Carbendazim sensitivity in populations of Colletotrichum gloeosporioides complex infecting strawberry and yams in Hubei Province of China
2018-06-06HANYongchaoZENGXiangguoXIANGFayunZHANGQinghuaGUOCongCHENFengyingGUYuchen
HAN Yong-chao , ZENG Xiang-guo XIANG Fa-yun ZHANG Qing-hua GUO Cong CHEN Fengying GU Yu-chen
1 Institute of Industrial Crops, Hubei Academy of Agricultural Sciences, Wuhan 430064, P.R.China
2 Hubei Key Laboratory of Crop Diseases, Insect Pests and Corp Weeds Control/Key Laboratory of Integrated Pest Management on Crops in Central China, Wuhan 430064, P.R.China
1. Introduction
Strawberry (Fragaria×ananassa) is an important fruit crop in the world. The cultivated area of strawberry in China has increased rapidly in recent years. In 2015, China became the largest producer of strawberry, with a total planting area of 130 000 hectares and a production of 3.5 million tons per year (MOA 2017). Anthracnose can occur throughout the entire production cycle, particularly during the seedling and transplanting stages of strawberry plants. Strawberry infected withColletotrichumspecies may develop lesions on all parts of plants, resulting in the collapse and death of the plant in serious cases (Freemanet al. 1998). The etiology of strawberry anthracnose is complex, and 12Colletotrichumspecies have been regarded as causal agents (Weiret al. 2012; Hanet al.2016). In a preliminary study,Colletotrichum siamense(in synonymy withColletotrichum murrayae) (Liuet al. 2016),Colletotrichum fructicola,Colletotrichum gloeosporioides,andColletotrichum aenigmainC. gloeosporioidescomplex were identified to cause anthracnose on strawberry in Hubei Province of China. Isolates ofC.siamenseandC.fructicolashowed strong virulence to petioles of strawberry.C.gloeosporioidesandC.aenigmashowed weak virulence to petioles.C.siamensewas the most frequently isolated species, accounting for 81% of all isolates (Hanet al. 2016).
Yams (Dioscoreaspp.) are widely cultivated in China.The area used for yams production has increased rapidly in recent years. Yam infected withColletotrichumspecies may develop lesions on leaves and stems, resulting in reduction of the effective photosynthetic surface area of the plants and seriously affecting tuber production (Abanget al. 2002).Anthracnose has become an important disease in most of the yam-growing regions, particularly in Hubei Province.
Anthracnose caused byColletotrichumspecies affects a wide variety of crops and fruit trees, particularly in tropical and subtropical areas (Hydeet al. 2009; Wikeeet al. 2011;Cannonet al. 2012). The genus has been denoted the world’s eighth most important group of plant-pathogenic fungi based on perceived economic and scientific importance (Deanet al. 2012).Colletotrichumspecies may exhibit significant differences in their response to fungicides.For example, most wild-type isolates ofC.gloeosporioidesare sensitive to methyl-benzimidazole carbamate (MBC)fungicides. Thus MBC fungicides are still recommended for the control of anthracnose (Brannen and Smith 2015).However,C.acutatumis tolerant to MBC fungicides (Pereset al. 2014). Chunget al. (2006) and Maymonet al. (2006)discussed the MBC resistance ofC. gloeosporioides, the phylogenetic tree obtained from the internal transcribed spacer (ITS) sequences indicated that the sensitive and resistant genotypes are two separate and independent populations. Single-gene phylogenetic analyses have not been successful in delineatingColletotrichumspecies.C.gloeosporioidesis a species complex that consists of 34 taxa (Weiret al. 2012; Hydeet al. 2014). The separate populations in the phylogenetic tree based on ITS sequences in Maymonet al. (2006) may belong to different species.
Carbendazim is one of the most commonly used fungicides to control strawberry and yam anthracnose in Hubei Province. Linet al. (2016) have evaluated the benzimidazole resistance ofColletotrichum gloeosporioidescomplex populations from strawberry in Zhejiang Province of China and found 99.1% isolates were resistant to benzimidazoles. But, little was known about the carbendazim resistance of species in theC.gloeosporioidescomplex from strawberry and yam in Hubei Province.Therefore, the objectives of this study were (i) to detect the occurrence of resistance to carbendazim withinC.gloeosporioidescomplex isolates obtained from strawberry and yam and (ii) to further understand the genetic evolution of fungicide resistance in the plant pathogen.
2. Materials and methods
2.1. Isolate collection

Fig. 1 Field symptoms of anthracnose caused by Colletotrichum spp. in strawberry (A) and yam (B).
From 2012 to 2016, symptomatic leaves and stems with diagnostic symptoms (Fig. 1-A and B) were collected from 17 fields in eight cities across Hubei Province of China. A 5 mm×5 mm block of tissue was cut from the edge of the lesion and surface disinfested by rinsing in 0.5% sodium hypochlorite for 3 min, and then washed three times with sterilized distilled water. The disinfested tissue was placed in the center of potato dextrose agar (PDA) plate acidified with lactic acid at a final concentration of 0.05% to inhibit bacterial growth. The plates were incubated for 3 to 7 days at 28°C in the dark. After incubation, a 5 mm×5 mm block of agar taken from the growing edges of any fungal hyphae developing on the PDA plates was then transferred aseptically to new PDA plates amended with lactic acid at 0.05%. Monoconidial cultures were subsequently obtained through serial dilution, and isolates without conidia on the PDA plates were purified by transferring samples from the advancing mycelial edge of each colony to new PDA plates three to five times. AllColletotrichumisolates were identified to the species using a multilocus phylogenetic analysis identification method (Hanet al. 2016).
