Molecular methods for detection of pathogenic viruses of respiratory tract-A review
2018-06-29MdShahidulKabir
Md. Shahidul Kabir
Department of Microbiology, Institutional Quality Assurance Cell, Stamford University, Bangladesh
1. Introduction
With the development of scientific knowledge and technical skills, scientists are now able to detect a number of pathogenic viruses using variety of methods e.g. serology, culture and molecular methods. Serology has been used as the major diagnostic approach for the detection of pathogenic viruses associated with the respiratory system. The most common serological tests used for such viral pathogens are the enzyme immunoassay,haemagglutination inhibition, and complement fixation tests.Efficiency of cell culture was improved in the early 1990s with the development of shell vial culture with addition of monoclonal antibodies. This approach reduced the cell culture time from 8-10 days to 1-2 days. Direct fluorescent antibody staining is another technique commonly used for rapid diagnosis of viral pathogens present in nasopharyngeal samples. Enzyme immunoassays are still used for virus detection but appear to be less sensitive than molecular methods. Consequently, molecular techniques such as,nucleic acid sequence based amplification (NASBA), PCR and loopmediated isothermal amplification (LAMP) were developed as more sensitive and alternative methods for diagnosis of such pathogens of the respiratory system. A number of molecular techniques that have been developed and utilized during last two decades for the detection of pathogenic viruses of the respiratory system are summarized in this article.
2. Molecular methods used for detection of virus
2.1. PCR and RT-PCR
PCR, a popular molecular method, is frequently used for detection of any nucleotide sequence of viral genome[1]. Appropriate primer design is a prerequisite for successful amplification and detection of a specific or multiple targets of nucleic acid sequences. Primers are generally designed targeting conserved sequences of viruses e.g. replication genes and matrix protein coding genes for detection of multiple members falling under same species or family. Primers designed for detection of the hexone gene of adenovirus were used to detect 18 genotypes in urine samples[2]. Multiple sets of primers designed for different viruses were used for the detection of several viruses together in multiplex PCR assay. It is therefore, necessary to optimize the thermo cycling conditions, for example, annealing temperature and extension time suitable for all primers to be used in such PCR amplification. PCR amplified DNA can be detected after separating them on agarose gel. Appropriate size of DNA band can be identified after hybridizing with sequence specific probes[3].Alternatively, amplified DNA can be sequenced for confirmation.
Both one-step and two-step Reverse Transcriptase (RT) PCR methods are used for detecting RNA viruses in clinical samples.Complementary DNA is initially prepared from the viral RNA which is subsequently used as template for further detection of target sequences by PCR amplification in two-step RT-PCR. Viruses can be detected directly in the extracted RNA by one-step RT-PCR amplification. This approach is more sensitive than two-step RTPCR because it includes relatively larger amount of RNA template for transcription of cDNA and amplification of sequence by PCR.Simultaneous reverse transcription of RNA and amplification in the same reaction in one-step RT-PCR excludes the possibility of pipetting errors and carryover contaminations. One-step RT-PCR was used to amplify the nucleoprotein, haemagglutinin and matrix genes of influenza A and B viruses[4]. Multiplex RT-PCR can be designed after optimizing amplification conditions of different primers specified for target viruses[5,6]. Human respiratory syncytial virus and human metapneumovirus were detected by a similar multiplex RT-PCR assay from clinical specimens[7].
2.2. Real-time PCR
Quantification of the viral load is a challenge for conventional methods while it can be addressed by molecular approaches[8]. Realtime quantitative PCR was used for rapid detection and quantitation of 13 common respiratory tract viruses using fluorescence resonance energy transfer hybridisation probes[9]. Such an approach was reported to be useful for determining the load and shedding of viruses in different stages of infection and for monitoring therapy[10]. Special clinical management policies and infection control procedures should be followed for patients, especially young children, showing high viral load and shedding for extended periods of time[11]. It was also possible to determine the concentrations of influenza A subtypes of H3N2 virus using multiplex primers for both types in a real-time PCR[12]. A new approach, high-speed dropletreal time PCR method, was used for detecting bovine respiratory syncytial virus in less than 10 minutes[13].
2.3. LAMP and reverse transcription loop-mediated isothermal amplification (RT-LAMP)
LAMP is an isothermal amplification method used for detecting nucleotide sequence. Two pairs of primers are used in both inner and outer sides flanking the target sequence. Specific DNA sequence is amplified through strand-displacement and synthesis of Bst DNA polymerase[14,15]. Bst DNA polymerase is the large fragment of DNA polymerase derived from Bacillus stearothermophilus which contains 5’→3’ polymerase activity but no 5’→3’ exonuclease activity[16].As a result, the LAMP reaction produces a blend of stem-loop DNAs of different sizes comprising of multiple loops. A positive amplification can be easily monitored either by gel electrophoresis or by measuring the concentration of magnesium pyrophosphate[17].RNA samples can also be amplified through such amplification after adding an initial reverse transcription step and additional pair of primers called RT-LAMP[18,19]. LAMP appeared to be 100 times more than standard PCR while detecting tomato yellow leaf curl virus DNA[20]. Sensitivity of RT-LAMP also appeared to be 10 times more than RT-PCR while detecting Taura syndrome virus RNA[21].
2.4. NASBA
In this technique, an RNA sequence is amplified to multiple copies using DNA-dependent RNA polymerase[22] (Figure 1). Initially a DNA primer binds to the T7 promoter which is subsequently extended by reverse transcriptase. RNA from the cDNA/RNA hybrid is degraded by RNase H and double-stranded DNA is synthesized by reverse transcriptase using a second primer. T7 polymerase synthesizes multiple copies of RNA from this DNA template.Amplified RNA can be detected by either electrochemiluminiscence or RT-PCR. This method was applied for detection of several viruses e.g. West Nile and St. Louis encephalitis virus[23].
