Isolation and identification of microorganism affecting eating-qualityin fermented fresh rice noodle
2018-05-02陈兰煊杨有望
陈兰煊 - 杨有望 - 周 慧
佟立涛2TONG Li-tao2 周素梅2ZHOU Su-mei2 易翠平1YI Cui-ping1
Fermented rice noodles are a popular staple food in South China and Southeast Asia. They are generally manufactured by fermentingindicarice naturally. This is then ground to a slurry, steamed to form a gel then finally extruded into different types of vermicelli (Fu, 2008). For traditional production, fermentation occurs spontaneously when whole polished rice grains are steeped in water at ambient temperature (10~40 ℃) for 2~6 d then fermented by the indigenous microbiota. The physicochemical changes in the rice induced by microbial fermentation have been shown to improve their texture and mouthfeel (Tan, Li, Tan, 2009). Rice fermentation relies on a complex microflora where both functional microorganisms and spoilage or pathogenic microorganisms, such as lactic acid bacteria (LAB) and yeasts, can breed (Lu, Peng, Cao, Tatsumi, & Li, 2008). This multiple ecosystem leads to variable and unstable qualities and thus a short shelf-life for the rice noodles.
Using a starter culture to initiate the rice fermentation process is a well-practiced and attractive method for produ-cing food with consistent qualities on a large scale. A starter culture should contain at least one definite microbial preparation with a high cell density, which should adapt well to the environment of rice fermentation and thereby create an unfriendly environment for undesirable spoilage and pathogenic microorganisms (Hong, Chen, Liu, Wu, Tan, Xie, Xu, Zou, Yu, Wang, & Qin, 2016). The starter culture should also improve the shelf-life, sensory quality, and safety of the product (Rubio, Jofré, Martín, Aymerich, & Garriga, 2014). The development of effective starter cultures relies on isolating and identifying the predominant microorganisms having the best technological, probiotic, and economic characteristics (Zanirati, Abatemarco, Sandes, Nicoli, & Nunes, 2015). Yeasts and LAB both have a long history of safety and technological use in fermented foods. For exam-ple, yeasts are widely used as a starter in manufacturing pancakes, bread, and in amylolytic fermentation (Aidoo, Rob Nout, & Sarkar, 2006). LAB are also used as starter cult-ures for manufacturing many fermented foods, particularly dairy products such as cheese and fermented milks. LAB fermentation can generate organic acids, carbon dioxide, exopolysaccharides and diacetyl that contribute to the texture, aroma and shelf-life of fermented foods (Leroy & De Vuyst, 2004; Settanni, Ventimiglia, Alfonzo, Corona, Miceli, & Moschetti, 2013).
The aim of the study is to evaluate the effect of specific microorganisms after isolation on the quality of rice noodles, to identify the predominant microorganisms present in the spontaneously fermented liquid, and to characterize these microorganisms using their technical properties. This will enable the selection of the most suitable starter cultures for producing fermented rice noodles.
1 Material and methods
1.1 Isolation of microorganisms
In three factories (Hunan, China), untreated rice grains were steeped in water at 40 ℃ for 3.5 d to ferment spontaneously. We sampled supernatants from the process after 0, 1, 2, 3, and 3.5 d of fermentation. The samples were then serially diluted with a sterile saline solution (0.9% NaCl) then plated on two types of medium:
(1) Yeast extract peptone dextrose (YPD) agar plates (yeast extract, 10 g/L; peptone, 20 g/L; dextrose, 20 g/L; agar, 18 g/L), with 0.2 g/L chloramphenicol (Ruibio, Darmstadt, Germany), were incubated at 30 ℃ for 48 h before isolation and evaluation of the yeasts. Yeast colonies showing the typical cell appearance under a microscope were randomly selected and labeled with a code (CSY 01 to CSY 41). After purifying five times with successive sub-culturing, the yeasts were maintained on YPD agar slant culture-medium, stored at 4 ℃ and transferred monthly.
(2) MRS agar plates, with 0.1 g/L cycloheximide (Sigma-Aldrich, St Louis, MO, USA), were incubated at 30 ℃ for 48 h before isolation and evaluation of lactic acid bacteria. LAB were randomly selected and labeled with a code (CSL 01 to CSL 60). After purifying five times with successive sub-culturing, LAB were maintained on MRS agar slant culture-medium with added CaCO3at 20 g/L then stored at 4 ℃ and transferred monthly.
Before each test, the isolated LAB and yeasts were grown up to 7 lg CFU/mL in MRS and YPD broth, respectively.
