Cell Transport of DOFH10 in Arachis hypogaea L. Is Possibly via Vesicle and Specifically Recognized in Clathrin Related Pathway
2018-11-01YANHaiyanHUANGZhenlingSUOHuiHUANGJiaquanLIAOBoshou
YAN Haiyan,HUANG Zhenling,SUO Hui,HUANG Jiaquan,LIAO Boshou
(1.College of Life Sciences,South-Central University for Nationalities,Wuhan 430074,China;2.Chinese Academy of Agricultural Sciences,Key Laboratory of Oil Crop Biology of the Ministry of Agriculture,Wuhan 430062,China)
Abstract:Study unknown DOFH10 (DNA binding with one finger) genes in peanuts,cloning of DOFH10 genes with PCR methods according to cDNA of DOF transcription factor GW391728 with primers s162-a1469 and primers s30-a1469 from 16 Different peanut cultivars of Arachis hypogaea L.,sequences comparison,analysis on enzyme site and 3D structure and Western Blot on DOFH10 proteins were conducted. Two types of DOFH10 genes were separated from genome DNA of 5 cultivars with primers s162-a1469,type A DOFH10 was only existed in four cultivars with multiple seeds per fruit and one cultivar with two seeds per fruit. They were identical to XM_016100960.2 from Arachis duranensis chromosome A03 and to XM_016334585.2 from Arachis ipaensis chromosome B03 respectively. With primers s30-a1469,only type B DOFH10 was cloned from all the 16 cultivars used including all the six cultivars with multiple seeds per fruit. Type B DOFH10 had identical RNA and protein sequences at DOF zinc finger region to that of putitive clathrin assembly protein XP_016199412.2 encoded by gene on chromosome B01 of Arachis ipaensis. Multiple possible myristoylation sites scattered on DOFH10 protein,both results suggested that transportation of DOFH10 was probable via vesicle membrane traffic related to clathrin. DOFH10 protein was specifically expressed in developing cotyledons displayed by Western Blot. A and B two types of DOFH10 genes existed in cultivated peanut. The type B was present in all 16 peanut cultivars,and was an essential gene,was possibly transported into nucleus through clathrin dependant vesicle pathway. The type A DOFH10 tended more appeared in cultivars with multiple seeds per fruit,probably related to cell division in early fruit development.
Key words:DOF; Transcription factor; Peanut; Clathrin related; Fruit development
DOF (DNA binding with one finger) is a large transcription factor family in plant. There are 28 inG.Max,36 inArabidopsis. It has not been thoroughly studied inArachishypogaeaL.. From the cDNA library of developing seed,total 8 DOF transcripts were found,7 of them were sequenced at whole cDNA level[1]. One of them,DOFH10 (GW391728) is mostly similar to pea DOF3,and designed as ahDOF3. Recently through comparison to similar sequences from genomic libraries ofArachisduranensisandArachisipaensis,which is the source of AA and BB genomes of cultured peanut respectively[1-2],GW391728 is mostly similar to DOF 1.4 on chromosome A03 ofArachisduranensisand identical to DOF 1.4 on chromosome B03 ofArachisipaensis[3]. GW391728 belongs to class VⅡ DOF protein,which is in the same class as maize PBF and GmDOF5[1]. Maize and barley PBF bind to P box in promoters of endosperm specific genes of storage proteins[4-5]. They coregulate gene expression by interacting with other proteins[4,6]. The target of DOF proteins may be set of genes. GmDOF11 and 4 regulate seed lipid content by network regulation of gene expression[7]. Thus the structure information is important for learning the regulation mechanism of DOF proteins. For study differences on GW391728 between cultivars with Different traits,we cloned and compared GW391728 similar genes from genomic DNA of 16 peanut cultivars with multiple repeat clones and with Different primers,predicted,analyzed and compared three dimensional structures and regulation sites of typical types of DOFH10 proteins. We have also studied the expression pattern of DOFH10 protein by Western Blot,provided the basic information for further study on function of DOFH10 protein.
1 Materials and Methods
1.1 Material
DNA templates used for cloning ofDOFH10 are extracted from leave of 20 peanut cultivars with CTAB method. The list the of the cultivars used as following:①Danfeng (DF),2 seeds per fruit (SPF);②Laofandou(LFD),2SPF;③ICG6326,2SPF;④Nc6,2SPF;⑤Shengxian xiaohongmao (Shengr),2SPF;⑥ICG12370,3SPF;⑦NcA927,3SPF;⑧Jienong (JN),2SPF;⑨Meiyin 6 (MY6),3SPF;⑩Indored (Indr),3SPF;Zhonghua 6 (ZH6),2SPF;Meiyin 8 (MY8),3SPF;Xuhua 3(XH3),2SPF;Nek16940,2SPF;Meiyin 4(MY4),3SPF;Silihong (SLH),3SPF;Yuhua 10 (YH10),3SPF;Yuhua 15 (YH15),2SPF;073110,2SPF;073103,2SPF. Proteins are extracted from developing cotyledons from cultivars of Heyouduoli (Heyou multiple seed,HYMS),Najingbaipi (White coat,NJWC,2 SPF),Yuhua 10 (YH10),Yuhua 15 (YH15),and from leave,stem,root,fruit shell,fruit needle,small seeds of YH15. All plant material are provided by Key Laboratory of Oil Crop Biology of the Ministry of Agriculture,Oil Crops Research Institute of Chinese Academy of Agricultural Sciences,Wuhan.
1.2 Cloning of DOFH10 genes
Using genomic DNA as template,PCR primers are designed according toGW391728,which is from whole cDNA library of developing seed. Two sets of cloning primers are used:the first set is:Forward primers30 5′-GGGGACCTTCTTTCTTCATT-3′,Reverse primera1469 5′-ATAACCAACTTACTCAAC-3′. The second set of primers are:Forward primers162 5′-AAATTCTG CTACTACAAC-3′,Reverse primera1469 5′-ATAACCA ACTTACTCAAC-3′. Each reaction solution containing ddH2O 36.4 μL,10×PCR Buffer 5 μL,10 μmol/L dNTP 2 μL,10 μmol/L Forward primer 2 μL,10 μmol/L Reverse primer 2 μL,10-50 μmol/L DNA template 2 μL,TaqDNA polymerase 0.6 μL. PCR procedures used for both sets of primers are degrading PCR methods. They are:For first set,95 ℃ 1 min,95 ℃ 40 s,60 ℃ 30 s,(0.6 ℃ degrade per cycle),72 ℃ 40 s for 23 cycles,then 95 ℃ 40 s,46.2 ℃ 30 s,72 ℃ 40 s for 18 cycles,72 ℃ for 5 mins. For second set primer:95 ℃ 1 min,95 ℃ 30 s,51 ℃ 30 s,(0.3 ℃ degrade per cycle),72 ℃ 40 s for 16 cycles,then 95 ℃ 40 s,46.2 ℃ 30 s,72 ℃ 40 s for 18 cycles,72 ℃ for 7 min.