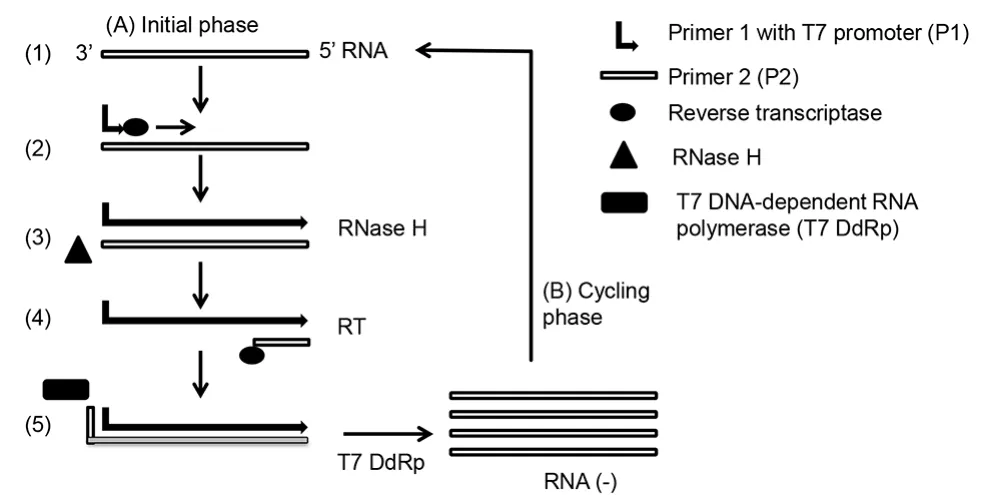
Figure 1. NASBA.
2.5. Nucleic acid hybridization
This technique is used to detect specific sequence of nucleic acid present within a viral genome. A short labeled nucleic acid probe of 20-30 bases is hybridized to a complementary sequence present in viral genome. This short probe can be either DNA or RNA (riboprobe). Signals from the hybridized probes can be amplified through chemiluminiscence or a hybrid-capture assay for quantitation of viruses. This nucleic acid hybridisation technique was applied on plasma sample to monitor human immunodeficiency virus type 1 (HIV-1) from patients enrolled in clinical trials for antiretroviral and immune-based therapies[25].
2.6. Microarray analysis of virus sequences
Nucleic acids hybridization technique is used in microarray analysis for the detection and characterization of genomic sequences.A range of single-stranded DNA oligonucleotide probes are spotted on a small glass slide or similar surfaces coated with quartz or membrane for hybridizing and revealing complementary sequence of nucleic acid. Specific sequences of genomic DNA or RNA are amplified using appropriate molecular techniques and labeled with fluorescent markers. Labeled sequences are allowed to hybridize with complementary sequences of probes placed in the array.Subsequently, fluorescence is measured to determine the presence of particular sequence. Microarrays have already been applied for the detection and typing of viral pathogens from the respiratory system[26-28]. A commercial microarray device CLART®PneumoVir(Genomica, Coslada, Madrid, Spain) was used for the detection of several pathogenic viruses of the respiratory tract. It is also considered to be cheaper than multiplexed conventional PCR-based assays[29]. There is an enormous prospect for adopting microarray platforms for cheaper detection of multiple infectious agents such as bacteria, virus and fungi in clinical samples. Microarray analysis coupled with sequence independent amplification was applied for detection of genotypes of noroviruses[30].
2.7. Multiplex PCR and emerging technologies
The application of molecular techniques is becoming widespread due to their efficiency e.g. rapidity and coverage for detecting different pathogens, compared to other conventional techniques.It is also more convenient to set up multiplex assays following a multiplex PCR amplification of the metagenome using modern techniques e.g. ResPlexTMtechnology and InfinityTMsystem which are described in the following sections.
2.7.1. Micro-bead suspension array (EraGen Biosciences)multiplex PCR
Multiple targets are amplified using multiplex PCR in this technique. Amplified products are subsequently hybridized to targetspecific capture probes which are covalently bound to color-coded beads. Hybridization of colored beads is detected using a dual-laser detection device for determining the presence of sequences related to pathogenic viruses. This technology was successfully applied for detecting common pathogenic viruses of the respiratory tract[31] and typing human papillomaviruses (HPV)[32]. This technology offers detection of 100 different types of sequences in the same reaction.
2.7.2. ResPlexTMtechnology (Qiagen, UK)
This technology involves multiplex PCR amplification followed by bead-based detection such as Luminex xMAP. This technology is suitable for detecting more than 15 pathogens present in a single test. ResPlexTMⅠ and ResPlexTMⅡ are developed for detecting bacteria and viruses associated with respiratory system, respectively.The Res PlexTMsystem was applied for detecting 21 pathogenic bacteria and viruses from patients having acute respiratory infections[33]. Res PlexTMtest was found to have a sensitivity of 84%-100% compared to direct fluorescent antibody test for detection of respiratory syncytial virus (RSV), parainfluenza 1-3 and influenza A and B. However, Res PlexTMwas found to be less than 10% sensitive while compared to direct fluorescent antibody for detection of adenovirus.
2.7.3. InfinityTMsystem
In the InfinityTMsystem (AutoGenomics) target sequences are amplified initially using multiplex PCR for subsequent detection using automated microarray hybridization in InfinityTManalyser.Unincorporated nucleotides are removed through enzymatic digestion and amplicons are labeled using fluorescent nucleotides.This microarray approach was compared with quantitative realtime PCR assay and yielded concordant result in a previous study for detecting 23 common viruses in children ≤≤3 years of age[34].Microarray analysis was found to be ≥≥90% sensitive for all viruses other than adenoviruses and coronavirus NL63. Although throughput was higher for microarray analysis for detection of >15 targets, a singleplex real-time PCR was reported to be superior over the other for detecting an individual virus[34].