2.2. In vitro bioassay
Carbendazim fungicide (Shenyang Academy of Chemistry and Industry, Shengyang, China) was dissolved in hydrochloric acid (HCl; 0.1 mol L–1) at 5 mg mL–1as a stock solution. The carbendazim stock solution was added to PDA after it had been cooled to approximately 50°C. To test the sensitivity of isolates to carbendazim,PDA media were amended with the active ingredient (a.i.)at 0, 1, 5, 50, and 100 μg mL–1.Colletotrichumisolates(62 from strawberry and 63 from yam) were screened for resistance to carbendazim. The 1 μg mL–1concentration of carbendazim was used as the discriminatory dose at which sensitive (S) or moderately resistant (MR) isolates were identified (Pereset al. 2004; Ramdialet al. 2016). The 50 μg mL–1concentration of carbendazim was used as the discriminatory dose at which MR or highly resistant (HR)isolates were identified. Sensitive isolates did not grow on PDA amended with carbendazim at 1, 5, 50 or 100 μg mL–1.MR isolates did not grow at either 50 or 100 μg mL–1. HR isolates could grow even at 100 μg mL–1. Four replicates of each carbendazim concentration were used for each isolate,and the entire experiment was performed twice.
Mycelial plugs were removed with a 5-mm cork borer from the leading edge of an actively growing 5-day-old colony and placed mycelium-side-down on the centers of a Petri dishes (9 cm in diameter) containing fungicide-amended or non-amended PDA. The plates were incubated at 28°C for 5 days in darkness. Colony diameters (minus the diameter of mycelial disc) of each plate were measured and is expressed as the maximum growth concentration of carbendazim.
2.3. Sequence analysis of β-tubulin 2 genes
Representative isolates of different resistant phenotypes andColletotrichumspecies were selected for sequencing of their β-tubulin 2 (TUB2) genes. The mycelia ofColletotrichumisolates were obtained by using a previously published protocol (Louet al. 2015). The mycelia of each isolate were ground to a fine powder in liquid nitrogen with a mortar and pestle. Genomic DNA (gDNA) was extracted and purified by using a previously published protocol (Hanet al.2016). Purified DNA was dissolved in 200 μL of TE buffer(10 mmol L–1Tris-HCl, 1 mmol L–1EDTA, pH 8.0), and diluted to approximately 10 to 100 ng μL–1for polymerase chain reaction (PCR). DNA samples were stored at –20°C until use.
For sequence analysis,TUB2genes were amplified from gDNA ofColletotrichumisolates with primers in Table 1. The DNA ofTUB2was amplified in two parts:part 1 (fragment size, 870 bp) of theTUB2gene was amplified with primer pair T1 (O’Donnell and Cigelnik 1997)and TUB2-R2 (Hanet al. 2016), and part 2 (fragment size,1 328 bp) of theTUB2gene was amplified with primer pair TUB2-F3 and TUB2-R4 (Table 1). The primers TUB2-R2(part 1), TUB2-F3 (part 2) and TUB2-R4 (part 2) were used for sequencing. The PCR amplification mixtures were set up in a final volume of 60 μL containing 6 μL of PCR buffer, 1 μL of each primer (50 μmol L–1), 1 μL of genomic DNA (approximately 50 ng), 1 μL ofTaqDNA polymerase(5 U per reaction mixture; TaKaRa Biotechnology Co.,Ltd., Dalian, China), 2 μL of dNTP mix (2.5 mmol L–1), and 49 μL of deionized water. The PCR assays were carried out in a thermal cycler (TaKaRa PCR Thermal Cycler, China).The cycling parameters for parts 1 and 2 consisted of a denaturation step at 94°C for 5 min, followed by 30 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 90 s and a final cycle at 72°C for 10 min.
PCR products were separated by electrophoresis in 1.5%agarose gels in 1.0× Tris-acetate EDTA (TAE) buffer; the gels were photographed under UV light after stained with gelred nucleic acid gel stain (Biotium, Hayward, USA) for30 min. dNTPs and unincorporated primers remaining in PCR products were removed with AxyPrep™ PCR Cleanup Kit (Axygen Biological Technology Co., Ltd., Hangzhou,Zhejiang Province, China) according to the manufacturer’s instructions. Sequencing of PCR products was conducted at Beijing AuGCT Biotechnology Co., Ltd. (Beijing, China).The nucleotide sequences ofTUB2gene generated in this study were deposited in GenBank (Table 2).