PCR products are purified from 1% agarose gel with purification reagent (New yangzi river) after electrophoresis,and ligated with PMD-18T vector,and transformed intoE.coli. DH5α. The positive clones were picked from LM solid medium with 80 mg/L ampicillin into 1 mL liquid LM medium with 80 mg/L ampicillin shaked over night at 37 ℃,then 2 μL cultured liquid medium was used as template for PCR selection as cloning reaction. The reaction volume could be both 10 or 25 μL. PCR positive clones were sent for sequence analysis by Sangon Biotech Co.,Ltd..
1.3 Bioinformatic analysis of cloned genes
DNA was translated into proteins with Augustus online service[8]. Prediction of protein three dimension structures are performed with I-TASSER ONLINE[9-10]. The analysis of regulation sites,function and structure analysis were conducted with Antheprot 3D viewer,Antheprot 2000,ExPASy PeptideCutter,ExPASy ProSITE.
1.4 PCR examination of transcripts in developing cotyledon from YH15
RNA was extracted from developing cotyledon of YH15 with CTAB method,then reversed transcripted with enzyme set from TaKaRa Company. The cDNA was used as template for PCR examination. Primer used were:Forward primer:S72 5′-TAGGGTTATGGAAAAACAAGGTCAA-3′ ,andS162,Reverse primer used wasa466 5′-CAGGGAAAGAAGGTAGGT-3′ ,control primers designed according to peanutactinDQ873525 sequence,Forward primer wasactinF 5′-CACACACATTCCCCGTTT-3′,andactinR 5′-CACCGTCTCCAGAGTCCA-3′. PCR reaction:the 95 ℃ 2 min;95 ℃ 1 min,50 ℃ 30 s,72 ℃ 1 min for 30 cycles;72 ℃ for 5 mins. PCR products were separated with electrophoresis on 1% agarose gel.
1.5 Western Blot
The tenmer peptides were synthesized by Wuhan Neweast Biotech. Inc. as:NTKFCYYNNY and EMQMRERREC,each was conjugated with KLH (Glutaraldehyde crosslink). After emusification,1 mL was injected into rabbit under skin on back close to neck at two weeks interval,4 times later,10 days from the last inject,collect blood,the blood were sit still for 10 min,then centrifuged at 6 000 r/min for 10 min,supernatant was collected as antisera,add glycerol to 20%,sodium azide to 0.05%,stored at-20 ℃.
The protein were prepared from tissues grounded in extraction Buffer (25 mmol/L Tris-HCl,pH 8.0,1 mmol/L EDTA,150 mmol/L NaCl,0.05% sodium azide) at ratio of 0.1 g/mL. The mixture were centrifuged at 6 000 r/min for 10 mins,the supernatant was collected. Add sample and loading Buffer at 1∶1 ratio,heat at 95 ℃ for 10 min,then load 20 μL sample in each lane on 12% SDS acrylamide gel. Run the gel at 80 V for 4 h,and then blot the gel onto nitrocellulose membrane. The proteins on NC membrane are blotted with antisera and then with rabbit anti IgG labeled with alkaline phosphatase,reacted with BCIP and NBT. The Molecular Marker lane on the same blot was cut away before blotting,and stained with Coomassie brilliant blue G250.
2 Results and Analysis