2.7.4. Jaguar system
The Jaguar system (BD Diagnostic) is an automated system for nucleic acid extraction and real-time PCR detection. This system is used for identification of multiple pathogens of respiratory system. Nucleic acid extraction occurs in individual cartridges.The amplification cartridge contains 12 reactions of 4 μL each comprising multiplexing capabilities of 2-6 targets. While compared with tissue culture detection, sensitivities of this system varied for the detection of influenza A (100%), influenza B (90%) and RSV(100%)[35].
2.7.5. Film ArrayTMtechnology
The Film ArrayTMsystem (Idaho Technologies) is an integrated automated system used for extraction of nucleic acid and amplification by nested PCR. Numerous viral and bacterial pathogens are identified through data analysis in a single-use pouch. A PCR reaction is performed in two steps; the first step is a multiplex PCR containing primers of a variety of the target sequences and the second step targets a specific pathogen. This technique has the potential for detecting >15 target sequences within an hour from a sample[36].
2.7.6. Scalable target analysis routine technology
Scalable target analysis routine technology (PrimeraDx) is a blend of PCR with a capillary electrophoresis system. Different targets are identified according to the sizes of the amplified products. It is possible to detect more than 20 pathogens in a single reaction.This system is still under investigation for further improvement in detection and quantification of pathogenic viruses[37].
2.7.7. PLEX-IDTMtechnology
In the PLEX-IDTMsystem (Abbott Molecular), target sequences are initially amplified by PCR using primers designed on the highly conserved sequences of pathogenic microorganisms that flank sequences variable regions. Nucleotide base compositions of amplified PCR products are analyzed using electronspray ionization mass spectrometry. Sequence information is matched with a known sequence database to confirm the presence of any organism. PLEXIDTMsystem showed 88% sensitivity for RSV and 95% sensitivity for influenza A and B when compared with culture, PCR and antigen based detection methods. However, the PLEX-IDTMsystem was shown to be 100% sensitive for detection of adenovirus,parain fluenzavirus and coronavirus[38].
Pro-Flu-1 (Pro-Flu+; Prodesse Inc.) is a FDA approved multiplex real-time RT-PCR system. This system is used for the detection of RSV, influenza A and influenza B viruses. According to two previous studies, sensitivity and specificity of this assay varied for the detection of influenza A (93%-100%) and influenza B (98%-99%)while compared to immuno fluorescence and culture assay[39,40].
2.8. Non-PCR based isothermal amplification of DNA/RNA
Isothermal amplification of nucleic acids in molecular biology has enabled scientists to amplify nucleic acids without using a thermocycler. Isothermal amplification of nucleic acid is relatively easy, cheap and can be done in remote areas where there is no access to a thermocycler. Amplification of nucleic acids is essential for the detection and characterization of genomic sequences in any research related to diagnosis and treatment[41]. Although PCR-based amplification of target sequences is widespread, several non-PCR based amplification techniques have been introduced in the past time.This new isothermal amplification technique has been developed by providing necessary proteins, dNTPs, primers and templates for in vitro synthesis of DNA/RNA.
2.8.1. Transcription mediated amplification
Transcription mediated amplification method amplifies the number of template RNA which is a very similar approach to NASBA[42,43].Three enzymes are used in this technique including reverse transcriptase, DNA dependent RNA polymerase and RNase H for reverse transcription and synthesis of multiple copies of RNA from the starting template RNA. Transcription mediated amplification can amplify 1 billion copy of the RNA from one RNA copy in 1.5 h at 41 ℃. Amplified RNA can be visualized after gel electrophoresis or detected using fluorescence probes in real-time PCR amplification or colorimetric assays[44].
2.8.2. Signal mediated amplification of RNA technology(SMART)
SMART technology (Figure 2) was developed by Cytocell Ltd.,Banbury, UK for diagnosing pathogens in clinical samples[45].SMART technology is also known as CytAMP (British Bio Cell International, Cardiff, UK). Performance of CytAMP for the detection of methicillin resistant Staphylococcus aureus was shown to be comparable to those of PCR in a previous study[46,47]. It was also found that many copies of an RNA signal were produced by SMART technology at 41 ℃ from nucleic acid templates. A threeway junction is formed after hybridization between the target and two probes. One of the probes contains RNA signal sequence and the other contains T7 promoter sequence[48]. Double stranded T7 RNA polymerase promoter is synthesized by Bst DNA polymerase by extending the short probe. Multiple copies of RNA are subsequently synthesized using T7 RNA polymerase. Synthesized RNA then binds to the second probe and is extended by Bst DNA polymerase generating double stranded promoter to facilitate transcription of RNA. Amplified RNA copies are detected by enzyme linked oligosorbent assay or real-time PCR assay[49].