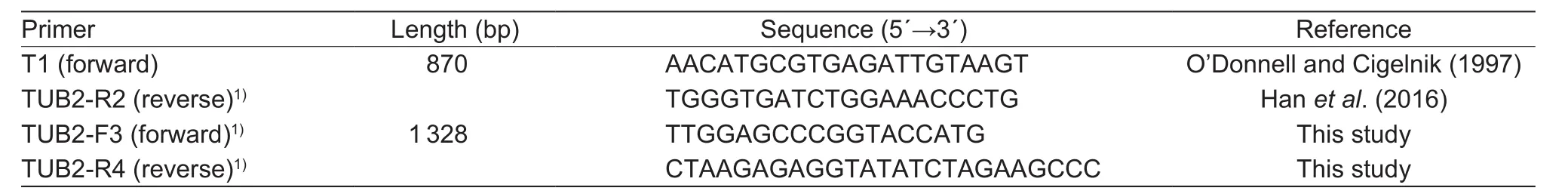
Table 1 PCR primers used in this study
Gene structure analysis ofTUB2gene sequences was conducted to determine whether carbendazim-sensitive or carbendazim-resistantColletotrichumspecies were structured specially enough to form separate resistant species. The exon, intron, and codon mutation positions in theTUB2genes ofC.fructicola,C.siamense,C.aenigma,andC.gloeosporioideswere determined in alignments with theTUB2gene sequence ofC.gloeosporioidesf. sp.aeschynomene(GenBank accession number:CGU14138).
2.4. Phylogenetic analysis
Genetic analysis of partial DNA sequences ofTUB2, actin(ACT), calmodulin(CAL), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and chitin synthase (CHS-1) gene was conducted to determine whether carbendazim-sensitiveColletotrichumisolates were genetically similar enough to form a separate cluster from resistant isolates. Partial DNA sequences ofTUB2(mutations at codons 6, 50, 167, 198,200, and 240 included) from the representative isolates from strawberry and yam (Table 2) were used in the first Bayesian inference (A) analysis. Partial DNA sequences ofTUB2(mutations at codons 198 and 200 were not included),ACT,CAL,GAPDHandCHS-1from representative isolates of strawberry (Table 2) were used to establish a five-locus combined dataset (TUB2+ACT+CAL+GAPDH+CHS-1) in the second Bayesian inference (B) analysis. The sequences for the second Bayesian inference (B) analysis were from our previous research (Hanet al. 2016).
The sequences generated for theTUB2genes in this study were manually truncated to align for constructing phylogenetic trees. The sequence of each gene was subjected to multiple sequence alignment with ClustalW as implemented in MEGA v.5 (Tamuraet al. 2011). Akaike information criteria (AICc) were used to select the best-fit models of nucleotide substitution forTUB2(GTR+G),ACT(K80+G),CAL(GTR+G),GAPDH(HKY+I) andCHS-1(K80+G) with MrModelTest v. 2.3 (Nylander 2004).
