive locus in the tetraploid cotton species,which further have been mapped by linkage analysis(Duncan and Pate 1967;Endrizzi et al.1984).Genes v1and v7were found to be homoeoallelic,which are functionally similar and located on homoeologous chromosomes(Turcotte and Feaste 1973).Therefore,cloning of gene v1is helpful to clone the other gene v7and to analyze their interaction in cotton.
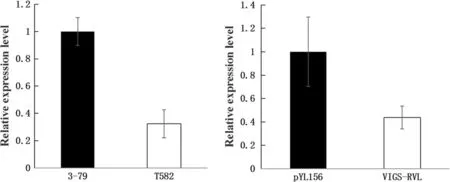
Fig.4 Results of qRT-PCR.a Expression analysis of the candidate gene in 3–79 and T582,respectively.Y-axes indicates the relative expression level of the gene and X-axes represents the candidate gene in the 3–79 and T582,respectively.b:The results of the pYL156 and VIGS-RVL in CCRI 12-Dgl,respectively.Y-axes indicates the relative expression level of the gene and X-axes represents the pYL156 and VIGS-RVL in the CCRI 12-Dgl,respectively

Fig.5 Silencing of the v1candidate gene by VIGS resulted in CCRI 12-Dgl.pYL156 and pYL156-PDS were used as negative and positive controls,respectively.Leaves of VIGS plants displayed mutant phenotype.a Normal green phenotype in negative control plant;b yellowish phenotype in the plant infected by pYL156-RVL;c Photo-bleaching phenotype in positive control plant
Previous study reported that gene v1was located in the interval between CIR094 and BNL2570 markers on chromosome 20 of the D sub-genome(Hu and Zhou 2006).In this study,we took advantage of the databases of cotton genome and genome-wide molecular markers(Wang et al.2012;Li et al.2014a,;Li et al.2015;Lu et al.2015)to develop new SSR markers and arbitrary sequences for fine mapping of the v1gene.The polymorphism and recombination events were detected in three populations,which identified one candidate gene in a 20 kb interval between VS18 and VS19 markers.Results showed that v1is a single recessive gene in cotton which is homologous to the magnesium chelatase I(ChlI)gene,contains a P-loop NTPase domain and is a member of the AAA+protein family(Fodje et al.2001;Iyer et al.2004).It plays an important role in chlorophyll biosynthesis by motivating the inclusion of Mg2+into protoporphyrin IX.Chlorina mutant aci5–3 in Arabidopsis and one semi-dominant Oil yellow 1(Oy1)mutants in maize are caused by missense mutations in the highly conserved AAA+domain of ChlI subunits(Soldatova et al.2005;Sawers et al.2006).However,CHLI is encoded by two genes in Arabidopsis compared with a single copy gene of barley and tobacco,which shows 82%similarity between CHLI1 and CHLI2(Kobayashi et al.2008).The expression level of CHLI2 which contributes to the assembly of the Mg-chelatase complex is much lower than that of CHLI1(Kobayashi et al.2008).But,a transgene of CHLI2 motivated by the promoter of CHLI1 can be functionally equivalent to CHLI1(Huang and Li 2009).In transgenic tobacco,either a decreased or increased expression of the CHLI subunit would diminish Mg chelatase activity and significantly reduce chlorophyll content(Papenbrock et al.2000).In current study,the candidate gene GhRVL was homologous to the magnesium chelatase I gene(ChlI,AAM98163)in Arabidopsis.The candidate gene GhRVL just has single base change at 1 082 bp position which caused the change of the 361st amino acid residue from Arg(3–79)to Lys(T582).And the results of qRT-PCR showed that the relative expression level of GhRVL in virescent plants was much lower than that in non-virescent plants.We hypothesized that the different phenotype of the virescent-1 mutant in T582 compared with normal plant in 3–79 is probably due to the promoter difference between them(Zhu et al.2017;Mao et al.2018).
Virescent character is a useful morphological indicator which is controlled by recessive genes(Benedict et al.1972).However,more than 30 virescent mutants were not found their target genes in cotton(Song et al.2012).The virescent gene v1can serve as a valuable resource for heterosis utilization(Duncan and Pate 1967;Ma et al.2013),for exploring the mechanism of photosynthesis as well as for a better understanding of genetic interactions.
Conclusions
This report reveals fine mapping and cloning of the candidate gene GhRVL of virescence in cotton which significantly turned the green leaf color of normal cotton plants into yellow by VIGS.
Acknowledgments
We thank State Key Laboratory of Cotton Biology,Institute of Cotton Research of Chinese Academy of Agricultural Sciences in China.
Funding
The National Key Research and Development Program of China(2016YFD0101401).
Availability of data and materials
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
Authors’contributions
Song GL managed the project and designed the research.Zhang YP,Wang QL,Zuo DY,Cheng HL,Liu K,Ashraf J,Li SM,Feng XX,and Yu JZ performed the experiments and prepared figures and tables.Zhang YP,Song GL and Wang QL wrote and revised the paper.All authors reviewed the manuscript.All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author details
1State Key Laboratory of Cotton Biology,Institute of Cotton Research of Chinese Academy of Agricultural Sciences,Anyang 455000,China.2U.S.Department of Agriculture-Agricultural Research Service(USDA-ARS),Southern Plains Agricultural Research Center,College Station,TX 77845,USA.
Received:7 June 2018 Accepted:20 August 2018

杂志排行
Journal of Cotton Research的其它文章
- Identification and screening of nitrogenefficient cotton genotypes under low and normal nitrogen environments at the seedling stage
- Hypoxia tolerance studies for yield,fiber and physiological traits in cotton(Gossypium hirsutum L.)
- A genome-wide analysis of SWEET gene family in cotton and their expressions under different stresses
- Comparative transcriptome study provides insights into acquisition of embryogenic ability in upland cotton during somatic embryogenesis
