Evaluation of a new qPCR test to identify the organisms causing high total bacterial count in bulk tank milk
2018-06-06rgenKatholmLeneTrierOlesenAndersPetersenSnorriSigurdsson
Jørgen Katholm, Lene Trier Olesen, Anders Petersen, Snorri Sigurdsson
1 DNA Diagnostic A/S, Risskov 8240, Denmark
2 ARLA Foods amba, Viby 8260, Denmark
3 SEGES, Aarhus N 8200, Denmark
1. Introduction
Milk quality in bulk tank milk (BTM) is measured by flow cytometry technology as total bacterial count (TBC) and somatic cell count (SCC). There has been a long tradition for using cultivation of BTM samples to identify different bacteria causing high SCC in the milk. Also qPCR tests, e.g., Mastit 4,a commercially available qPCR test (DNA Diagnostic,Risskov, Denmark), can be used to detect mastitis bacteria in BTM (Rattenborget al. 2015).
Tests for milk quality and bacteria in BTM include standard plate count (SPC), coliform count (CC), and laboratory pasteurization count (LPC) (Guterbock and Blackmer 1984; Murphy 1997). Estimation of the type and number of bacteria in BTM is valuable in understanding and troubleshooting issues related to udder health, milk harvest hygiene, cleaning practices, and milk storage conditions(Elmoslemanyet al. 2009). They investigated risk factors for bacteriological quality of bulk tank milk, highlighting the importance of udder hygiene and milking system washing factors on hygienic quality of bulk tank milk. Milking machine wash failures is strongly associated with in-line CC, which suggests that proper and consistent washes play a fundamental role in minimizing BTM contamination with coliforms (Pantojaet al. 2011). Increases in SPC (Costelloet al. 2003) and slightly higher CC (Pantojaet al. 2011) were found after transfer of raw milk from farm tanks to dairy processor bulk tanks. Environmental contamination is an important factor for the bacterial content of milk. Vacheyrouet al. (2011) investigated the bacterial content in air, dust,hay, and cow teat surface and found that milk contamination by the stable environment was considerable, although it was lower in farms with a milking parlor compared to tie stalls.The study of Lucaliet al. (2015) underlined the correlation between forage quality, dairy farm management practices and the presence of milk and cheese anaerobic sporeforming bacteria.
It is well known thatStreptococcifrom mastitis cows can cause high TBC.Streptococcus agalactiaeandStreptococcus uberishave been found to be shed in very high numbers (up to 109bacteria mL–1from infected quarters(Guterbock and Blackmer 1984; Schukkenet al. 2011).Zadokset al. (2004) found thatStreptococci,Staphylococci,and Gram-negative bacteria accounted for 69, 3, and 3%of TBC variability, in 48 BTM samples from New York State dairy farms. Keefeet al. (1997) found that herds infected withStrep.agalactiaewere 5.48 times more likely to be penalized for a high SPC. Also Gillespieet al. (2012) found strong correlations between SPC andStreptococcusspp.counts (0.72).
Detection of bacterial DNA can be used for analyses of bacterial content in BTM. Katholmet al. (2012) tested Danish BTM samples with qPCR and found the highest correlation to TBC forEnterococcus,Strep.uberisandStrep.agalactiaeof the bacteria investigated. Analysis with community 16S rRNA gene sequence was used by Kableet al. (2016) to identify bacteria in raw bovine milk samples from tanker trucks arriving to two dairy processors in California, USA. They found the highest total cell numbers and the highest proportions being those ofActinobacteria.Even with this complexity, a core microbiota was present,consisting of 29 taxonomic groups and high proportions ofStreptococcusandStaphylococcusand unidentified members ofClostridiales. To our knowledge, thus far no qPCR addressing the bacteria involved in TBC has been commercially introduced. The aim of this study is to evaluate a recently introduced 3-h qPCR test, TBC 4 (DNA Diagnostic, Denmark). We will continue to compare the TBC 4 qPCR test with traditional culture. The TBC 4 qPCR gives a Ct-value for four targets,Pseudomonas,Streptococci,Enterobacteriacea/Enterococcus, andBacillus/Clostridia.These four targets correlates to the problems on the farm related to cooling, mastitis, environment, or silage. We were not able to compare the TBC 4 qPCR to culture in this trial but further trials will evaluate this.