A Markov Chain Monte Carlo (MCMC) algorithm was used to reconstruct phylogenetic trees with Bayesian probabilities using MrBayes v. 3.2.1 (Ronquistet al. 2012).The MCMC chains analysis was performed twice base on the full data set for 5×107generations and sampled every 1 000 generations. The convergence of all parameters was verified using the Tracer Program (Rambaut and Drummond 2007). On this basis, the first 25% of generations were discarded as burn-in. The concatenated alignment and trees were submitted to TreeBASE (www.treebase.org) with a submission number of 21211.
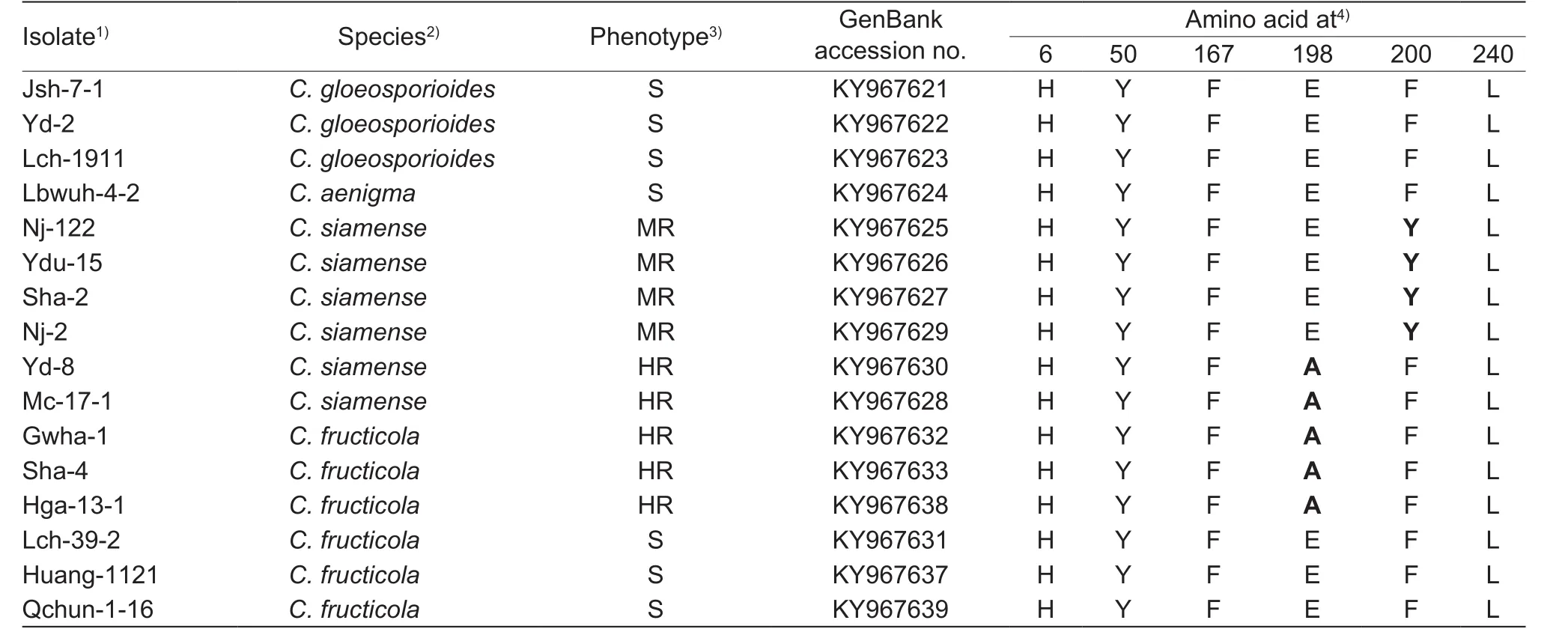
Table 2 Altered amino acids encoded by the β-tubulin 2 (TUB2) gene in carbendazim-resistant and -sensitive isolates of Colletotrichum
3. Results
3.1. Fungicide sensitivity assessment in vitro
A total of 125 isolates were collected, 62 from strawberry and 63 from yam, 51.6% of the isolates were determined to be resistant to carbendazim (Table 3). Forty-eight isolates from strawberry were resistant to carbendazim, with a frequency of 77.4%. Within these 48 resistant isolates, the frequency of MR and HR isolates were 25.0 and 75.0% respectively(Table 3). Eighteen of the isolates from yam were resistant to carbendazim, with a resistance frequency of 28.6%.Among these 18 resistant isolates, the frequencies of MR and HR isolates were 22.2 and 77.8% respectively (Table 3).
Overall, the frequencies of resistance differed by region: the highest carbendazim resistance frequencieswere observed in Xiaogan (100%), Jingzhou (100%) and Xianning (85.7%), and the lowest frequencies were detected in Xiangyang (11.1%) and Enshi (15.4%) (Fig. 2). Isolates resistant to carbendazim were distributed throughout the province, but resistance frequency varied depending on the sampled sites.
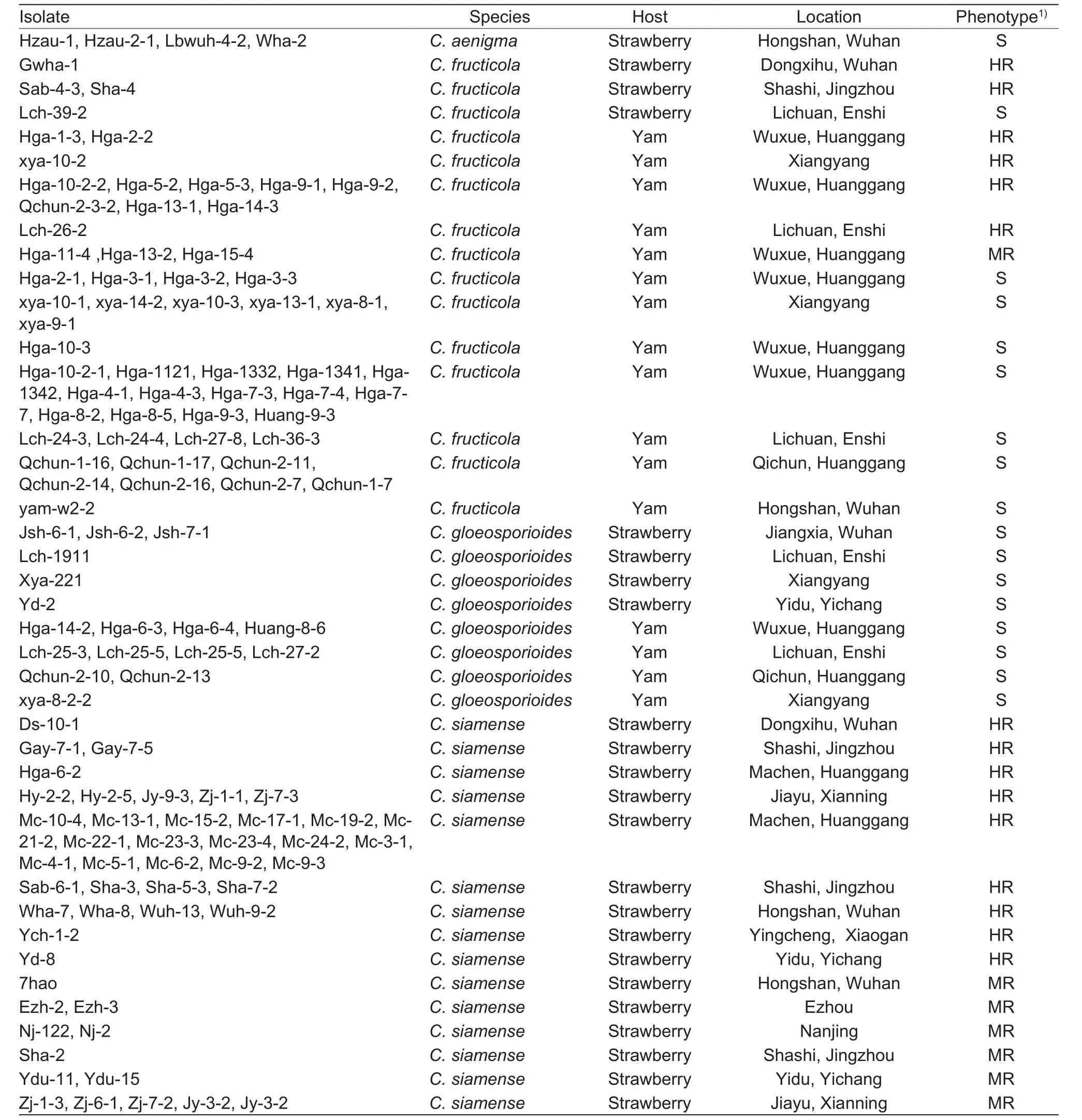
Table 3 Isolates of Colletotrichum used in this study

Fig. 2 Relative frequencies of carbendazim-resistant phenotypes. Each of these pie charts shows the relative frequency of isolates resistant to carbendazim. The relative frequencies were calculated based on the combined population from both strawberry and yam.