1.2 Technological characterization
1.2.1 Screening for enzyme activity The activities of the enzymes were tested as follows: catalase as described by Gamero-Sandemetrio, Gómez-Pastor, & Matallana (2013),β-glucosidase as described by Caridi, Pulvirenti, Restuccia, Sidari, Mediterranea, Francesco, Agrarie, & Emilia (2005), lipase as described by Kouker & Jaeger (1987),α-amylase as described by Strauss, Jolly, Lambrechts, & Van Rensburg(2001), and proteolytic enzymes as described by Saran, Isar, & Saxena (2007). The enzymatic activity was expressed as: “-” (no activity), “+” (weak activity) and “++” (strong activity).
1.2.2 Growth at different pH values and temperatures
Sterile tubes, containing 10 mL broth, were adjusted to pH values of 3.0, 4.0, 5.0, 6.0, and 7.0 using HCl, inoculated with approximately 5 lg CFU/mL of yeasts or LAB then incubated at 30 ℃ for 48 h. In addition, sterile tubes containing 10 mL of broth (pH 6.0) were inoculated with approximately 5 lg CFU/mL of yeasts or LAB then incubated at 10, 25, 40 ℃ for 48 h.
Microbial growth at 48 h was measured after being diluted 100 times at 600 nm using an ultraviolet-visible (UV-Vis) spectrophotometer (UV-2800; Unico, Shanghai, China). The absorbance value was used to define the growth ability (GA) of isolated isolates under different conditions, with GA values classified as follows:
GA< 0.1: no growth ability;
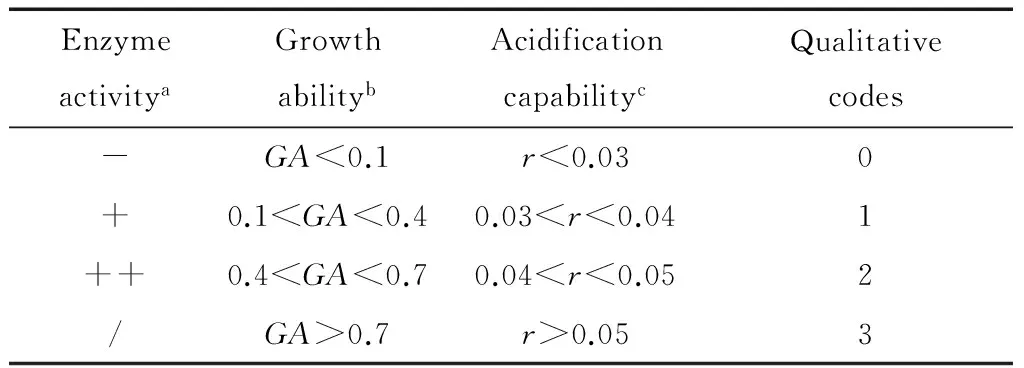
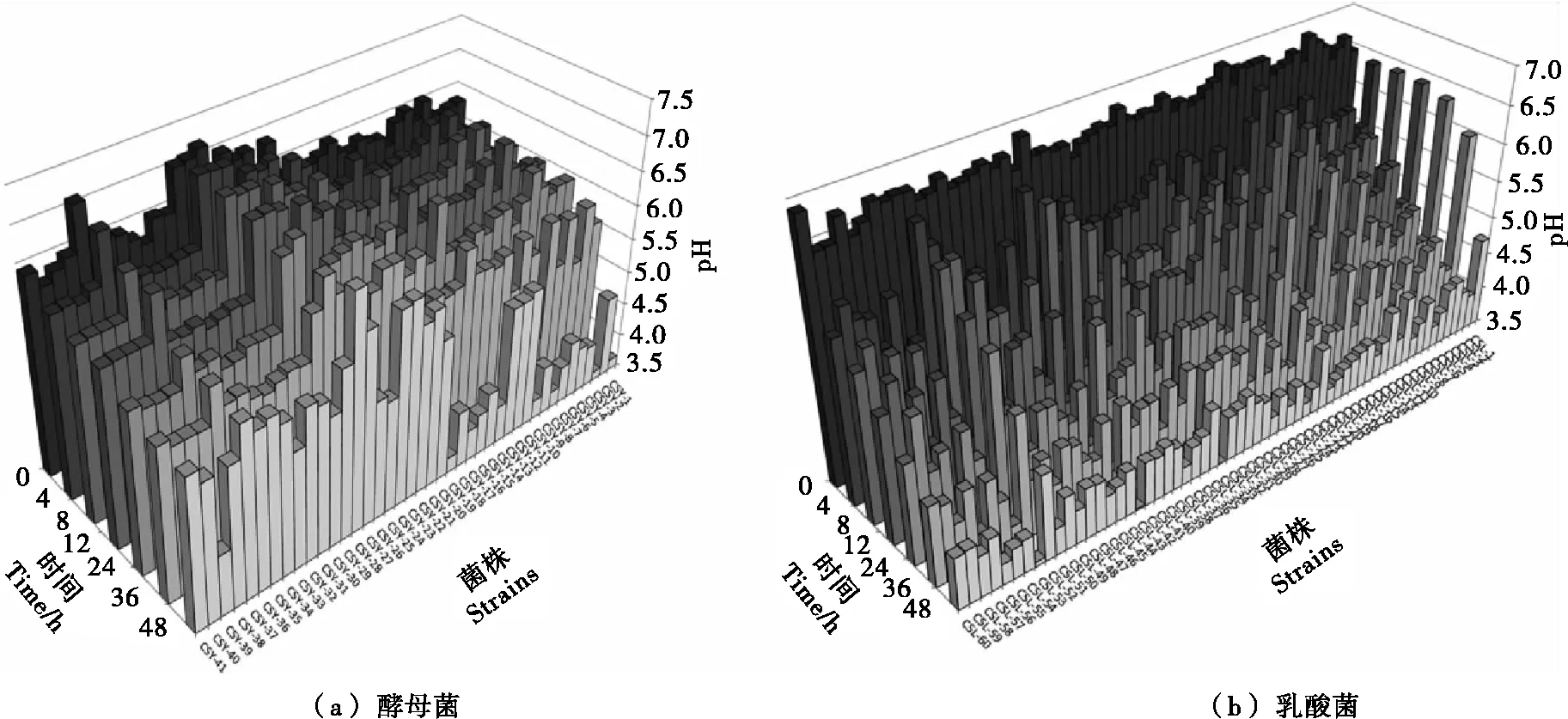
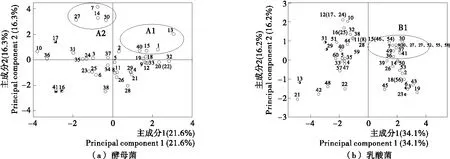
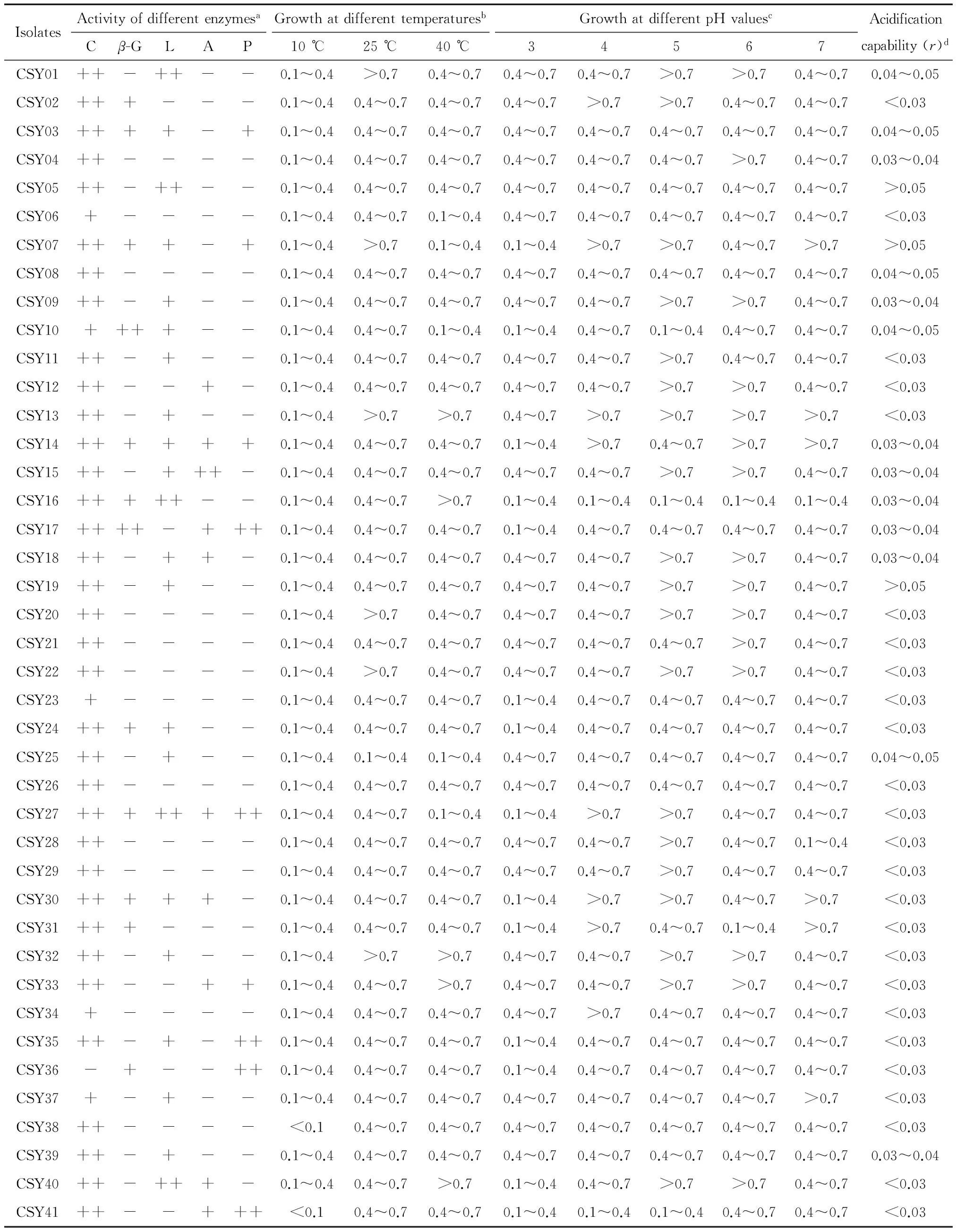
0.1 0.4 GA> 0.7: strong growth ability. 1.2.3 Acidification capability in sterile rice extracts (SRE) Sterile rice extract (SRE) broth was prepared as follows: 200 gindicarice was added to 800 mL sterile water, boiled for 20 min so that the rice was well cooked, then filtered using eight layers of gauze. The filtered liquor was diluted with sterile water to 1 000 mL then sterilized at 121 ℃ for 20 min for use in the subsequent experiments. The yeasts and LAB cultures, grown overnight in YPD and MRS broth, respectively, were collected by centrifuga-tion at 4 500 ×gfor 5 min, washed with Ringer’s solution, then re-suspended in the same solution to