2. Materials and methods
In the period between 7th March and 5th April, 2017, BTM samples obtained from Eurofins laboratory (Vejen, Denmark)were measured for TBC by routine flow cytrometry with BactoCount IBC (Bentley instruments, Inc., Chaska, USA).For this study, we selected 346 milk samples from different TBC intervals for qPCR test with TBC 4. The samples were selected among all Danish dairy herds. The general descriptive data for these Danish herds are 172 cows/herd, yield is 10 008 kg per cow, almost all cows are feed total mixed ratio, average geometric mean BTM SCC is 205 800 cells mL–1and geometric mean TBC is 7 690 bacterial count mL–1(Danish Agriculture and Food Council 2016). Mastitis treatments per cow year was 0.33 in yield control herds withStrep.uberisandStaphylococcus aureusas the dominant pathogens.
After the result from the flow cytometry TBC test was obtained, the samples were immediately transported on ice to the laboratory of DNA Diagnostic A/S, Risskov, Denmark and tested by the TBC 4 qPCR test within 24 h.
3. Results
The results from the TBC 4 test of the 346 BTM samples in different groups of CFU mL–1is shown in Table 1.
In total 158 (46%) samples were positive forPseudomonas, 157 (45%) forStreptococci, 128 (37%)forEnterobacteriacea/Enterococcus,and 122 (35%) forBacillus/Clostridia.
In each of the different intervals of TBC, the percent of positive samples, average Ct-value of positive samples,percent samples with Ct<30 and percent samples with Ct<25 were calculated for all four targets of the test (Fig. 1).
ThePseudomonas,Streptococciand theEntero bacteriacea/Enterococcustarget showed increasing percent positive samples with higher CFU and also reduced Ct-value at higher CFU, indicating more of these bacteria is present at higher CFU. ForBacillus/Clostridia, the increase in positive samples stopped at 30 000 CFU mL–1and the average Ctvalue were above 30 in all groups of CFU (Fig. 1-A and B).The percent positive samples with Ct-value below 30 and 25 is shown in Fig. 1-C and D. As it can be seen, we did not find manyBacillus/Clostridiapositive samples with really low Ct-values. For theStreptococci,they have the highestpercent of samples with low Ct-values in the samples up to 100 000 CFU mL–1, whereas both thePseudomonasand theEnterobacteriacea/Enterococcustarget have the highest percent of samples with low Ct-values in the samples above 100 000 CFU mL–1.

Table 1 Number of bulk tank milk samples tested in each group of colony forming unit (CFU) mL–1
4. Discussion
The new qPCR test TBC 4 enables the user to classify high TBC in BTM to four different groups of problems related to cooling, mastitis, environment or silage. Not all problems with high TBC are solved by optimizing cooling and the washing procedures, as we found 46% of samples positive forPseudomonasand 37% forEnterobacteriacea/Enterococcus.
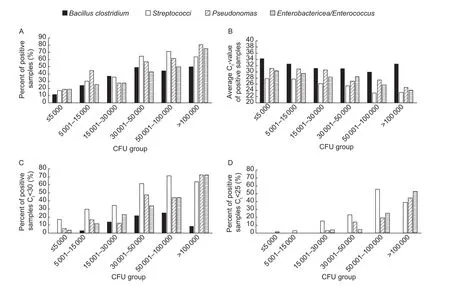
Fig. 1 A, percent of positive samples. B, average Ct-value for positive samples. C, percent Ct-value under 30. D, percent Ct-value under 25 for the different groups of colony forming units (CFUs) for each of the four different targets in the qPCR test the TBC 4(DNA Diagnostic, Denmark).