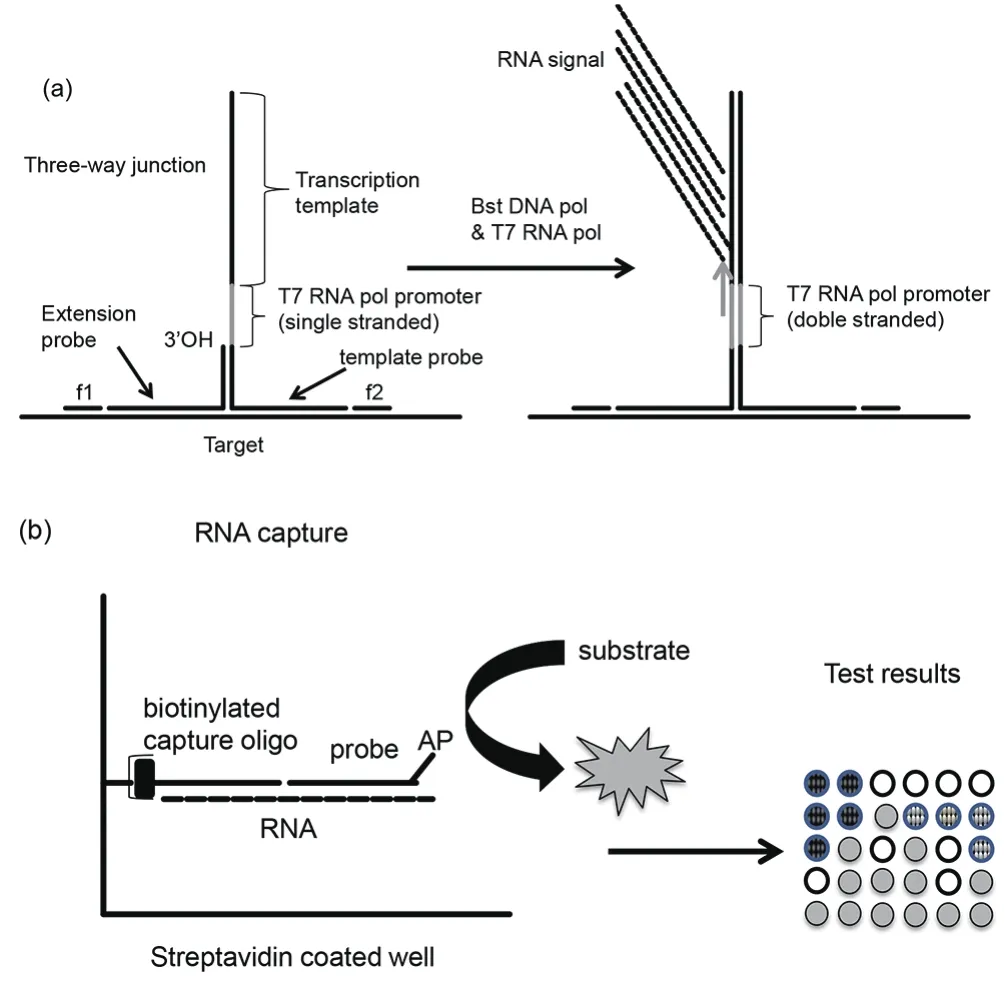
Figure 2. SMART.
2.8.3. Strand displacement amplification (SDA)
This method uses an endonuclease to produce nick in the DNA strands. A second enzyme DNA polymerase (exonuclease deficient)extends 3’-ends at the nick and displaces the downstream DNA strand. After an initial heat denaturation, four primers (B1, B2, S1 and S2) bind to the both sides of target sequence to be amplified.The four primers are extended simultaneously by exonuclease deficient DNA polymerase. Extension of B1 displaces the S1 and B2 displaces S2 primers. SDA primers contain restriction sites and new DNA strands flanked by the nickable sites which are synthesized through their extension and strand displacement activity. The newly synthesized DNA is nicked again, the 3’-end is extended displacing a single stranded copy and regenerating double stranded DNA with the available primers. Each single stranded DNA can be amplified 1010-fold using this technology. SDA amplicons can also be used for real time assay[51]. This technology was used for detection of pathogenic microorganisms e.g. Chlamydia trachomatis and Neisseria gonorrhoeae using the Becton Dickinson Probe Tec ET System (BD Biosciences, Sparks, Md.)[52]. Such amplification can also be used on RNA template but that will need additional amount of reverse transcriptase in the reaction mix[51].
从用钢量方面来讲,用钢量受较多因素影响,如边界条件、建筑平面尺寸等,但一般而言,对于建筑长宽比较大的建筑,类似于单向板受力体系,采用于空间桁架结构较为经济,而建筑长宽比较小的建筑,类似于双向板采,用多向传力的均匀的网架结构更为经济,本工程阀厅长宽比接近1,显然更适合采用网架结构体系。
2.9. Amp-PCR: Combining phi29 amplification with RTPCR
Performance of real-time PCR for detection of any pathogen depends on several factors, for example, binding of the primers to desired sequences, variation in the genome sequence where primers are going to bind, copy number of genome and efficiency of the reaction. Sometimes, it is not possible to amplify any target sequence by PCR when it is present in very low concentration from samples containing a variety of templates[53]. This limitation of PCR can produce false negative results. Such inefficiency in diagnosis may cause delay in treatment of patients. It was found in a study that the detection of herpes simplex virus was often missed in PCR detection when present in very low levels in cerebrospinal fluid samples[54]. A new approach (Amp-PCR) has been developed to improve detection of samples containing as little as a single copy of a template. The Amp-PCR technique combines phi29-amplification technique and a real-time PCR together. The first one amplifies the template randomly and unbiased manner which is subsequently amplified by virus specific primers. This approach shows 100×106-fold enhancement of PCR signals. Phi29 amplification enriches the template concentration preceding the PCR in a single tube reaction which aids detecting amounts of template below the detection limit of PCR[55].
2.10. RT-Bst: Combining Bst amplification with reverse transcription
An integrated approach for reverse transcription along with Bst DNA polymerase amplification was reported by other researchers for isothermal amplification of RNA sequences of the pathogenic viruses of the respiratory system[56]. The advantage of this method is that it initially enriches the cDNA concentration in the test sample which improves subsequent detection of virus specific sequence by PCR. This approach was applied on nasopharyngeal samples and demonstrated improved detection of RSV which could not be detected by RT-PCR alone[57]. This approach has potential to be used as a commercial kit for enrichment of RNA templates to cDNA copies especially when they are below detection level for RT-PCR technique.
3. Important aspects of molecular detection for pathogenic viruses of the respiratory system
Interpretation on the results of molecular detection for pathogenic viruses of the respiratory tract is critical for accurate diagnosis of infections of the respiratory tract. Several important factors need to be considered before interpretation of the result of molecular tests such as, viral shedding and virus concentration.
