Prediction of protein structure of novel protein (116 kDa) from human sperm membrane
2018-06-04mieLestariWidodoSutimanBambangSumitro
U mie Lestari, Widodo, Sutiman Bambang Sumitro
1Laboratory of Molecular Biology, Department of Biology, State University of Malang, Malang, East Java, Indonesia
2Biology Department, Faculty of Mathematics and Natural Sciences, Brawijaya University, Malang, Indonesia
1. Introduction
Sperm membrane has proteins that are specific and immunogenic[1].Membrane proteins in the sperm anterior play a role in their interaction with the egg, especially with protein ZP3. Membrane proteins that are located on the equatorial head play a role in spermegg membrane fusion[2]. In general, a protein is located in the membrane largely as the result of biosynthesis by the spermatozoa themselves, so it is believed by some researchers that this specific protein is solely owned by a sperm[3,4]. Due to the properties of the sperm membrane protein as mentioned above, it has promising potential for immunocontraception.
The developed immunocontraception is a protein isolated from membranes of human sperm with the use of Tween-20 detergents,which is a protein with a molecular weight of 116 kDa. By using two dimensional electrophoresis to specify isoelectric point (pI) of the protein, it has been established that 116 kDa protein has a pI
of 4.4[5,6]. By staining with fluorescence rhodamine and through observation using a Confocal Laser Scanning Microscope (Olympus FV-1000, Argon Laser), protein 116 kDa-seen expressed in the inner acrosomal membrane, posterior head, neck and midpiece in human sperm-most likely plays a role in fertilization.
Information on the 116 kDa protein isolated from human sperm membrane is not yet available on a database. Hence, to understand the functional biology of protein molecules 116 kDa more accurately,especially its fertilization function, it is necessary to study the threedimensional structure of this protein. A three-dimensional structure will be able to provide an overview of proteins in the cell membrane[7,8].A technique with a web-based program that makes a comparison with the existing protein structure may predict the structure of 116 kDa protein.
2. Materials and methods
2.1. Protein identification
A protein on the sperm membrane with the characteristics of 116 kDa. pI 4.4 protein-kinase activity was previously identified[14]. And the protein-kinase activity was not performed. The protein was then predicted using “TagIdent tool” software[9]. Analysis was performed based on pI and molecular weight as well as the properties of nonkinase proteins in the UniProtKB/TrEMBL database.
2.2. Protein structure and function
Three-dimensional protein structures were modeled using SWISSMODEL, the software modeling for three-dimensional structure of proteins that runs using the principle of fully automated protein structure homology-modeling server, and is accessible via the ExPASy web server[10]. The protein model was visualized using Discovery Studio Viewer software. The function of the protein is subsequently tracked through UNIPROT database and Superfamily 1.7 in the Hidden Markov Model (HMM) library and genome assignment server. HMM is a software for predicting the biological function of proteins based on the domain function of the structure[11].
2.3. Orientation of cell membrane proteins
The orientation of proteins in a cell membrane was predicted with orientations of proteins in membranes server. This software calculates the rotational and translational positions of transmembrane and peripheral proteins in membranes based on the three-dimensional structure of the protein. It can be applied to newly determined experimental or theoretical models of protein structures. Many examples of proteins that exist in the Protein Data Bank have been recalculated using this orientations of proteins in membranes software[12]. Subsequently, protein models were visualized using Discovery Studio Viewer software.
3. Results
Predictions for the 116 kDa protein sequences were performed through comparative analysis of the proteins in the UniProtKB/TrEMBL (48,744,721) database. The 116 kDa protein had pI of 4.4;the molecular weight of 116 kDa and its properties as a non-protein kinase in human sperm membrane were expected to have similarities with CDH23, which had a pI of 4.38 and a molecular weight of 116.612. pI varies for different proteins, based on the difference in the constituent amino acids, whereas each amino acid had its own characteristics. Hence, the similarity of the pI owned by 116 kDa protein with that of CDH23 protein, indicated that there may be similarities in its physical and biochemical properties. Amino acid sequences of CDH23 were shown in Figure 1.