Of the four targets investigated by the TBC 4 qPCR test,Pseudomonas,StreptococciandEnterobacteriacea/Enterococcusseems to have the highest influence on the CFU in BTM collected during March and April, 2017 in Denmark. This is seen in the Fig. 1-B where the low Ct-values for these targets in samples with CFU mL–1>30 000 indicates higher number of bacteria. We foundEnterobacteriacea/Enterococcus,Pseudomonas, andStreptococciin high number of bacteria (Ct<25, Fig. 1-D) in 25, 19 and 56% of samples with CFU mL–1between 50 001–100 000 and 53, 44, and 39% in samples with CFU mL–1>100 000, respectively. Holmet al. (2004) found, in Danish BTM samples with >30 000 CFU mL–1, microorganisms primarily associated with poor hygiene dominated the microflora in 64% of the samples; bacteria also related to poor hygiene, but in addition associated with growth at low temperatures (psychrotrophic bacteria) dominated the microflora in 28% of the samples; and bacteria mainly associated with mastitis dominated the microflora in 8% of the samples. Their findings for microorganisms, primarily associated with poor hygiene and psychrotropic bacteria,corresponds with our findings forEnterobacteriacea/EnterococcusandPseudomonas, whereas our data indicate much more problems related to mastitis bacteria.In contrary to the data from Holmet al. (2004), our test do not detectStaphylococcibut the mastitis primer detectsStreptococci. On the other hand, theStreptococciprimer can also detectStreptococcinot so often related to mastitis as e.g.,Strep.bovis.
Our finding, thatStreptococciis an important factor in high TBC, is in accordance with the findings of Keefeet al. (1997). Also Gillespieet al. (2012) and Hayeset al. (2001) found a strong correlation between SPC andStreptococcusspp. counts. Katholmet al. (2012) found the best correlation between TBC in bulk tank milk and Ctvalues from real-time PCR assays specific forEnterococcus,Strep.uberisandStrep.agalactiae, less correlation to Ct-values forStrep.dysgalactiae,EscherichiacoliandKlebsiella, and no correlation toStaph.aureus.These findings are in agreement with Zadokset al. (2004), who found thatStreptococci,Staphylococci, and Gram-negative bacteria account for 69, 3, and 3% of total bacterial count,respectively. Our finding, that theEnterobacteriacea/Enterococcusis an important finding in milk samples with high CFU is in accordance with the results from Pyz-Lukasiket al. (2015), who tested the microbiological quality of milk sold directly from producers to consumers in Poland. They foundEnterobacteriaceaeranging from 6.4×10 to 1.7×106CFU mL–1.
5. Conclusion
The new TBC 4 qPCR test proved to be useful in indicating the major causes of high TBC in Danish BTM samples.We expect the test to be a strong and fast tool for farmers,advisors and service technicians to address problems with high TBC and ensuring the delivery of good quality milk to the dairy.
Acknowledgements
The authors thank ARLA Food Amba, Denmark for providing us with the bulk tank samples and the CFU count. The authors thank Sven Erik Sørensen, Eurofins, Denmark for collecting the bulk tank samples.
Costello M, Rhee M S, Bates M P, Clark S, Luedecke O, Kang D H. 2003. Eleven-year trends of microbiological quality in bulk tank milk.Food Protect Trends,23, 393–400.
Danish Agriculture and Food Council. 2017. Danish Dairy Statistics. 2016. [2017-04-21]. http://www.lf.dk/tal-oganalyser/statistik/mejeri/mejeristatistik/mejeristatistik-2016
Elmoslemany A M, Keefe G P, Dohoo I R, Jayarao B M. 2009.Risk factors for bacteriological quality of bulk tank milk in Prince Edward Island dairy herds. Part 1: Overall risk factors.Journal of Dairy Science,92, 2634–2643.
Gillespie B E, Lewis M J, Boonyayatra S, Maxwell M L, Saxton A, Oliver S P, Almeida R A. 2012. Short communication:Evaluation of bulk tank milk microbiological quality of nine dairy farms in Tennessee.Journal of Dairy Science,95,4275–4279.