give an absorbance of 1.00 at 600 nm measured using a UV-Vis spectrophotometer (UV-2800) (Alfonzo, Ventimiglia, Corona, Di Gerlando, Gaglio, Francesca, Moschetti, & Settanni, 2013). A 2 mL sample of the cell suspension was incubated in 150 mL of SRE at 30 ℃. The pH was measured after 0, 4, 8, 12, 24, 36, and 48 h of incubation. The acidification capability of each microorganism was calculated using one-variable linear regression to assess the relationship between the time and the pH value. The absolute values of the regression coefficient (r) represent the acidification capability as follows: r< 0.03: no acidification capability; 0.03 0.04 r> 0.05: strong acidification capability. 1.3.1 ITS (Internal Transcribed Spacer) sequence analysis of yeasts The genomic DNA of the selected yeasts was extracted using an extraction kit (Sangon Biotec, Shanghai, China) according to the manufacturer’s procedure. The ITS-5.8s-ITS regions of the genomic DNA were amplified from the total DNA using the universal forward primer ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and the reverse primer ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) (Bellemain, Carlsen, Brochmann, Coissac, Taberlet, & Kauserud, 2010). The PCR was performed in a 50 μL reaction system containing 5 μL 10×PCR buffer, 0.5 μL dNTPs (10 mmol/L each), 10 ng genomic DNA, 0.5 μL Forward Primer (50 mmol/L), 0.5 μL Reverse Primer (50 mmol/L), 0.5 μL 5 U/μL Taq DNA Polymerase (Thermo, Shanghai, China). The PCR conditions were as follows: 95 ℃ for 5 min; 35 cycles at 95 ℃ for 30 s, 54 ℃ for 30 s, 72 ℃ for 40 s; and finally, 72 ℃ for 10 min. The PCR product was verified by agarose gel electrophoresis and purified using the Gel Extraction Kit (Sangon Biotec). The PCR-amplified DNA for each sample was sent to a commercial company (Sangon Biotec) for sequencing. The sequences obtained were compared with reference sequences from the GenBank Database using the BLAST program so that the most similar relative sequence could be determined. All sequences were aligned using Clustal X2.1 software (www.clustal.org). The phylogenetic tree was constructed by the MEGA program (version 6.06) using the maximum likelihood method with the Tamura-Nei model (www.megasoftware.net). 