The frequencies of resistant isolates were different for differentColletotrichumspecies. The highest frequency of carbendazim resistance was observed inC.siamense,allC.siamenseisolates tested in this study were resistant to carbendazim. In all, 12 of 48C.siamenseisolates from strawberry were MR to carbendazim, and 36 of 48C.siamenseisolates from strawberry were HR to carbendazim. In contrast, allC.gloeosporioides(this species has been epitypified by Weiret al. (2012)) andC.aenigmaisolates used in this study were sensitive to carbendazim. Fifty-six isolates ofC.fructicolawere collected, where the number of S, MR and HR isolates was 38, 3 and 15, respectively (Table 3).
3.2. Nucleotide sequence analysis of TUB2 gene
Sixteen representative isolates of different resistance phenotypes and species (Table 2) were selected forTUB2gene analysis. Two bands of the expected sizes of 870 and 1 328 bp were obtained for all DNA samples, respectively.Parts 1 and 2 were assembled into one sequence of 1 842 bp with DNAMAN (version 7.0), which, when translated,corresponds to amino acids 5 to 447. The typical amino acid codon mutations at 6, 50, 167, 198, 200 and 240 associated with carbendazim resistance were included (Ma and Michailides 2005).
Nucleotide sequences from representative isolates from strawberry and yam in Hubei Province, were submitted to GenBank (Table 2). TheTUB2genes from 16 randomly selected representativeColletotrichumisolates showing the resistant phenotype were sequenced to identify point mutations leading to amino acid substitutions associated with carbendazim resistance. Comparison of the nucleotide sequences of theTUB2genes in differentColletotrichumisolates showed that two types of point mutations were found in all the resistant isolates. Among these isolates, a change from phenylalanine to tyrosine (TTC (Phe)→TAC(Tyr)) at codon 200 (F200Y) was found in MR isolates. All HR isolates from both strawberry and yam contained the point mutation GAG (Glu)→GCG (Ala) at codon 198 (E198A)in theirTUB2gene (Table 2).
The results ofTUB2gene structure analysis showed that seven exons and six introns were included in theTUB2genes ofC.fructicola,C.siamense,C.aenigma,C.gloeosporioides, andC.nymphaeae.The numbers of introns and exons and the lengths of exons in theTUB2genes of the five species were highly conserved (Fig. 3).The lengths of the introns in theTUB2genes ofC.fructicola,C.siamense, andC.aenigmawere also conserved. The third intron in theTUB2gene ofC.gloeosporioideswas 72 bp, which is different fromC.fructicola,C.siamense,andC.aenigma(Fig. 3).
3.3. Phylogenetic analysis of sensitive and resistant isolates
The phylogenetic tree obtained from the partialTUB2sequence is shown in Fig. 4-A, and the phylogenetic tree obtained from a five-locus combined dataset(TUB2+ACT+CAL+GAPDH+CHS-1) is shown in Fig. 4-B. The five populations identified by both theTUB2sequence and the five-locus combined dataset(TUB2+ACT+CAL+GAPDH+CHS-1) analyses showed significant correlations with species. The representative isolates (Table 2) were grouped into five clusters in both Fig. 4-A and B.
The populations identified by partialTUB2(mutations at codon 198 and 200 included) sequence (1 656 bp) analysis showed significant correlations with the carbendazimresistant phenotype. S, MR and HR isolates were grouped within separate clusters regardless of species, with strong bootstrap support (Fig. 4-A). Within the resistance cluster ofC.siamense, there was an apparent differentiation among those isolates with the F200Y and E198A genotypes, with strong support (100%) for clustering of the F200Y and E198A genotypes (Fig. 4-A). Within theC.fructicolacluster,there was an apparent differentiation among sensitive and resistance genotypes, with strong support (99%) for clustering of sensitive and resistance genotypes (Fig. 4-A).The populations identified by five-locus dataset (3 373 bp)sequence analysis combining the partial sequences ofTUB2(mutations at codon 198 and 200 not included) withACT,CAL,GAPDHandCHS-1showed no correlation with the carbendazim-resistant phenotype (Fig. 4-B). Phylogenetic analysis revealed that MR and HR isolates ofC.siamensewere grouped into one group. Within the species ofC.fructicola, the HR isolate Gwha-1 was clustered in one group with sensitive isolate Lch-39-2, but distinct from another HR isolate Sha-4 (Fig. 4-B).