2.1 DOFH10 similar proteins are highly conserved among 16 cultivars with primers s30-a1469
We had cloned GW391728 similar sequences with two sets of primers from total 16 peanut cultivars. These two sets of primers had one common 3′ primera1469,and 5′primers are Different. The first set of 5′ primer begins from 30 bp (s30),and the second 5′ primer begins from 162 bp (s162),which was at the conserved DOF ZIP domain. With the first set of primers,all PCR sequences from all the cultivars are basically the same,except at four positions:in six cultivars there were three bases CAG more than others at position 179-180,and a base A at position of 1 311 Different from the base G with other sequences,at position 1 249-1 251,lack the TTC in sequences of cultivars of IFD and MY8,a base T was replaced by C in sequence of cultivar MY6 at position of 1 323 (Fig.1,Tab.1),analysis on protein sequences indicated only one Q amino acid residue more in DOFH10 similar proteins from these six cultivars than that from other cultivars (Fig.2). Only DOFH10 protein C terminal from cultivar NcA927 had a Different domain as gvAESDDGTyd ,which together with 33 residues before it had a heat shock hsp20 proteins family profile (BL01031B,PEVEGERGGVMGcSSlKeVKgELLgEErGRErLgaveSDDGTyd). The predicated DOF protein molecular weight fors30-a1469 was 30 ku.
Note:*. TheDOFH10 cloned from the cultivars and with sprimers with multiple seeds per fruit labeled with bold.
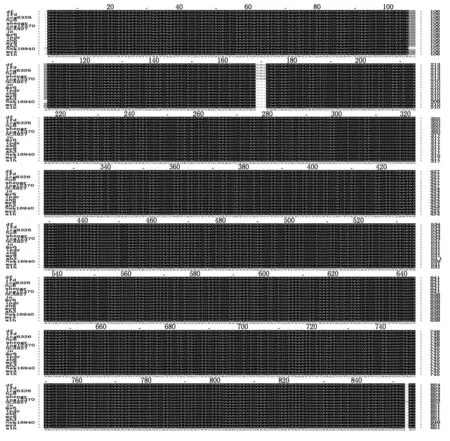
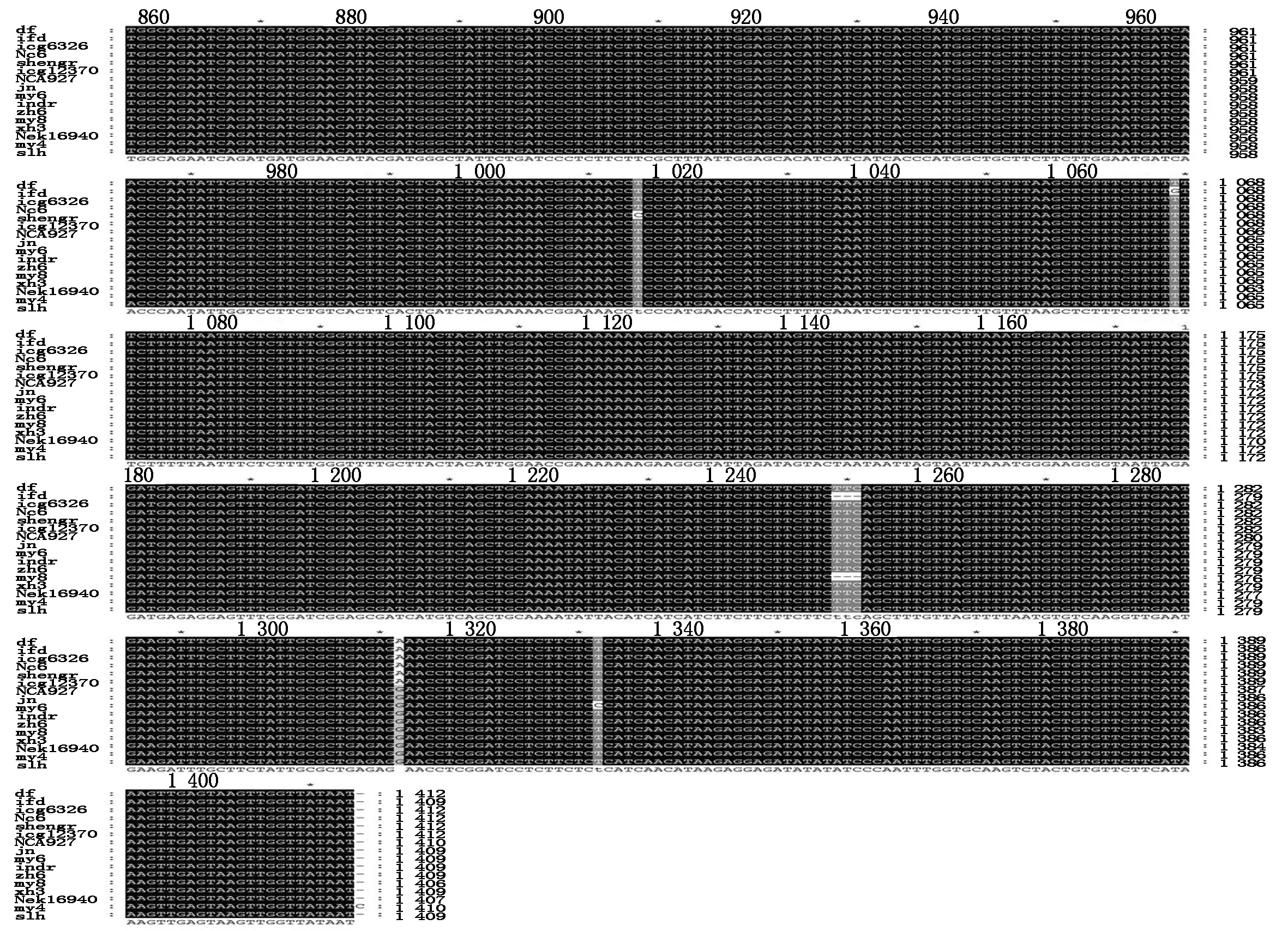
Fig.1 Alignment of sequences of DOFH10 from 16 cultivars of peanut Arachis hypogaea with primers s30-a1469
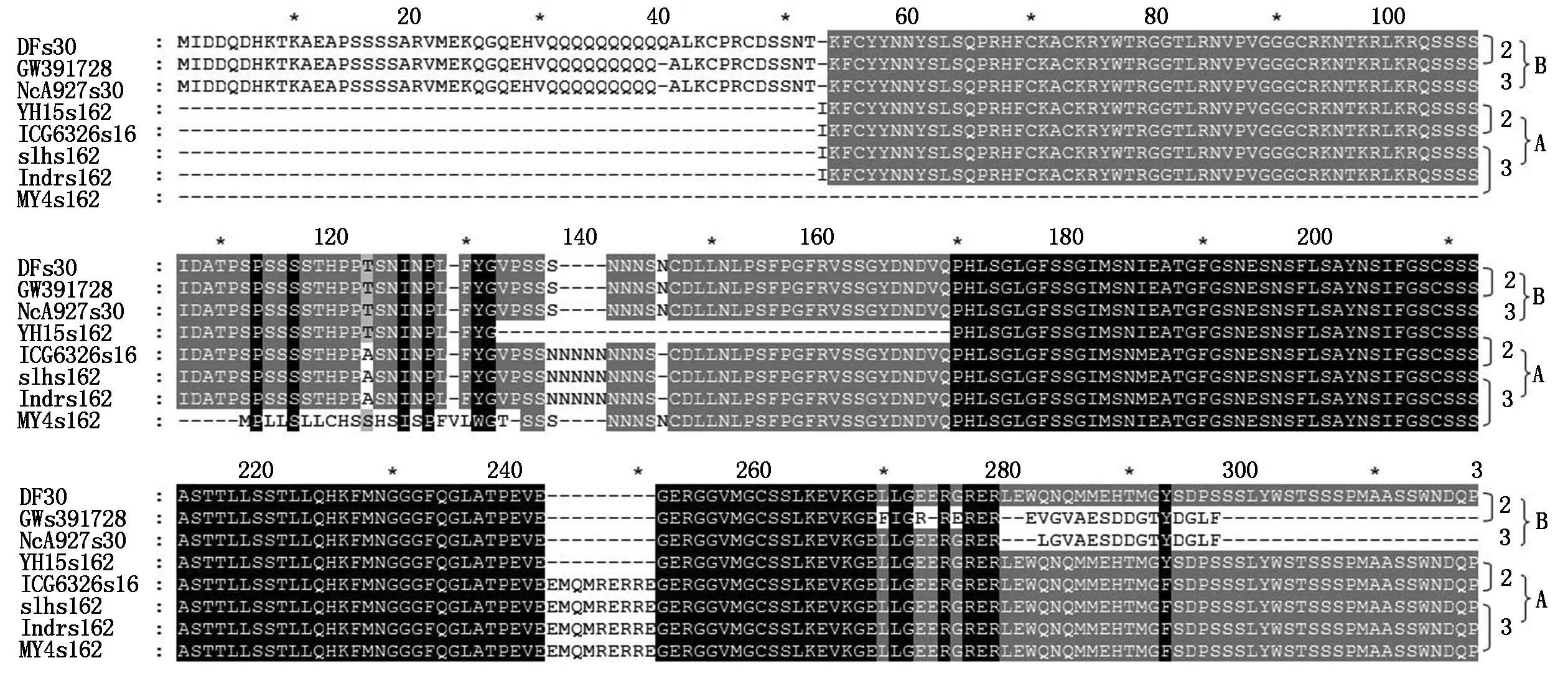
A. Proteins translated from s162-a1469 PCR products with two insert peptides;B. Proteins translated from s30-a1460 PCR products and some PCR products of s162-a1469 lacking two pipetides;2.2 seeds per fruit;3.3 seeds per fruit.
2.2 Two types of PCR products were obtained from different cultivars with primers s162-a1469
Withs162-a1469 primers,two types of sequences were obtained from Different cultivars (Fig.3,Tab.1). One types of sequence was the same as the products withs30-a1469 primers (Type B),the other type had three fragments of insert with 9,15,26 bp respectively (Type A,in Different cultivars might have slightly Different length),and had multiple single bases mutation. The type A DOFH10 were from four cultivars of YH10,SLH,IDR,and MY4 with multiple seeds per fruit and one cultivar of ICG6326 with 2 seeds per fruit,while the B type of DOFH10 were from cultivars with 2 seeds per fruit and two cultivars of ICG12370 and MY8 with multiple seeds per fruit. The protein sequences of type A had less mutated single amino acid residues,but had two insert peptides (Fig.2). The molecular weight of protein of type B without insert was predicated as 24.74 ku,A with insert was 29.86 ku.