3.1. Viral infection and shedding
Collection of patient samples during the early stage of infection is a prerequisite for reliable diagnosis of the respiratory tract infection[58].The incubation period, infection period and mode of transmission of respiratory tract viruses can vary for different viruses. Strong association between presence of 15 viruses and the duration of symptoms was observed in a previous study on analysis of respiratory tract samples of patients[59]. Patients having symptoms of six days or less could be detected more easily (51%) compared to those having symptoms of seven days or more (30%, P<0.01)[60]. In another study on symptoms of respiratory tract infection, it was found that the threshold cycle (CT) values increased in real-time PCR detection as the duration of symptoms persisted in such patients[60]. CTvalue is a cut off value where the fluorescence generated within a reaction crosses the background fluorescence. Background fluorescence is determined from the fluorescence produced by a no template control. The CTvalue is inversely proportional to the template concentrations in quantitative real-time PCR reaction. These results of real-time PCR detection suggest that concentrations of viruses decrease gradually after the onset of infection[60]. In a similar study it was found that concentration hRV declined slowly and could still be detected after 10 days of infection. In other studies, hRV and hEV were found to shed from respiratory mucous for several weeks and bocaviruses were found to shed for several months[61].
3.2. Significance of a positive result in a molecular method
Molecular diagnostic tests were reported to be more sensitive than old techniques, such as culture and immunofluorescence[58,62].However, a positive result may indicate a carrier state and misinterpret active infection state. In a previous study, hRV and influenza A virus infected cases showed 2% and 6% carrier state, respectively[63].Presence of hRV in healthy children was also found to be associated with asymptomatic carriage or prolonged shedding of the virus[64].In another study, hRV could be diagnosed by PCR for two weeks following infection or more than two weeks after appearance of symptoms[60]. Immunocompromised patients may not show any clinical symptoms but shed a detectable amount of respiratory tract viruses, for example, RSV[65]. Some other viruses such as human bocavirus can be secreted for several months[66,67]. Quantification of pathogenic viruses in clinical specimen can help determine the true causative agents by determining and comparing the CTvalues of different agents[68]. Use of a panel of viruses can be more helpful for detection of multiple infections or for reconfirmation of a negative result. In such studies, Influenza A and human respiratory syncytial virus are strongly associated with clinical symptoms compared to other viruses, thereby detection of low level of these viruses may be of clinical relevance. However, it is necessary to pursue more clinical studies in order to standardize a quantitative molecular method to define cut-off level for different clinical situations. Quantitative assay using molecular methods will be more useful if specimens are collected from nasopharynx and/or oropharynx in the beginning of infection containing high titre of viruses[59].
4. Unbiased approach for detection of a panel of viruses
Respiratory system acts a transit for a number of viruses including pathogenic ones. A total of 200 viruses have been identified and reported to be associated with the infections of respiratory system[69].A number of new virus types have been reported since 2000, for example, human metapneumovirus, avian influenza viruses (H5N1,H7N7 and H7N3), human CoVs NL63 and HKU1 and severe acute respiratory syndrome CoV[15,69]. Respiratory tract infections can cause mild infection such as, self-limiting upper respiratory tract infection to more serious lower respiratory tract infections.Conventional methods e.g. culture and immunofluorescences are still used in diagnostic laboratories for diagnosis of viruses of the respiratory tract but they are often laborious, time consuming and less sensitive than molecular methods[70,71]. With the development of molecular techniques, it is now possible to detect 15-20 different viral agents simultaneously through multiplexing. This advantage of diagnostic capacity not only increased the detection of viruses but also expanded the horizon of our knowledge on aetiology of such infections. For example, human respiratory virus and human CoV were previously recognized for mild causes of infections in the upper respiratory tract infections but have now been found to be responsible for severe infections of lower respiratory tract.However, most of the viruses of such kinds can show similar clinical manifestations thereby showing difficulty in diagnosis of such infection based on their symptoms[72]. Multiple viral infections in the respiratory system could be detected in 10% of the respiratory specimens. These infections were found in patients demonstrating severe clinical symptoms[68]. Inclusion of a panel of pathogenic viruses of respiratory tract will not only help diagnose infections more accurately for appropriate treatment but also reduce associated expenses.
5. Conclusion
Improvement in the molecular detection methods has revolutionized diagnosis of respiratory pathogens compared to those of conventional techniques. Variety of viruses in certain concentrations can be detected by multiplex detection systems. However, it is essential that the diagnostic data are in concordance with the patient symptoms and state of infection. Further studies will be required for determining the threshold for detecting a healthy carrier state and shedding of different viruses from human hosts. Accurate and sensitive diagnosis of respiratory viruses is crucial in identification of causes,investigation of outbreaks, transmission, control and prevention of viral infections. Proper management of patients and reduction of costs for diagnosis of pathogenic viruses will depend on the development and application of novel molecular approaches.
Conflict of interest statement
We declare that we have no conflict of interest.
[1] Zumla A, Al-Tawfiq JA, Enne VI, Kidd M, Drosten C, Breuer J, et al.Rapid point of care diagnostic tests for viral and bacterial respiratory tract infections-needs, advances, and future prospects. Lancet Infect Dis 2014;14(11): 1123-1135.
[2] Echavarria M, Forman M, Ticehurst J, Dumler JS, Charache P. PCR method for detection of adenovirus in urine of healthy and human immunodeficiency virus-infected individuals. J Clin Microbiol 1998; 36:3323-3326.
[3] Osiowy C. Direct detection of respiratory syncytial virus, parainfluenza virus, and adenovirus in clinical respiratory specimens by a multiplex reverse transcription-PCR assay. J Clin Microbiol 1998; 36(11): 3149-3154.
[4] Poddar SK. Influenza virus types and subtypes detection by single step single tube multiplex reverse transcription-polymerase chain reaction (RTPCR) and agarose gel electrophoresis. J Virol Methods 2002; 99: 63-70.
[5] Sanghavi SK, Bullotta A, Husain S, Rinaldo CR. Clinical evaluation of multiplex real-time PCR panels for rapid detection of respiratory viral infections. J Med Virol 2011; 84(1): 162-169.