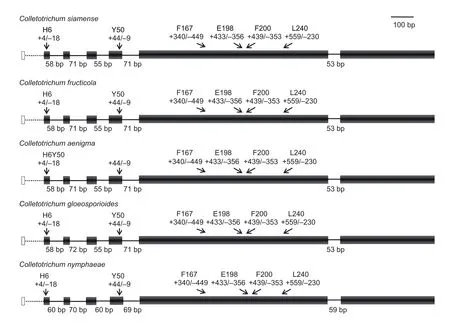
Fig. 3 Comparison of nucleotide sequences at exon/intron junctions in the β-tubulin 2 (TUB2) genes of different plant pathogen species. Empty boxes indicate exons, and lines indicate introns. Dashed boxes and lines represent non-sequenced parts of the gene. The relative positions of cordon at the exon are shown with number followed by “+” and “–”, which mean base pair downstream the initiation of exon and upstream the end of exon, respectively.

Fig. 4 Bayesian inference phylogenetic tree of 14 isolates of Colletotrichum spp. The two trees were built using sequences of the β-tubulin 2 (TUB2) gene (A) and concatenated sequences of the TUB2, ACT, CAL, GPDH and CHS-1 genes (B) respectively, each gene with a separate model of nucleotide substitution. C. nymphaeae was used as an outgroup. Bayesian posterior probability values based on 500 000 generations and above 0.5 are shown at the nodes of the tree. Moderately resistance (MR) isolates are marked with a square frame, and highly resistance (HR) isolates are marked with a black background.
4. Discussion
In China, fungicide application is a necessary measure for the control of anthracnose on both strawberry and yam productions. Since 30 years ago, benzimidazole fungicides,including carbendazim and thiophanate-methyl, have been the most popular fungicides for anthracnose control.As many other examples indicate, repeated application of the MBC fungicides likely enhances the selection of carbendazim-resistant isolates (Kumaret al. 2007). The frequencies of resistance isolates differed by region (Fig. 2).This evidence possibly explains the difference in fungicide usage. Most growers experience poor management of anthracnose in fields subjected to repeated use of the benzimidazole fungicides.
Benzimidazole resistance has been reported inC.acutatum,C.cereale,C.gloeosporioides, andC.siamense(Pereset al. 2004; Wonget al. 2008; Huet al. 2015). Benzimidazole fungicide resistance has been reported associated with point mutations in the β-tubulin gene, whereby the benzimidazole fungicide is unable to bind to the mutated β-tubulin protein with the altered amino acid sequence (Maet al. 2003; Maymonet al. 2006). In most fungi studied, a single β-tubulin gene exists within the genome (Maet al. 2003; Maymonet al. 2006). However, an additional gene with a divergent sequence is present in a few fungi. The presence of two such genes has been reported inColletotrichumspp. (Panaccione and Hanau 1990). InC.gloeosporioidesandC.graminicola, the two β-tubulin genes,TUB1andTUB2, are highly divergent (Panaccione and Hanau 1990; Maymonet al. 2006). TheTUB2fragment is more closely related in sequence tobenAfromB.cinereaand β-tubulin genes from other fungi (with a single β-tubulin gene) thanTUB1.
Several mutations in the β-tubulin gene can lead to MBC fungicide resistance. The codon changes of most field resistant isolates that have arisen in several phytopathogenic fungal species seem to be restricted to positions 6, 50, 167, 198, 200 and 240 (Zhanet al. 2014).Primers were designed from the conserved domain of theC.gloeosporioidescomplexTUB2gene (Table 1).DNA sequences of 1 842 bp were assembled that, when translated, it corresponded to amino acids 5 to 447 (Table 2).Sequence analysis of theTUB2region that is responsible for carbendazim resistance showed that two types of point mutations were present in theTUB2genes of resistant isolates in this study. Among these resistant isolates, a change from phenylalanine to tyrosine at codon 200 (F200Y)was found in MR isolates. Isolates containing the point mutation GAG (Glu) to GCG (Ala) at codon 198 (E198A) in their β-tubulin 2 gene were highly resistant to carbendazim(Table 2). Mutations at positions 6, 50, 167, and 240 were not observed in this study. As in our study, the level of MBC fungicide resistance depends on the type of mutation; the mutation GAG (Glu) to GCG (Ala) at codon 198 (E198A)is the dorminant type of all mutations and is able to confer high resistance to MBC fungicides in theC.gloeosporioidescomplex (Chunget al. 2006, 2010).