The difference at multiple positions throughout the whole molecule between two types of protein sequences were coordinated located on the same set of sequence (Fig.3). The results revealed that the type ADOFH10 gene was not locate on the same chromosome as GW391728,and indicated they were not the same genes. Compared the traits of fruits with 2 and multiple seeds,A type ofDOFH10 existed in four cultivars with 3 seeds per fruit,and one cultivar with 2 seeds per fruit,whereas was lacked in all other cultivars with 2 seeds per fruit and in two cultivars with multiple seeds per fruit (Tab.1). Type A DOFH10 tends to affect the seed number per fruit,possibly function together with other factors.
Blasting results with NCBI tool showed that the type ADOFH10 DNA sequence was identical to theDOF1.4 (XM_016100960.2/NC_029774.2) on chromosome A03 ofArachisduranensiswhile the type BDOFH10 DNA sequence was identical to theDOF1.4 (XM_016334585.2/NC_029787.2) on chromosome B03 ofArachisipaensis. Both types ofDOFH10 DNA sequences were also highly similar to mRNA of a putative clathrin assembly protein on chromosome B01 (XM_016343926.2/) ofArachisipaensisat DOF zinc finger region,type B of DOF 1.4 protein had identical sequence at DOF zinc finger region with that of putative clathrin XP_016199412.2 which encoded by XM_016343926.2/NC_029785.2 at chromosome B01 at LOC107640400 (Fig.4),but not similarity to the same type of clathrins XP_0159358 and XP_0209808 at choromosome A01(Fig 4).

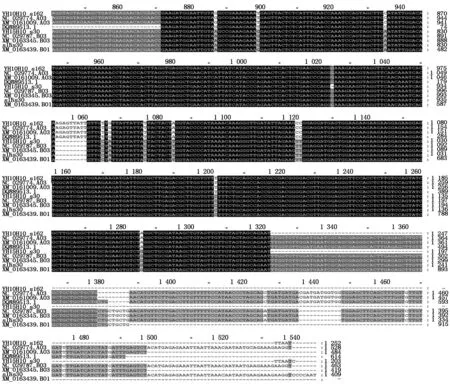
Fig.3 Blasting alignment of DOFH10 DNA sequences with that from genomic library of Arachis duranensis and Arachis ipaensis
s30-a1469 products are almost identical in all cultivars studied(Fig.1),was the same as type B DOFH10 (Fig.3). Thus,it was possible thats30-a1469 products wereDOFH10 genes in BB genome of cultivatedArachishypogaea,and type A PCR product withs162-a1469 primers and some of type B PCR product withs162-a1469 primers were genes located on AA genome of cultivatedArachishypogaea.
Two cultivars with multiple SPF do not had type A DOFH10. But since most of the cultivars with the type A DOFH10 had traits with multiple seeds per fruit,it was possibly related to the seed number per fruit in peanut,which was caused in multiple cell division of zygote.
2.3 DOFH10 is specifically expressed in cotyledon of developing seed
PCR examination with cDNA from developing cotyledon confirmed that boths72-a466,ands162-a466 products exist,but fors162-a466,there were certain smaller fragment exist,probably they were the degraded microRNA (Fig.5).
Conserved ten amino acids peptide NTKFCYYNNY from DOF domain,and conserved insert EMQMRERREC after modified with KLH,was used as antigens immunizing rabbit,antisera were used to conduct Western Blot experiment,to examine the presence of two types of DOF proteins. Dof domains from all 7 DOF proteins expressed in developing seeds are highly similar,the ten peptide NTKFCYYNNY are 100% similar in 5 DOF proteins (Fig.6-7),but these DOF proteins had Different molecular weight (Fig.6-7). After optimized experiments condition,we got a positive band with molecular weight close to the GW391728 (DOFH10) (Fig.7). And among four cultivars examined,the concentration of DOF level was highest in cultivar HYMS with 3 seeds per fruit. The cultivar YH10 which grown in Wuhan had more fruits with two seeds,less fruits with three seeds. Other two cultivars had 2 seeds per fruit (Fig.8-B).
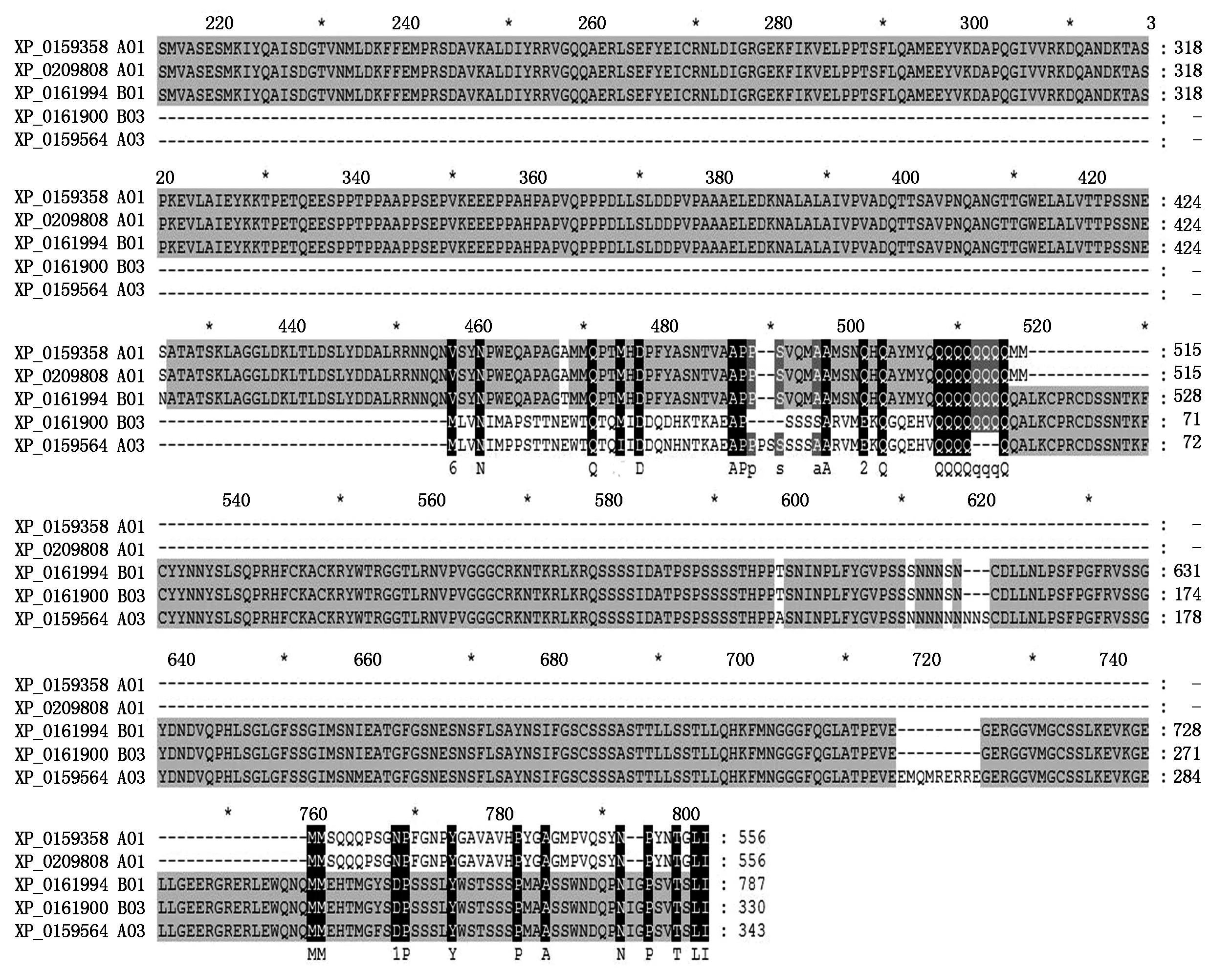
Fig.4 Comparison of putative clathrin assembly proteins to DOF 1.4 from Arachis duranensis and Arachis ipaensis
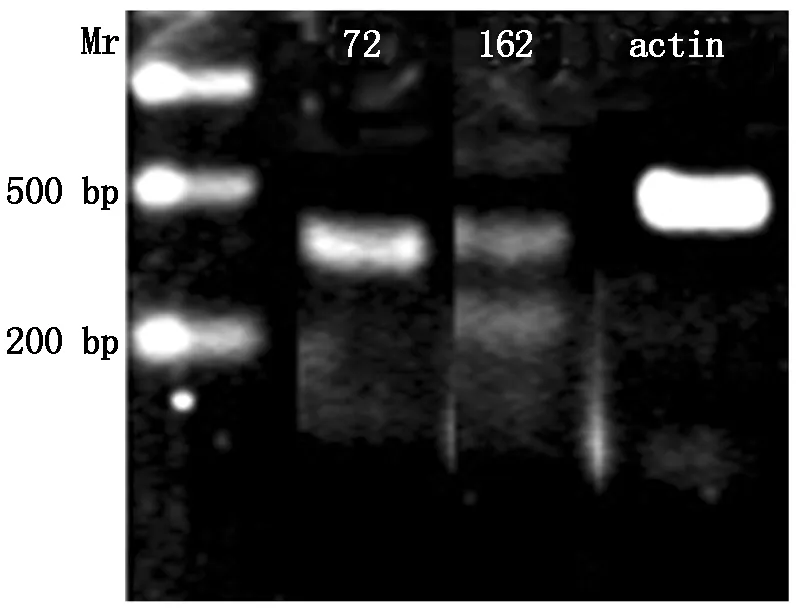
Withs72-a466,only one positive band presents (72),but withs162-a466,addition one distinct band appeared below the predicted band (162).