[6] Wang C, Wang Q, Hu J, Sun H, Pu J, Liu J, et al. A multiplex RT-PCR assay for detection and differentiation of avian-origin canine H3N2,equine-origin H3N8, human-origin H3N2, and H1N1/2009 canine influenza viruses. PLoS One 2017; 12(1): e0170374.
[7] Dayakar S, Pillai HR, Thulasi VP, Nair RR. Development of a multiplex RT-PCR for simultaneous diagnosis of human metapneumovirus (HMPV)and human respiratory syncytial virus (HRSV) from clinical specimens.Virus Disease 2016; 27(4): 375-381.
[8] Wathuo M, Medley GF, Nokes DJ, Munywoki PK. Quantification and determinants of the amount of respiratory syncytial virus (RSV) shed using real time PCR data from a longitudinal household study. Wellcome Open Res 2016; 1(27): 27.
[9] Lassauniere R, Kresfelder T, Venter M. A novel multiplex realtime RT-PCR assay with FRET hybridization probes for the detection and quantitation of 13 respiratory viruses. J Virol Methods 2010; 165: 254-260.
[10] Takeyama A, Hashimoto K, Sato M, Kawashima R, Kawasaki Y, Hosoya M. Respiratory syncytial virus shedding by children hospitalized with lower respiratory tract infection. J Med Virol 2016; 88(6): 938-946.
[11] To KK, Chan KH, Li IW, Tsang TY, Tse H, Chan JF, et al. Viral load in patients infected with pandemic H1N1 2009 influenza A virus. J Med Virol 2010; 82: 1-7.
[12] Cui D, Zhao D, Xie G, Yang X, Huo Z, Zheng S, et al. Simultaneous detection of influenza A subtypes of H3N2 virus, pandemic (H1N1) 2009 virus and reassortant avian H7N9 virus in humans by multiplex one-step real-time RT-PCR assay. Springerplus 2016; 5(1): 2054.
[13] Uehara M, Matsuda K, Sugano M, Honda T. A new high-speed dropletreal-time polymerase chain reaction method can detect bovine respiratory syncytial virus in less than 10 min. J Vet Med Sci 2014; 76(3): 477-480.
[14] Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 2000; 28(12): E63.
[15] Bhadra S, Jiang YS, Kumar MR, Johnson RF, Hensley LE, Ellington AD.Real-time sequence-validated loop-mediated isothermal amplification assays for detection of Middle East respiratory syndrome coronavirus(MERS-CoV). PLoS One 2015; 10: e0123126.
[16] Aliotta JM, Pelletier JJ, Ware JL, Moran LS, Benner JS, Kong H.Thermostable Bst DNA polymerase I lacks a 3’→5’ proofreading exonuclease activity. Genet Anal 1996; 12(5-6): 185-195.
[17] Mori Y, Kitao M, Tomita N, Notomi T. Real-time turbidimetry of LAMP reaction for quantifying template DNA. J Biochem Biophys Methods 2004;59(2): 145-157.
[18] Lee SH, Baek YH, Kim YH, Choi YK, Song MS, Ahn JY. One-pot reverse transcriptional loop-mediated isothermal amplification (RTLAMP) for detecting MERS-CoV. Front Microbiol 2017; 9(7): 2166.
[19] Hoos J, Peters RM, Tabatabai J, Grulich-Henn J, Schnitzler P, Pfeil J.Reverse-transcription loop-mediated isothermal amplification for rapid detection of respiratory syncytial virus directly from nasopharyngeal swabs. J Virol Methods 2017; 242: 53-57.
[20] Qu GG, Fu SJ, Shen N, Wang JL, Xiao YQ, Guan Y, et al. Rapid and sensitive diagnosis of porcine parvovirus by loop-mediated isothermal amplification (LAMP) method. J Appl Animal Res 2010; 37: 113-116.
[21] Kiatpathomchai W, Jareonram W, Jitrapakdee S, Flegel TW. Rapid and sensitive detection of Taura syndrome virus by reverse transcription loopmediated isothermal amplification. J Virol Methods 2007; 146(1-2): 125-128.
[22] Guatelli JC, Whitfield KM, Kwoh DY, Barringer KJ, Richman DD,Gingeras TR. Isothermal, in vitro amplification of nucleic acids by a multienzyme reaction modeled after retroviral replication. Proc Nat Acad Sci USA 1990; 87(19): 7797.
[23] Lu X, Shi X, Wu G, Wu T, Qin R, Wang Y. Visual detection and differentiation of Classic Swine Fever Virus strains using nucleic acid sequence-based amplification (NASBA) and G-quadruplexDNAzyme assay. Sci Rep 2017; 13(7): 44211.
[24] Asiello PJ, Baeumner AJ. Miniaturized isothermal nucleic acid amplification, a review. Lab Chip 2011; 11(8): 1420-1430.
[25] Dewar RL, Highbarger HC, Sarmiento MD, Todd JA, Vasudevachari MB,Davey RT, et al. Application of branched DNA signal amplification to monitor human immunodeficiency virus type 1 burden in human plasma.J Infect Dis 1994; 170(5): 1172-1179.
[26] Zhang Y, Liu Q, Wang D, Chen S, Wang X, Wang S. Genotyping and detection of common avian and human origin-influenza viruses using a portable chemiluminescence imaging microarray. Springerplus 2016;5(1): 1871.
[27] Wang LC, Huang D, Chen HW. Simultaneous subtyping and pathotyping of avian influenza viruses in chickens in Taiwan using reverse transcription loop-mediated isothermal amplification and microarray. J Vet Med Sci 2016; 78(8): 1223-1228. Doi: 10.1292/jvms.15-0602.
[28] Chen XH, Wang JH, Yao XH. Clinical utility of a near patient care microarray based diagnostic test for influenza and respiratory syncytial virus infections. Int J ClinExp Med 2015; 8(9): 16504-16511.
[29] Frobert E, Escuret V, Javouhey E, Casalegno JS, Bouscambert-Duchamp M, Moulinier C, et al. Respiratory viruses in children admitted to hospital intensive care units: Evaluating the CLART® Pneumovir DNA array. J Med Virol 2011; 83(1): 150-155.