All MR or HR isolates in this study have same mutations in theTUB2gene but sensitive isolates do not (Table 2), the result of phylogenetic tree is strongly affected by sequences selected for phylogenetic analysis. S, MR and HR isolates were grouped within separate clusters regardless of species if the mutated codon positions were included in the phylogenetic analysis (Fig. 4-A). If the mutated codon positions inTUB2gene were not included in the phylogenetic analysis, S, MR and HR isolates could not be grouped within separate clusters (Fig. 4-B). It is generally accepted that phylogenetic tree based on multiple polygenic locus can better show the phylogenetic relationship between differentColletotrichumstrains. So, the result of phylogenetic tree analysis (Fig. 4-B) revealed that carbendazim-resistantColletotrichumisolates in this study could not form separate clusters from sensitive isolates.
In this study, 48 isolates belonging toC.siamensewere collected, where 12 MR isolates and 36 HR isolates, with a resistance frequency of 100% (Table 3). All isolates ofC.gloeosporioidesandC.aenigmain this study were sensitive to carbendazim (Table 3). But, there were no known differences among the five species inTUB2gene that would lead to species-specific resistance to carbendazim(Fig. 3). Previous results showed that mutation of a base pair at the intron location will affect self-splicing in the mitochondrial cytochromeb(CYTB) gene, which can protect G143 from being mutated into G143A (Luoet al.2010). However,TUB2is a nuclear gene, intron splicing will not be affected by mutations of codons at exons. Even though 100% of theC.siamenseisolates in this study were resistant to carbendazim, there was no correlation betweenC.siamenseas a species and fungicide sensitivity because both S and HR isolates have been isolated from peach and blueberry (Huet al. 2015).C.siamenseshowed strong virulence to petioles of strawberry and is the major pathogen causing anthracnose of strawberry in Hubei Province.Other species, includingC.fructicola,C.aenigmaandC.gloeosporioideswere rarely isolated from strawberry(Hanet al. 2016).
5. Conclusion
Resistance survey of theC.gloeosporioidescomplex allowed the discovery of species-specific resistance to MBC fungicides. From the standpoint of a disease management, the result of this study emphasizes the importance of knowing whichColletotrichumsp. is present,when strategies for disease control are made.
Acknowledgements
This study was financially supported by the National Natural Science Foundation of China (31701882), the Competitive Nature Project of the Hubei Academy of Agricultural Sciences, China (2016JZXJH006), and the Agricultural Science and Technology Innovation Center Program of Hubei Province, China (2016-620-000-001-014).
Abang M M, Winter S, Green K R, Hoffmann P, Mignouna H D,Wolf G A. 2002. Molecular identification ofColletotrichum gloeosporioidescausing yam anthracnose in Nigeria.Plant Pathology,51, 63–71.
Brannen P, Smith P. 2015. Southeast regional strawberry integrated pest management guide. [2017-03-11]. http://www.smallfruits.org/SmallFruitsRegGuide/(2014)
Cannon P F, Damm U, Johnston P R, Weir B S. 2012.Colletotrichum- Current status and future directions.Studies in Mycology,73, 181–213.
Chung W H, Chung W C, Peng M T, Yang H R, Huang J W.2010. Specific detection of benzimidazole resistance inColletotrichum gloeosporioidesfrom fruit crops by PCRRFLP.New Biotechnology,27, 17–24.
Chung W H, Ishii H, Nishimura K, Fukaya M, Yano K, Kajitani Y.2006. Fungicide sensitivity and phylogenetic relationship of anthracnose fungi isolated from various fruit crops in Japan.Plant Disease,90, 506–512.
Dean R, Van Kan J A, Pretorius Z A, Hammond-Kosack K E, Di Pietro A, Spanu P D, Foster G D. 2012. The top 10 fungal pathogens in molecular plant pathology.Molecular Plant Pathology,13, 414–430.
Freeman S, Katan T, Shabi E. 1998. Characterization ofColletotrichumspecies responsible for anthracnose disease of various fruits.Plant Disease,82, 596–605.
Han Y C, Zeng X G, Xiang F Y, Ren L, Chen F Y, Gu Y C.2016. Distribution and characteristics ofColletotrichumspp.associated with anthracnose of strawberry in Hubei, China.Plant Disease,100, 996–1006.
Hu M J, Grabke A, Dowling M E, Holstein H J, Schnabel G.2015. Resistance inColletotrichum siamensefrom peach and blueberry to thiophanate-methyl and azoxystrobin.Plant Disease,99, 806–814.
Hyde K D, Cai L, Cannon P F, Crouch J A, Crous P W, Damm U, Zhang J Z. 2009.Colletotrichum- Names in current use.Fungal Diversity,39, 147–182.
Hyde K D, Nilsson R H, Alias S A, Ariyawansa H A, Blair J E,Cai L, de Cock A W A M, Dissanayake A H, Glockling S L, Goonasekara I D, Gorczak M, Hahn M, Jayawardena R S, van Kan J A L, Laurence M H, Lévesque C A, Li X H,Liu J K, Maharachchikumbura S S N, Manamgoda D S,et al. 2014. One stop shop: Backbone trees for important phytopathogenic genera: I (2014).Fungal Diversity,67,21–125.