Fig.5PCRfromtranscriptsofDOFH10geneindevelopingpeanutcotyledonwithprimerss72-a466ands162-a466
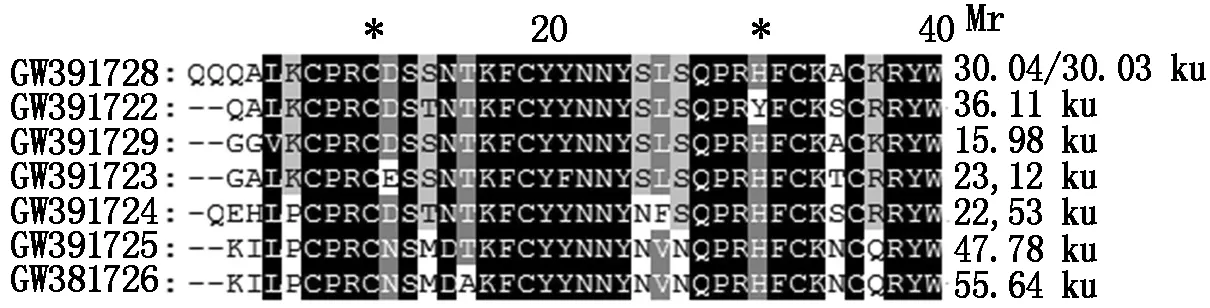
GW391728 is the DOFH10 protein,it′s predicted molecular weight is about 30 ku,different from other DOF proteins expressed in peanut seeds.
Fig.6ComparisonofDOFdomainsbetweensevenDOFproteinsexpressedindevelopingpeanutseed

DOFH10 protein is detected as most obviously band; Other types of DOF proteins were also detected as weak bands; The band is heavier in SLH with multiple seeds per fruit than that in YH15 with 2 seeds per fruit.
Fig.7WesternBlotofDOFproteinsfromdevelopingcotyledonsofYH15andSLHwithantiseratoconservedtenmerNTKFCYYNNY
The antisera to the insert peptide hybridized to the blot does not have positive hybridization band,but with a negative clear band at the position close to DOFH10 among the smears of light background (Fig.8-A). Antibody binds with the insert,if the insert contained protein was removed during the blotting processing,it would be a clear band in the position,it indicated that the protein was there before blotting and they were possibly were dissolved in the solution after binding to the antibody.
When antisera the insert blotted to protein extracted from various tissues, noreaction happens Fig.9-A.When antisera to conserved DOF ten peptide domains blotted to proteins extracted from Different tissues, only proteins from developing cotyledons have positive hybridization bands (Fig.9-B).This demonstrates the expression specificity of DOFH10 in developing cotyledon.

HYMS has multiple seeds per fruit,YH10 occasionally has three seed per fruit. Others are two seeds per fruit.
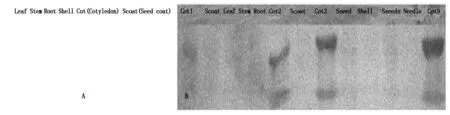
DOFH10 proteins is detected only in proteins from cotyledons (Cot1,2,3,9).
2.4 N-terminal peptide before DOF domain is possible early regulation site for DOF activity
The protein encoded byGW391728 could be divided into four region,N-terminal peptide,DOF domain,connection domain between DOF domain and C-region,and C region. In the predicted three dimension structure of GW391728 protein,DOF domain located at one side of the globoid structure,connected with C-domain and form a hollow channel between these two domain,N-terminal peptide formed helix insert into this channel. In the DOF domain of GW391728 with N-terminal peptide,four Cys (1,2,4,5) forming Zn-C4 structure in active DOF zinc finger were separated by N-terminal peptide in the channel,thus made the whole DOF protein inactivated (Fig.10-A,B). And when cut away the N-terminal peptide,the predicated three dimension structure changed the conformation (Fig.10-F). The four Cys located at closed small space,provide more opportunity for the formation of Cys2-Zn-Cys2structure,became the activate form of the DOF domain. The channel between DOF domain and C-region in this processed form could be a place for target DNA. From Western Blotting result with antibody,we could know that multiple middle processing products with Different molecular weight could exist. We didn′t know if the N-terminal was removed to outside or cut away by other protein or enzymes,but the position change of N-terminal peptides was possible important for DOF activity.
The removal of N-terminal had also changed conformation and enzyme sites in C-region. With N-terminal peptide inserts inside of the structure of protein,the C-region of the GW391728 was in a more compacted and irregular conformation,most enzyme regulation sites were at positions where were hard to access or closed to each other (Fig.10-E). But in the form of structure without N-terminal peptide,C region was more loosed and layered with scattered loops,on some tip of loops located with enzyme regulation sites,some other enzyme regulation sites became more easily accessed (Fig.10-F).