1.3.2 16S rRNA gene sequence analysis of LAB The 16S rRNA gene regions of the selected LAB were amplified from the genomic DNA using the universal forward primer 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and reverse primer 1492R (5′-TACGGTTACCTTGTTACGACTT-3′) (Heilig, Zoetendal, Vaughan, Marteau, Akkermans, & DE VOS, 2002). The PCR conditions were as follows: 95 ℃ for 5 min; 35 cycles at 95 ℃ for 30 s, 51 ℃ for 30 s, 72 ℃ for 90 s; and finally, 72 ℃ for 10 min. The other procedures were the same as those for the ITS sequence analysis assay of yeasts. 1.3.3 Nucleotide sequence accession numbers The sequences in this study were deposited in the GenBank database: 16S rRNA sequences under the accession numbers, KT800388-KT800406 and ITS sequences under the accession numbers, KT800407-KT800417. 1.4.1 Preparation of rice noodles Yeasts and LAB were inoculated in YPD and MRS broth, respectively, then incubated overnight at 40 ℃. Under aseptic condition, 5 mL of the overnight cultures were transferred into 500 mL Erlenmeyer flasks containing 300 mL sterile water and 250 g earlyindicarice. The flasks were sealed with eight layers of gauze and maintained at 40 ℃ for 48 h. Meanwhile, rice was steeped in tap water with no starter culture and held at 40 ℃ for 48 h to serve as the control group. The fermented rice granules were then freeze dried in a vacuum freeze-drying oven (Beijing Boyikang Instruments Co., Ltd., Beijing, China), milled using a universal grinder (Taisite, Tianjin, China), then passed through a 100 mesh sieve. The rice noodles were prepared as described by Singh & Kaur (2010) with minor modifications. Briefly, 5 g of the dry rice flour was mixed with water at a ratio of 1∶7 (g/mL), then cooked for 5 min in a water bath at 95 ℃. Then, 95 g of dry rice flour was mixed with the cooked flour and kneaded thoroughly by hand for 5 min to obtain a uniform dough with a moisture content of about 40%. The surface of the dough was gelatinized by steaming for 90 s, then extruded into vermicelli (1.5 mm diameter) using a cylindrical hand extruder. The extruded noodles were immersed in boiling water for 30~40 s, cooled by immersion