Figure 1. The amino acid sequences of CDH23 (Q8N5B3_HUMAN) from UniProtKB/TrEMBL database that similar with the isolated protein from head of sperm.
Three-dimensional model of the protein was built using SWISSMODEL (Basel, Switzerland) based on amino acid sequences of CDH23 (Figure 2). The protein structure showed the transmembrane form with the extracellular part which was the N-terminal, while the part in the cytoplasm was the C-terminal. The extracellular N-terminal played a role in the receptor. Based on the biological function of CDH23, there were similarities with the function of 116 kDa protein of sperm as shown in Table 1.
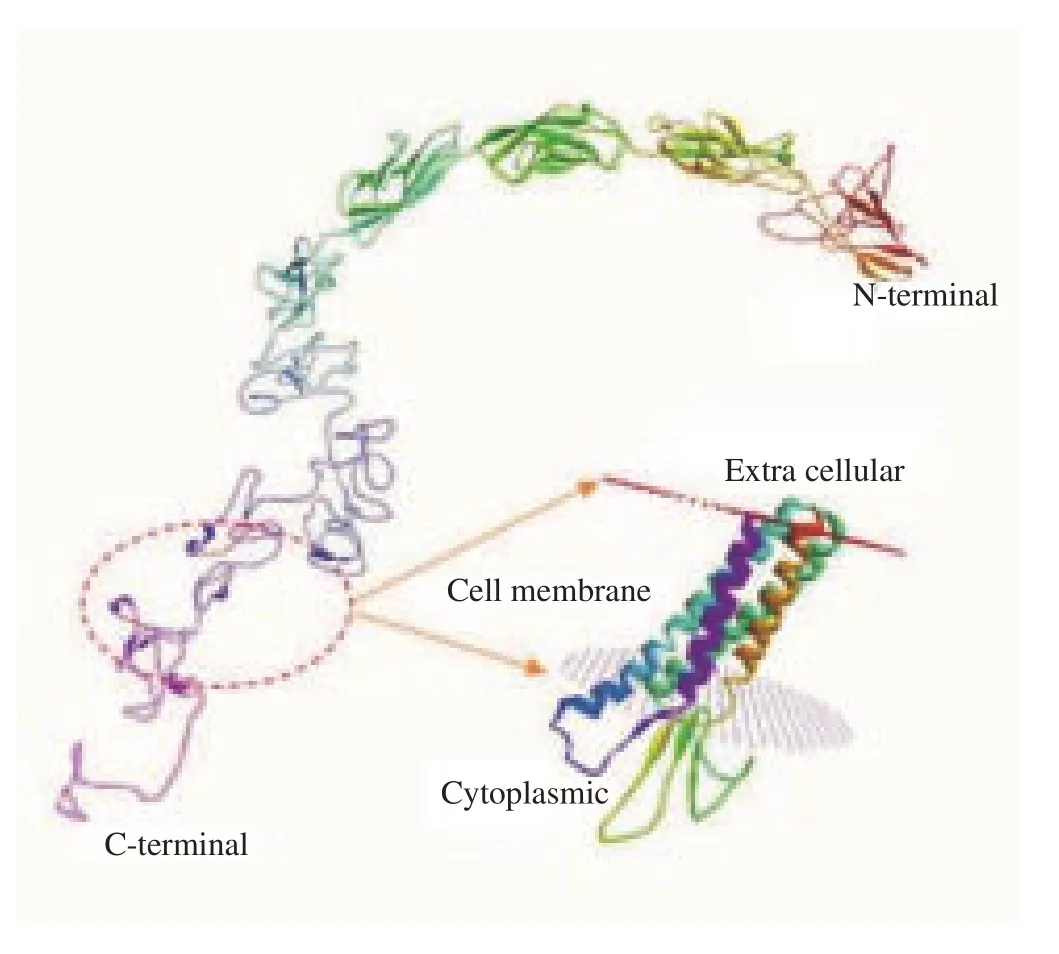
Figure 2. The 3D structure of CDH23 protein made using SWISS-MODEL.