Guterbock M, Blackmer P E. 1984. Veterinary interpretation of bulk-tank milk.Veterinary Clinnics of North America: Large Animal Practice,6, 257–268.
Hayes M C, Ralyea R D, Murphy S C, Carey N R, Scarlett J M, Boor K J. 2001. Identification and characterization of elevated microbial counts in bulk tank raw milk.Journal of Dairy Science,84, 292–298.
Holm C, Jepsen L, Larsen M, Jespersen L. 2004. Predominant microflora of downgraded danish bulk tank milk.Journal of Dairy Science,87, 1151–1157.
Kable M E, Srisengfa Y, Laird M, Zaragoza J, McLeod J,Heidenreich J, Marco M L. 2016. The core and seasonal microbiota of raw bovine milk in tanker trucks and the impact of transfer to a milk processing facility.Mbio,7, e00836-16.
Katholm J, Bennedsgaard T W, Koskinen M T, Rattenborg E. 2012. Quality of bulk tank milk samples from Danish dairy herds based on real-time polymerase chain reaction identification of mastitis pathogens.Journal of Dairy Science,95, 5702–5708.
Keefe G P, Dohoo I R, Spangler E. 1997. Herd prevalence and incidence ofStreptococcus agalactiaein the dairy industry of prince edward island.Journal of Dairy Science,80, 464–470.
Murphy S C. 1997. Raw milk bacteria tests: Standard plate count, preliminary incubation count, lab, pasteurization count and coliform count. What do they mean for your farm? In:Regional Meeting Proceedings. National Mastitis Council, Syracuse, NY, Madison, WI. pp. 34–41.
Pantoja J C F, Reinemann D J, Ruegg P L. 2011. Factors associated with coliform count in unpasteurized bulk milk.Journal of Dairy Science,94, 2680–2691.
Pyz-Lukasik R, Paszkiewicz W, Tatara M R, Brodzki P, Belkot Z. 2015. Microbiological quality of milk sold directly from producers to consumers.Journal of Dairy Science,98,4294–4301.
Rattenborg E, Paulrud C O, Jensen S K S, Katholm J. 2015. Bulk tank milk surveillance for contagious mastitis pathogens:Comparison of two commercial real-time PCR test kits. In:Proceedings of the 54th Annual Meeting of the National Mastitis Council. National Mastitis Council In., Madison,WI. pp. 215–216.
Schukken Y H, Gunther J, Fitzpatrick J, Fontaine M C, Goetze L, Holst O, Leigh J, Petzl W, Schuberth H J, Sipka A, Smith D G E, Quesnell R, Watts L, Yancey R, Zerbe H, Gurjar A,Zadoks R N, Seyfert H M, 2011. Pfizer mastitis research consortium. Host-response patterns of intramammary infections in dairy cows.Veterinary Immunology and Immunopathology,144, 270–289.
Vacheyrou M, Normand A C, Guyot P, Cassagne C, Piarroux R, Bouton Y. 2011. Cultivable microbial communities in raw cow milk and potential transfers from stables of sixteenFrench farms. International Journal of Food Microbiology,146, 253–262.
Zadoks R N, Gonzàlez R N, Boor K J, Schukken Y H. 2004.Mastitis-causingStreptococciare important contributors to bacterial counts in raw bulk tank milk.Journal of Food Protection,67, 2644–2650.
杂志排行
Journal of Integrative Agriculture的其它文章
- Improve access to the EU market by identifying French consumer preference for fresh fruit from China
- Management and prevention of mastitis: A multifactorial approach with a focus on milking, bedding and data-management
- Elimination of ceftiofur hydrochloride residue in postpartum cows’milk after intramammary infusing at dry-off
- Prevalence and characteristics of extended spectrum β-lactamaseproducing Escherichia coli from bovine mastitis cases in China
- Evolutionary analysis of plant jacalin-related lectins (JRLs) family and expression of rice JRLs in response to Magnaporthe oryzae
- Molecular mapping of YrTZ2, a stripe rust resistance gene in wild emmer accession TZ-2 and its comparative analyses with Aegilops tauschii