Kumar A S, Reddy N E, Reddy K H, Devi M C. 2007.Evaluation of fungicidal resistance amongColletotrichum gloeosporioidesisolates causing mango anthracnose in Agri Export Zone of Andhra Pradesh, India.Plant Pathology Bulletin,16, 157–160.
Lin T, Xu X F, Dai D J, Shi H J, Wang H D, Zhang C Q. 2016.Differentiation in development of benzimidazole resistance inColletotrichum gloeosporioidescomplex populations from strawberry and grape hosts.Australasian Plant Pathology,45, 241–249.
Liu F, Wang M, Damm U, Crous P W, Cai L. 2016. Species boundaries in plant pathogenic fungi: AColletotrichumcase study.BMC Evolutionary Biology,16, 81.
Lou Y, Han Y C, Yang L, Wu M D, Zhang J, Cheng J S, Wang M Y, Jiang D H, Chen W D, Li G Q. 2015.CmpacCregulates mycoparasitism, oxalate degradation and antifungal activity in the mycoparasitic fungusConiothyrium minitans.Environmental Microbiology,17, 4711–4729.
Luo C X, Hu M J, Jin X, Yin L F, Bryson P K, Schnabel G.2010. An intron in the cytochrome b gene ofMonilinia fructicolamitigates the risk of resistance development to QoI fungicides.Pest Management Science,66, 1308–1315.
Ma Z, Michailides T J. 2005, Advances in understanding molecular mechanisms of fungicide resistance and molecular detection of resistant genotypes in phytopathogenic fungi.Crop Protection,24, 853–863.
Ma Z, Yoshimura M A, Michailides T J. 2003. Identification and characterization of benzimidazole resistance inMonilinia fructicolafrom stone fruit orchards in California.Applied Environmental Microbiology,69, 7145–7152.
Maymon M, Zveibil A, Pivonia S, Minz D, Freeman S. 2006.Identification and characterization of carbendazim-resistant and -sensitive populations ofColletotrichum gloeosporioidesfrom statice (Limoniumspp.).Phytopathology,96, 542–548.
MOA (Ministry of Agriculture of China). 2017. Sown area and yield of vegetable, watermelon, melon, strawberry and potatoe throughout the country in 2015.ChineseVegetables,1, 18. (in Chinese)
Nylander J A A. 2004.MrModeltest v2.Program Distributed by the Author. Evolutionary Biology Centre, Uppsala University,Uppsala, Sweden.
O’Donnell K, Cigelnik E. 1997. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungusFusariumare nonorthologous.MoIecular Phylogenetics and Evolution,7, 103–116.
Panaccione D G, Hanau R M. 1990. Characterization of two divergent beta-tubulin genes fromColletotrichum graminicola.Gene,86, 163–170.
Peres N A R, Souza N L, Peever T L, Timmer L W. 2004.Benomyl sensitivity of isolates ofColletotrichum acutatumandC.gloeosporioidesfrom citrus.Plant Disease,88,125–130.
Rambaut A, Drummond A J. 2007. Tracer v1.4. University of Oxford, Oxfordshire, U.K. [2017-03-01]. http://beast.bio.ed.ac.uk/Tracer
Ramdial H, Hosein F N, Rampersad S N. 2016. Detection and molecular characterization of benzimidazole resistance amongColletotrichum truncatumisolates infecting bell pepper in Trinidad.Plant Disease,100, 1146–1152.
Ronquist F, Teslenko M, van der Mark P, Ayres D, Darling A,Höhna S, Larget B, Liu L, Suchard M, Huelsenbeck J. 2012.MrBayes v. 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space.Systematic Biology,61, 539–554.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M. 2011.MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods.Molecular Biology and Evolution,28,2731–2739.
Weir B S, Johnston P R, Damm U. 2012. TheColletotrichum gloeosporioidesspecies complex.Studies in Mycology,73, 115–180.
Wikee S, Cai L, Pairin N, McKenzie E H C, Su Y Y, Chukeatirote E, Thi H N, Bahkali A H, Moslem M A, Abdelsalam K, Hyde K D. 2011.Colletotrichumspecies from Jasmine (Jasminum sambac).Fungal Diversity,46, 171–182.
Wong F P, de la Cerda K A, Hernandez-Martinez R, Midland S L. 2008. Detection and characterization of benzimidazole resistance in California populations ofColletotrichum cereale.Plant Disease,92, 239–246.
Zhan J S, Wu E J, Liu X L, Chen F P. 2014. Molecular basis of resistance of phytopathogenic fungi to several site-specific fungicides.Scientia Agricultura Sinica,47, 3392–3404. (in Chinese)
杂志排行
Journal of Integrative Agriculture的其它文章
- Improve access to the EU market by identifying French consumer preference for fresh fruit from China
- Management and prevention of mastitis: A multifactorial approach with a focus on milking, bedding and data-management
- Elimination of ceftiofur hydrochloride residue in postpartum cows’milk after intramammary infusing at dry-off
- Evaluation of a new qPCR test to identify the organisms causing high total bacterial count in bulk tank milk
- Prevalence and characteristics of extended spectrum β-lactamaseproducing Escherichia coli from bovine mastitis cases in China
- Evolutionary analysis of plant jacalin-related lectins (JRLs) family and expression of rice JRLs in response to Magnaporthe oryzae