2.5 Regulation sites in GW391728 protein
In N-terminal peptide,positions of 9-12 and 18-20 helix part,exist a potential Casein kinase Ⅱ phosphorylation site and a protein kinase C phosphorylation site respectively,indicates the phosphorylation regulation possibility on N-terminal peptide (Fig.10-C). At
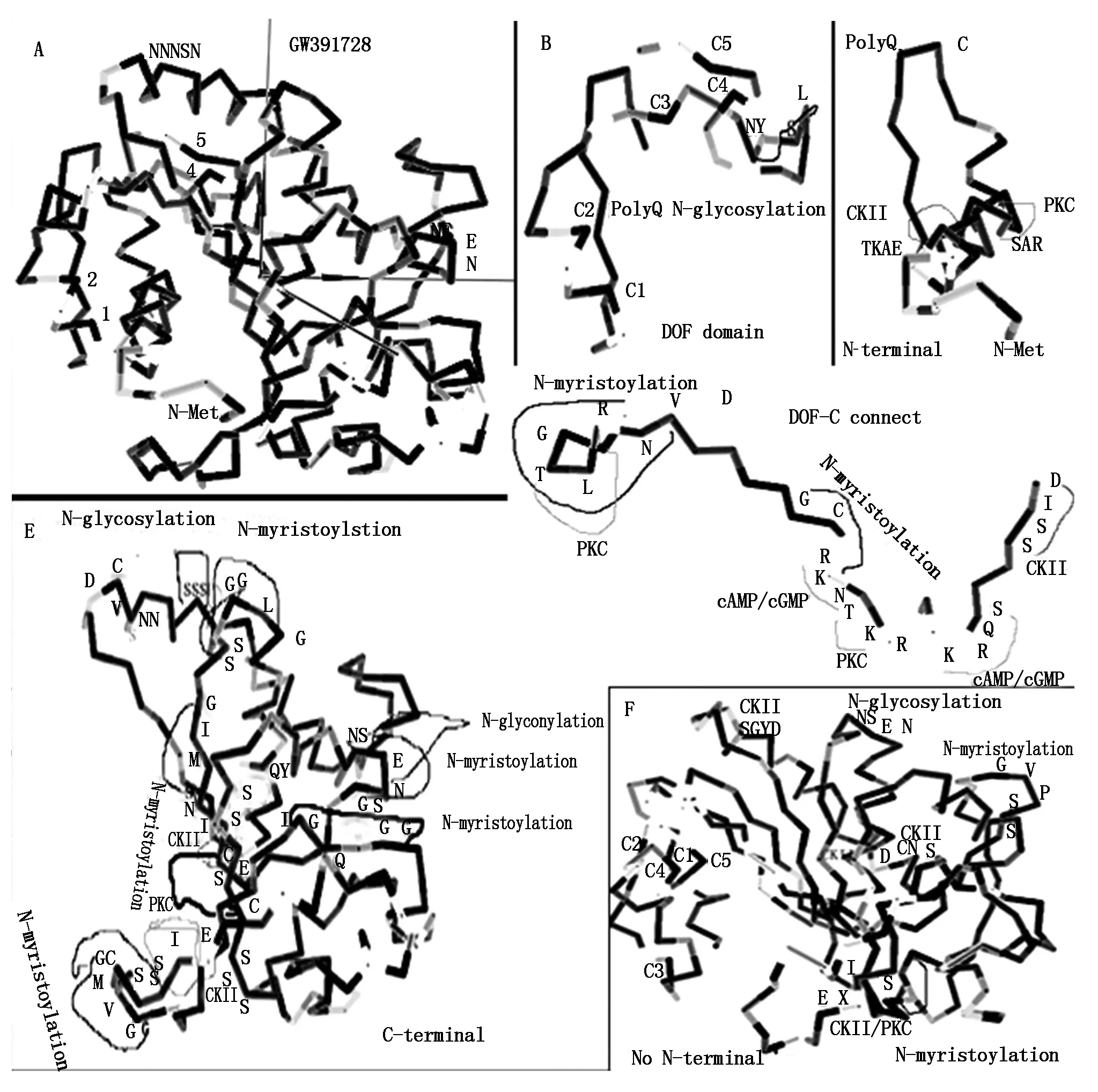
A.Predicted 3D structure of whole length protein of GW391728,four Cys residues in DOF conserved zinc binding domain are marked as 1,2,4,5. N terminal peptides insert into the hollow space between 1,2 and 4,5 Cys,block the formation of C2ZnC2;B.Detailed predicted 3D structure of conserved DOF zinc binding domain. Beside C4,C5 there is a glycosylation site. Glycosylation usually funcation in regulation of conformation of protein structure;C.Detailed predicated 3D structure of N-terminal of GW391728,two kinases sites located on the helical structure part;D.Detailed predicated 3D structure of peptide after DOF conserved zinc domain and before C-region,which connect DNA binding region and other types of regulation region. Nuclear locus site(NLS) locates on C side of this peptide(RK-KRKR). Symmetrically in NLS distribute two cAMP/CGMP dependent kinase sites and two kinases sites around. On N-terminal adjacent to DOF zinc domain,there is also a kinase site;E.Detailed predicated 3D structure of C region,show the two glycosylation sites,multipla myristoylation sites and multiple kinases sites;F.Detailed predicated 3D structure of GW391728 with N terminal removed. Four Cys residue get closed(C1,C2,C4,C5),ready to form C2ZnC2 structure. C region become more loosen,scattered and ordered as layered loops. Regulation sites are more exposured.
Fig.10Predicted3DstructuralandregulationsitesanalysisonGW391728DOFH10protein
2,3,5,11,22,27,46 positions were Asp-N endopeptidase sites,20,45 are Arg-C proteinase sites. Position 27 could be a good site for removal of N-helix. Position 2,3 could be a release site if the N-terminal attached to other structures in the cell.
In DOF domain,between C3 and C4,there was a N-glycosylation site at 58-62,which was a possible site for regulation on formation of Zinc finger (Fig.10-B).
On the peptide connecting DOF domain and C-region,there were five phosphorylation sites,they located on two sides of the connection peptide (Fig.10-D). Four phosphorylation sites located in the connection domain as two cAMP/cGMP dependant phosphorylation enzyme,CKII,and PKC. One was PKC site close to DOF domain. On the domain close to the C domain,a nucleus localization locus (NLS) site (RK-KR) motif exist,beside this was NLS motif,which was transportation signal into nucleus on both sides overlayed with cAMP/cGMP dependant phosphorylation sites and N-myristoylation sites. This indicated that transportation of this protein needs the energy provided by cAMP/cGMP and active phosphory bond. N-myristoylation provided the lipiphilic affinity for binding to the membrane,intermediate the transportation over membrane.
C region had more phosphorylation and N-myristoylation sites,two N-glycosylation sites. The accessibilities were changed by removal of the N-terminal (Fig.10-E,F).