[30] Hu Y, Yan H, Mammel M, Chen H. Sequence-independent amplification coupled with DNA microarray analysis for detection and genotyping of noroviruses. AMB Express 2015; 5(1): 69. Doi:10.1186/s13568-015-0156-x.
[31] Mahony J, Chong S, Merante F, Yaghoubian S, Sinha T, Lisle C,et al. Development of a respiratory virus panel test for detection of twenty human respiratory viruses by use of multiplex PCR and a fluid microbead-based assay. J Clin Microbiol 2007; 45: 2965-2970.
[32] Schmitt M, Bravo IG, Snijders PJ, Gissmann L, Pawlita M, Waterboer T. Bead-based multiplex genotyping of human papillomaviruses. J Clin Microbiol 2006; 44(2): 504-512.
[33] Brunstein JD, Cline CL, McKinney S, Thomas E. Evidence from multiplex molecular assays for complex multipathogen interactions in acute respiratory infections. J Clin Microbiol 2008; 46: 97-102.
[34] Raymond F, Carbonneau J, Boucher N, Robitaille L, Boisvert S, Wu W,et al. Comparison of automated microarray detection with real-time PCR assays for detection of respiratory viruses in specimens obtained from children. J Clin Microbiol 2009; 47: 743-750.
[35] Beck ET, Jurgens LA, Kehl SC, Bose ME, Patitucci T, LaGue E, et al. Development of a rapid automated influenza A, influenza B, and respiratory syncytial virus A/B multiplex real-time RT-PCR assay and its use during the 2009 H1N1 swine-origin influenza virus epidemic in Milwaukee, Wisconsin. J Mol Diagn 2010; 12: 74-81.
[36] Rand KH, Rampersaudm H, Houckm HJ. Comparison of two multiplex methods for detection of respiratory viruses: Film Array RP and xTAG RVP. J Clin Microbiol 2011; 49(7): 2449-2453.
[37] Garcia EP, Dowding LA, Stanton LW, Slepnev VI. Scalable transcriptional analysis routine-multiplexed quantitative real-time polymerase chain reaction platform for gene expression analysis and molecular diagnostics. J Mol Diagn 2005; 7(4): 444-454.
[38] Chen K, Hardick A, Blyn LB, Sampath R, Melton R, Matthews H, et al.PCR and electrospray ionization mass spectrometry (PCR/ ESI-MS) for identifying viral respiratory infections [abstract D-2256]. In: 48th Annual Interscience Conference on Antimicrobial Agents and Chemotherapy/46th Annual Meeting of the Infectious Diseases Society of America. Washington,DC; 2008.
[39] Legoff J, Kara R, Moulin F, Si-Mohamed A, Krivine A, Bélec L, et al.Evaluation of the one-step multiplex real-time reverse transcription-PCR ProFlu-1 assay for detection of influenza A and influenza B viruses and respiratory syncytial viruses in children. J Clin Microbiol 2008; 46(2):789-791.
[40] Liao RS, Tomalty LL, Majury A, Zoutman DE. Comparison of viral isolation and multiplex real-time reverse transcription-PCR for confirmation of respiratory syncytial virus and influenza virus detection by antigen immunoassays. J Clin Microbiol 2009; 47(3): 527-532.
[41] Moore P. Replicating success. Nature 2005; 435: 235-238.
[42] Compton J. Nucleic acid sequence-based amplification. Nature 1991;350(6313): 91-92.
[43] Gill P, Ramezani R, Amiri MVP, Ghaemi A, Hashempour T, Eshraghi N, et al. Enzyme-linked immunosorbent assay of nucleic acid sequencebased amplification for molecular detection of M. tuberculosis. Biochem Biophys Res Commun 2006; 347: 1151-1157.
[44] Dimov IK, Garcia-Cordero JL, O’Grady J, Poulsen CR, Viguier C,Kent L, et al. Integrated microfluidic tmRNA purification and real-time NASBA device for molecular diagnostics. Lab Chip 2008; 8(12): 2071-2078.
[45] Wharam SD, Marsh P, Lloyd JS, Ray TD, Mock GA, Assenberg R, et al. Specific detection of DNA and RNA targets using a novel isothermal nucleic acid amplification assay based on the formation of a three-way junction structure. Nucleic Acids Res 2001; 29(11): E54.
[46] Levi K, Bailey C, Bennett A, Marsh P, Cardy DL, Towner KJ. Evaluation of an isothermal signal amplification method for rapid detection of methicillin-resistant Staphylococcus aureus from patient-screening swabs.J Clin Microbiol 2003; 41(7): 3187-3191.
[47] Brown DF, Edwards DI, Hawkey PM, Morrison D, Ridgway GL, Towner KJ, et al. Joint Working Party of the British Society for Antimicrobial Chemotherapy; Hospital Infection Society; Infection Control Nurses Association. Guidelines for the laboratory diagnosis and susceptibility testing of methicillin-resistant Staphylococcus aureus (MRSA). J Antimicrob Chemother 2005; 56(6): 1000-1018.
[48] Hall MJ, Wharam SD, Weston A, Cardy DL, Wilson WH. Use of signalmediated amplification of RNA technology (SMART) to detect marine cyanophage DNA. Biotechniques 2002; 32(3): 604-606, 608-611.
[49] Tyagi S, Bratu DP, Kramer FR. Multicolor molecular beacons for allele discrimination. Nat Biotechnol 1998; 16(1): 49-53.
[50] Wharam SD, Hall MJ, Wilson WH. Detection of virus mRNA within infected host cells using an isothermal nucleic acid amplification assay:Marine cyanophage gene expression within Synechococcus sp. Virol J 2007; 4: 52.
[51] Nycz CM, Dean CH, Haaland PD, Spargo CA, Walker GT. Quantitative reverse transcription strand displacement amplification: Quantitation of nucleic acids using an isothermal amplification technique. Anal Biochem 1998; 259(2): 226-234.