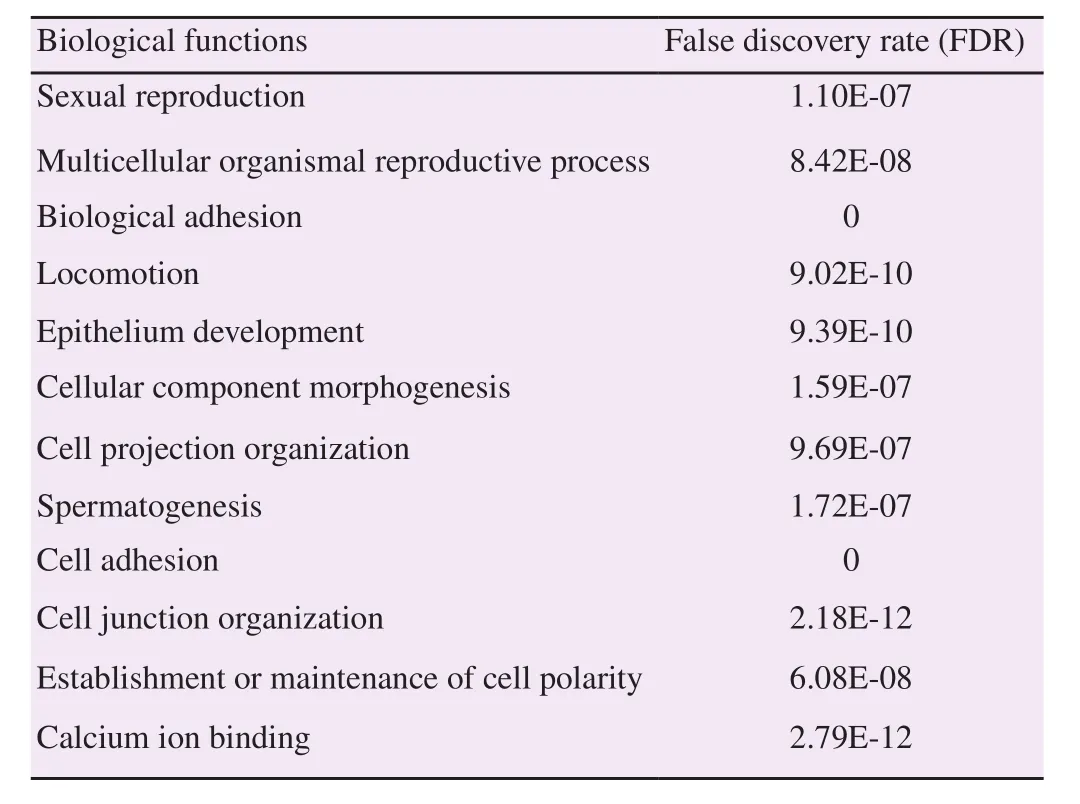
Table 1Biological functions of CDH23 (Q8N5B3).
4. Discussion
According to the UniProtKB/TrEMBL (48,744,721) database,CDH23 serves to bind with other proteins, or in cell recognition,and most likely plays a role in the fertilization process and the introduction of sperm with cells around it, either in the uterus or ovum. When compared with the 116 kDa protein, there is a similarity in the properties to the CDH23 protein. The 116 kDa protein might be expressed at the human sperm head (acrosome) and neck (midpiece). The 116 kDa protein is likely to play a role in the fertilization process. It is known that the membrane protein engages with the ZP3 protein in the anterior part during the interaction process, and that this process also likely involves outer acrosome membrane proteins, which will fuse with the membrane of the spermatozoa through exocytosis[13].
The CDH23 protein was found in cells that have mobility equal to 9.021 E-10, whereas 116 kDa protein found in human sperm has high mobility along the reproductive tract. The results of immunohistochemistry using a polyclonal antibody of 116 kDa protein showed that 116 kDa protein was not found in human cells that are immobile, such as the pancreas, kidney, spleen, prostate,heart, liver, brain and blood vessels[14]. This data suggested that the protein only expressed specifically in spermatozoa may be important for recognition or fertilization process. The CDH23 was found in the several tissues, but there has been no information concerning about the protein found in the spermatozoa. Therefore, in the study,this prediction is novel information that the 116 kDa protein from spermatozoa membrane is CDH23. Further research to elucidate the information that the 116 kDa is CDH23 is necessary to be done.