2.6 Two insertion in C region caused more compacted conformation and three more regulation sites
Compared two types of protein structures encoded bys162-1469 PCR products,the conformation of the type A with two inserts more compacted which changed the accessibility of most of the original regulation sites (Fig.11-A,B),but present three new regulation sites,all in accessible position. Among three new regulation sites,one was N-myristoylation sites at 79-84,one N-glycosylation sites at 90-93,they both located on the first insert peptide (Fig.11). The third one was phosphorylation site by CKII at 108-111 (Fig.11). The loosed structure of type B DOFH10 protein had pocket which could bind DNA motif,but the type A did not have enough space. This enzyme accession difference might cause the difference of degradation speed of two types of DOF protein. And this explain that heaver DOF protein band on Western Blot in SLH which had type ADOFH10 gene than that in YH15 which had type BDOFH10 gene.
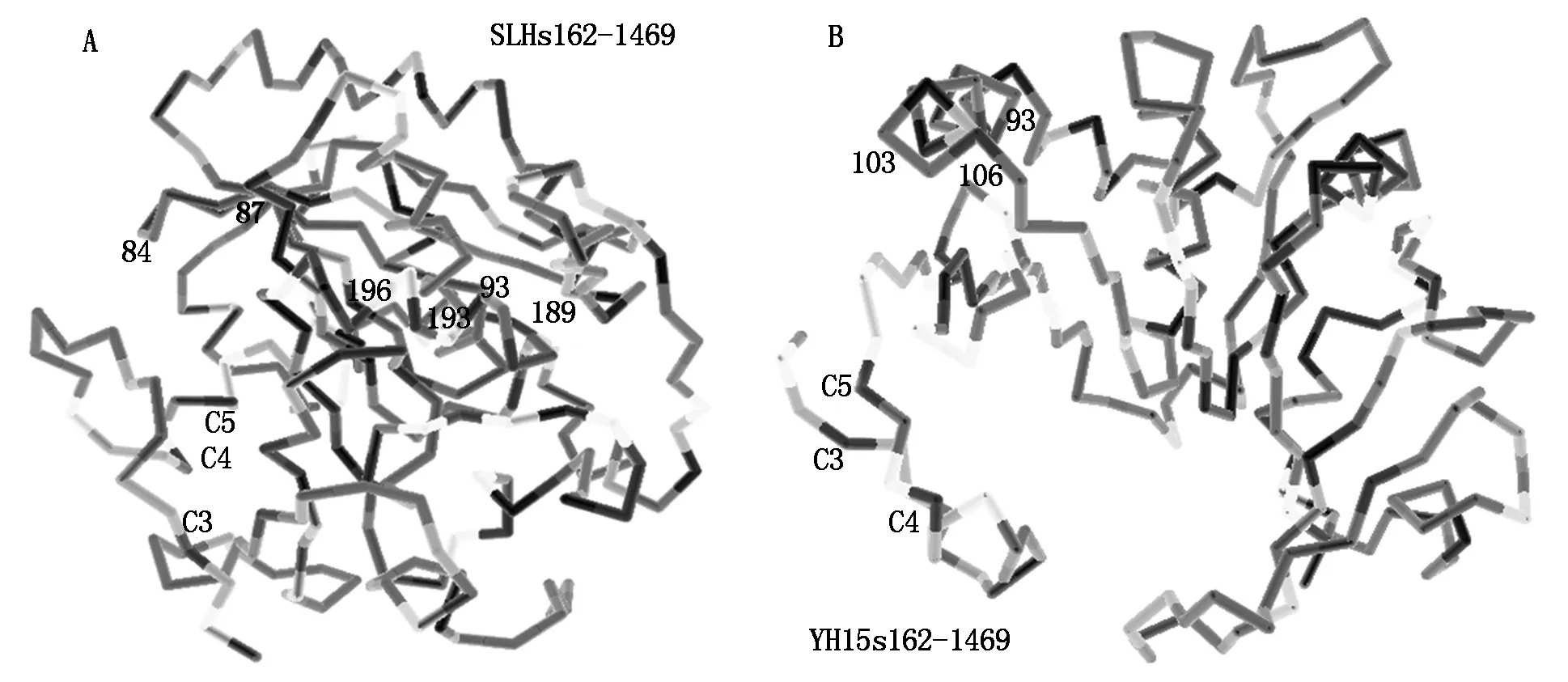
A.Predicated 3D structure of A type DOFH10 protein translated from PCR products with primerss162-a1469 from YH15 genome DNA with 2 seeds per fruit. The protein lack two small peptides,is more loosen,scattered and ordered;B.Predicated 3D structure of B type DOFH10 protein translated from PCR products with primerss162-a1469 from SLH genome DNA with 3 seeds per fruit. The protein structure is more compacted.
Fig.11Comparisonof3DstructuresbetweentypeAandBDOFH10proteins
3 Discussion
3.1 DOFH10 genes located on different chromosomes in cultivated peanut and different at N-terminal and insert
16 protein sequences encoded by PCR product with primer pair ofs30-a1469 from 16 cultivars were highly conserved,highly similar to that of GW391728,which was a cDNA sequence expressed in developing peanut seed of cultivar 06-3103. This indicated that DOFH10 protein encoded by PCR product with primer pair ofs30-a1469 was possible an essential functional transcription factor. Blasting result showed thats30-a1469 PCR products belong toDOF1.4 gene (XM_016334585.2/ NC_029787.2) located on B03 chromosome ofArachisipaensis. But with primers162-a1469,two types (A and B) of PCR products present in Different cultivars. Type B was identical toGW391728,as well as XM_016334585.2. This confirmed thatGW391728 was most similar toGW989805 fromA.ipaensiswhich was source of BB genome in peanut. Type A had two insert in the sequence. It was identical to theDOF1.4 (XM_016100960.2/NC_029774.2) on chromosome A03 ofArachisduranensis,which was source of AA genome in peanut[2]. Conserved type BDOFH10 was possibly located on BB genome of cultivated peanut. On AA genome of cultivated peanut,possibly two types ofDOFH10 were existed according Different cultivar. Two types ofDOFH10 genes duplicated from two genomes possibly evolved new functions and thus provided new variable properties for the peanut plant[11].
3.2 DOFH10 genes specifically expressed in developing cotyledons
In Western Blot assay with antisera to conserved DOFH10 domain peptides,only positive protein band appeared in developing cotyledons of peanut seed. This demonstrates that DOFH10 protein was specifically expressed in developing cotyledons of peanut. Transcripts analyzed with primerss72-a466 ands162-a466 by PCR method on cDNA from developing cotyledon indicate that withs72-a466,only one distinct band appeared,but withs162-a466,two distinct bands appeared,the difference of the Mrs between these two distinct bands was about 100 bp,larger than the difference between the products of type A and B,indicates the smaller one could be a microRNA or other gene fragment.