[52] Van Der Pol B, Ferrero DV, Buck-Barrington L, Hook E, Lenderman C,Quinn T, et al. Multicenter evaluation of the BDProbeTec ET System for detection of Chlamydia trachomatis and Neisseria gonorrhoeae in urine specimens, female endocervical swabs, and male urethral swabs. J Clin Microbiol 2001; 39(3): 1008-1016.
[53] Karrer EE, Lincoln JE, Hogenhout S, Bennett AB, Bostock RM,Martineau B, et al. In situ isolation of mRNA from individual plant cells:Creation of cell-specific cDNA libraries. Proc Natl Acad Sci USA 1995;92(9): 3814-3818.
[54] Schloss L, van Loon AM, Cinque P, Cleator G, Echevarria JM, Falk KI, et al. An international external quality assessment of nucleic acid amplification of herpes simplex virus. J Clin Virol 2003; 28: 175-185.
[55] Erlandsson L, Nielsen LP, Fomsgaard A. Amp-PCR: Combining a random unbiased Phi29-amplification with a specific real-time PCR,performed in one tube to increase PCR sensitivity. PLoS One 2010; 5(12):e15719.
[56] Kabir MS, Clements MO, Kimmitt PT. RT-Bst: An integrated approach for reverse transcription and enrichment of cDNA from viral RNA. Brit J Biomed Sci 2014; 72(1): 1-6.
[57] Kabir MS, Clements MO, Atkins M, Kimmitt PT. Application of RT-Bst to enhance detection of pathogenic viruses fo the respiratory tract. Brit J Biomed Sci 2015; 72(3): 128-134.
[58] Antonishyn NA, Levett PN. Molecular diagnostic assays for detection of viral respiratory pathogens in institutional outbreaks. Mol Diagn Ther 2010; 14(5): 283-293.
[59] Brittain-Long R, Nord S, Olofsson S, Westin J, Anderson LM, Lindh M.Multiplex real-time PCR for detection of respiratory tract infections. J Clin Virol 2008; 41: 53-56.
[60] Brittain-Long R, Westin J, Olofsson S, Lindh M, Anderson LM.Prospective evaluation of a novel multiplex real-time PCR assay for detection of fifteen respiratory pathogens – duration of symptoms significantly affects detection rate. J Clin Virol 2010; 47(3): 263-267.
[61] Von Linstow ML, Hogh M, Hogh B. Clinical and epidemiologic characteristics of human bocavirus in Danish infants: Results from a prospective birth cohort study. Pediatr Infect Dis J 2008; 27(10): 897-902.
[62] Beck ET, Henrickson KJ. Molecular diagnosis of respiratory viruses.Future Microbiol 2010; 5(6): 901-916.
[63] Creer DD, Dilworth JP, Gillespie SH, Johnston AR, Johnston SL, Ling C, et al. Aetiological role of viral and bacterial infections in acute adult lower respiratory tract infection (LRTI) in primary care. Thorax 2006;61(1): 75-79.
[64] Herberhold S, Eis-Hubinger AM, Panning M. Frequent detection of respiratory viruses by real-time PCR in adenoid samples from asymptomatic children. J Clin Microbiol 2009; 47(8): 2682-2683.
[65] Ison MG. Respiratory viral infections in transplant recipients. Antivir Ther 2007; 12(4 Pt B): 627-638.
[66] Don M, Soderlund-Venermo M, Hedman K, Ruuskanen O, Allander T, Korppi M. Don’t forget serum in the diagnosis of human bocavirus infection. J Infect Dis 2011; 203(7): 1031-1032.
[67] Martin ET, Fairchok MP, Kuypers J, Magaret A, Zerr DM, Wald A, et al. Frequent and prolonged shedding of bocavirus in young children attending daycare. J Infect Dis 2010; 201(11): 1625-1632.
[68] Utokaparch S, Marchant D, Gosselink JV, McDonough JE, Thomas EE, Hogg JC, et al. The relationship between respiratory viral loads and diagnosis in children presenting to a pediatric hospital emergency department. Pediatr Infect Dis J 2011; 30(2): e18-e23.
[69] Abed Y, Boivin G. Treatment of respiratory virus infections. Antiviral Res 2006; 70(2): 1-16.
[70] Mahony JB. Detection of respiratory viruses by molecular methods. Clin Microbiol Rev 2008; 21(4): 716-747.
[71] Tregoning JS, Schwarze J. Respiratory viral infections in infants: Causes,clinical symptoms, virology, and immunology. Clin Microbiol Rev 2010;23(1): 74-98.
[72] Li H, McCormac MA, Estes RW, Sefers SE, Dare RK, Chappell JD, et al. Simultaneous detection and highthroughput identification of a panel of RNA viruses causing respiratory tract infections. J Clin Microbiol 2007; 45(7): 2105-2109.
猜你喜欢
杂志排行
Asian Pacific Journal of Tropical Biomedicine的其它文章
- Co-detection and isolation of Leishmania and Crithidia among naturally infected Tatera indica (Rodentia: Muridae) in Fars province, southern Iran
- Larvicidal efficacy of crude and fractionated extracts of Dracaena loureiri Gagnep against Aedes aegypti, Aedes albopictus, Culex quinquefasciatus, and Anopheles minimus mosquito vectors
- Antiplasmodial activity of silver nanoparticles: A novel green synthesis approach
- Combination treatment of bisphosphonate (pamidronate) and Quercus infectoria semipurified fraction promotes proliferation and differentiation of osteoblast cell via expression of Osterix and Runx2 marker
- Hepatoprotective effect of Opuntia dillenii seed oil on CCl4 induced acute liver damage in rat
- Phytochemical bioprospecting, antioxidant, antimicrobial and cytotoxicity activities of saline extract from Tithonia diversifolia (Hemsl) A. Gray leaves