Based on the in silico analysis, it is suggested that 116 kDa protein found in sperm has similar characteristics to the CDH23 protein.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Acknowledgments
The authors thank the State University of Malang and Brawijaya University for facilitating this research.
[1] Naz RK. Antisperm contraceptive vaccines: Where we are and where we are going? Am J Reprod Immunol 2011; 66(1): 5-12.
[2] Young C, Grasa P, Coward K, Davis LC, Parrington J. Phospholipase C zeta undergoes dynamic changes in its pattern of localization in sperm during capacitation and the acrosome reaction. Fertil Steril 2009; 91(5):2230-2242.
[3] Bowles J, Koopman P. Retinoic acid, meiosis and germ cell fate in mammals. Development 2007; 134: 3401-3411.
[4] Dadoune JP, Siffroi JP, Alfonsi MF. Transcription in haploid male germ cells. Int Rev Cytol 2004; 237: 1-56.
[5] Umie L, Aulanni’am, Basuki BP, Sutiman BS. Human sperm protein 116 kDa: A candidate antingen for immunocontraception technology. J Biol Res 2013; 18(2): 86-90.
[6] Baker MA, Witherdin R, Hetherington L, Cunningham-Smith K, Aitken RJ. Identification of post-translational modifications that occur during sperm maturation using difference in two-dimensional gel electrophoresis.Proteomics 2005; 5(4): 1003-1012.
[7] Kemege KE, Hickey JM, Lovell S, Battaile KP, Zhang Y, Hefty PS. Ab initio structural modeling of and experiment validation for chlamydia trachomatis protein CT296 reveal structural similarity to Fe(II)2-oxoglutarate-dependent enzymes. J Bacteriol 2011; 193(23): 6517-6528.
[8] Bill RM, Henderson PJF, Iwata S, Kunji ERS, Michel H, Neutze R, et al.Overcoming barriers to membrane protein structure determination. Nat Biotechnol 2011; 29: 335-340.
[9] Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD,et al. Protein identification and analysis tools on the ExPASy server. In:Walker JM, editor. The proteomics protocols handbook. New York: Humana Press; 2005, p. 571-607.
[10] Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 2014; 42: 252-258.
[11] Cheema J, Basu G. MAPS: An interactive web server for membrane annotation of transmembrane protein structures. Indian J Biochem Biophys 2011; 48: 106-110.
[12] Lomize MA, Pogozheva ID, Joo H, Mosberg HI, Lomize AL. OPM database and PPM web server: Resources for positioning of proteins in membranes. Nucleic Acids Res 2012; 40: 370-376.
[13] Wassarman PM, Jovine L, Litscher ES. A profile of fertilization in mammals. Nat Cell Biol 2001; 3(2): 59-64.
[14] Lestari U. Spermatozoa binding constrains towards the goat oocyte zoona pellucida by antibodies induced from 116 kDa protein human spermatozoa membrane. In: Proccessings of the 3rd international conference and workshop on basic and applied science: Enabling research innovation on sciences and technology to meet global challenges. Indonesia; 2012.
杂志排行
Asian Pacific Journal of Reproduction的其它文章
- Missed estradiol determination resulting in oocyte retrieval and embryo development following controlled ovarian hyperstimulation at early pregnancy: Case report
- Role of preputial washing in reducing microbial load and improving bovine semen quality
- Effect of different concentration of fish oil in skim milk-egg yolk extenders on postthawed semen qualities of Kalang swamp buffalo bull
- Antiandrogenic activity of Calotropis procera latex in rats
- Improvement of cortical granules migration and in vitro embryo production of vitrified bovine oocyte by 9-cis retinoic acid
- Effect of alcohol infusion of Cissus populnea root on testicular function and serum hormone of male Wistar rats