3.3 Type A DOFH10 protein is possibly related to seed number per fruit indirectly
The more concentrated DOF H10 protein band appeared in cultivar with multiple seeds per fruit,may has following reason:Less protein be degraded,more protein synthesized. Since more cultivars with multiple seeds per fruit is associated with appearance of insert fragment,which cause compacted three dimension structure,and thus protecting the protein from degradation. The type A of DOFH10 proteins may function in regulation of level of the essential DOFH10 proteins by acting as protective competition for protease or inhibitor,or cofactor for its activation and function. The type B DOFH10 was highly conserved through all the cultivars,indicated that it might be an essential DOF protein. 3D structural analysis revealed that looser structure of type B DOFH10 may permit easier access by degrading enzymes. It might be a reason for higher concentration of DOFH10 protein in cultivar of SLH with multiple seeds per fruit,which had type A DOFH10 with more compacted protein conformation,and thus less degradation opportunity.
3.4 Transportation of DOFH10 to nucleus is possible via membrane traffic pathway
The DOFH10 had total 12 predicted N-myristoylation sites,scattered on the whole protein,and most of them closely localized adjacent to kinase sites. N-myristoylation on protein can help the protein binding to lipid monolayer,which is part of the membrane[12],and make the proteins joining the membrane traffic flow from synthesis site such as ER,to target membrane such as mitochondrial[13]. The identical fragment of the conserved DOF domain to that on clathrin assembly protein had also suggested that the DOFH10 was possibly transported via membrane system associated to clathrin from synthetic site to nucleus. The process of transportation was also the process of regulation,for the sites of N-myristoylation were often colocalized with kinase sites and on the surface of the globoid.
Clustered localization of N-myristoylation sites and two cAMP/cGMP dependant phosphorylation sites,one PKC and one CKII sites on NLS of connection domain between DOF domain and C region reveals that the mechanism of transportation into nucleus possibly involved the G-adaptor,GTP,and N-myristoylation regulation. ADP-ribosylation factors (ARFs),small monomeric GTPases which involved in cargo selection and vesicular budding in membrane trafficking process is myristoylated,and the myristoylation is related to regulation of guanine nucleotide exchange and stable membrane association[14]. It′s possible that for nucleus importation,the association of myristoylation to cAMP/cGMP dependant phosphorylation is a integrate structure for the process.
Two locations encoding three putative clathrin assembly proteins were found in chromosome A01 ofArachisduranensisand chromosome B01 ofArachisipaensis. All of them belong to At5g35200 type of protein,which is ANTH (AP180 N-Terminal Homology) family protein[15]. Two transcripts were located on A01,and one was located on B01. Only the transcript located on B01 had identical RNA sequence to DOF1.4 of type B on DOF zinc finger region,as well as protein sequence (XP_016199412.2). As predominant coat particles,clathrin involves in endocytosis and post-Golgi trafficking in coated vesicle transportation in cell. The coated vesicle transport process include sorting of cargo,coat assembly,budding and fissing under tight and complex regulation,with many proteins involved. ANTH family protein is in the class of ENTH/ANTH/VHS domain-containing proteins,which link the subdomain,clathrin and adaptor proteins in the early steps of clathrin coated vesicle formation[15]. TOLs (TARGET OF RAPAMYCIN LIKEs) is a group of proteins carries out the function of recognition of cargo intraluminal vesicle in generation and the formation of multivesiculate bodies (MVB),which contain small internal vesicles as prevacuole compartment (PVC) or lysomomal vesicle (LV)[15-16]. At5g35200 is coexpressed with TOL1[15],indicates that it is possible link of endocytosis lysosomal degradation or MVB formation[15]. Thus,the protein XP_016199412.2 which belongs to At5g35200 could has similar function as ANTH types,its recognition site could be the identical sequence with B type of DOFH10,and lead the B type of DOFH10 to degradation or to the MVB for storage. The much less protein from YH15 which had only B type of DOFH10 than that from SLH which had A type of DOFH10 in Western Blot blotted with antisera to conserved tenmer peptide in DOF zinc finger region was consistent with this. In the mean time,the possible formation of hetero complex between type A and B DOFH10 to form protected type B DOFH10 could be the reason for higher content of DOFH10 in SLH. The less compacted conformation of B type also gave more chance to degrade.
The prediction on the membrane traffic pathway of DOFH10 in above according to its protein structure was also consistent with the fact of identical sequence on both DOFH10 and XP_016199412.2,which was a ANTH type of putative clathrin assembly protein,function in vesicle trafficking. The common fragment of B type with XP_016199412.2 is on conserved DNA binding region. This suggested that the specificity of transportation of DOFH10 via the vesicle transport. It was suggested that the lateral diffusion of plasma proteins may depend on the identity and protein sequence of each individual cargo[17-18],and also depend on its ability to interact with other plasma components and on their aggregation with the plasma[19]. The type A of DOFH10 tends more associated to the trait of multiple seeds per fruit,it is possibly function in process of mitosis in early stage of fruit development. It was reported that the clathrin dependent vesicle pathway is involved in cell division[20-22]. The connections between type A DOFH10,clathrin-related protein,type B DOFH10,lysozyme and cell division are possibly through the interactions between them. From TGN through clathrin,proteins can be transported to lysome and cell plate[23],it may exist the targets switch of the clathrin dependant vesicle transport. Rencent review on membrane-bound transcription factors described routes of both PM and ER membrane-bound TFs can be both transported into nucleus[24]. The detailed routes and mechanism for DOFH10 need to be further studied.
3.5 N-terminal peptide may be important regulation domain for formation of functional DOF zinc finger
Comparison analysis on predicted 3D structure of GW391728 DOF protein with and without N-terminal peptide showed that the presence of N-terminal peptide separate 4 Cys residues apart far away,without hinder of N-terminal peptide,4 Cys residues in DOF domain could naturally approach together to provide for the suitable environment to formation of zinc finger structure. N-terminal could be removed by enzyme or drove out and reshaped by force outside of the globe structure. Among multiple endopeptidase sites at N-terminal peptides,Asp-N endopeptidase site on residue 27 could move away N-terminal peptide and give a functional integrate DOF domain,thus was mostly possible the activating site for enzyme process. And a glycosylation site on N-terminal could also function on formation of active DOF zinc finger.
In summary,we have conclusion as below:
In most cultivars ofArachishypogaeaL. with multiple seeds per fruit,there were two types of DOF1.4 B and A,they located on BB genome and AA genome respectively,they were highly similar but Different at multiple place. Type B DOF 1.4 was present in all cultivars. Types of A and B DOFH10 were possible function in early fruit development,and type of B DOF H10 was possibly a necessary factor in early development of peanut fruit.
DOFH10 was possibly transported via clathrin related vesicle pathway,and its amount was possibly regulated.
Compliance and ethics: The authors declare that they have no conflict of interest.
Acknowledgements:This work was supported by the Natural Science Foundation of Hubei Province (Bzy05011) and South-Central University for Nationalities. Undergraduate students Zhenling Huang and Hui Suo clonged five and eight DOF H10 sequences with primerss162-a1469,graduate student Qiuting Luo studied the suitable PCR method with primerss162-a1469. All other experiments conducted by Haiyan Yan and performed in Life Sciences College,South-Central University for Nationalities.