in tap water for 3 min, and then drained for 2 h. 1.4.2 Tensile strength and texture profile analysis (TPA) Semi-dry rice noodles were boiled in water for 3 min, cooled in tap water for 1 min, then drained and evaluated immediately. The tensile strength and texture profile analysis (TPA) of the noodles were evaluated using a TA-XT plus texture analyzer (Stable Micro Systems Ltd., Godalming, UK). The tensile strength of the noodles was tested at a speed of 3.0 mm/s. The textural properties of the cooked noodles were measured using a P/36R probe with the following parameters: deformed 50% at a speed of 1 mm/s then paused for 5 s. The textural characteristics, hardness, adhesiveness, and cohesiveness, were obtained. Ten tests were made on each sample. 1.4.3 Determination of cooking qualities The cooking qualities were determined as described by Cham & Suwannaporn (2010). Briefly, 5 g of semi-dry rice noodles were boiled in 150 mL distilled water for 6 min, drained for 5 min then weighed. The cooking water was collected then dried in an air oven at 105 ℃ to a constant weight of residue. The cooking loss was expressed as the ratio of the weight of residue to the original noodle weight. The cooked weight was expressed as the ratio of the increased weight of cooked rice noodle to the original weight of the noodle on a semi-dry basis. Three tests were made on each sample. The statistical analysis was conducted using the Statis-tical Package for Social Sciences (Version 19.0; SPSS Inc., Chicago, IL, USA). Principal component analysis (PCA) was used to investigate the differences in technological traits between the isolates. The results of enzyme activity, growth value, and acidification ability were converted into four qualitative codes, 0, 1, 2,and 3 (Table 1), then used for the multivariate analysis. The data on the quality of the rice noodles, expressed as mean±standard deviation, were analyzed using ANOVA. The Duncan test was used for investigating differences between means, being deemed significant where P<0.05. 表1 主成分分析定性参数†Table 1 Qualitative codes used for principal component analysis † a. Catalase,β-glucosidase, lipase,α-amylase, and proteolytic enzyme activities. b. Growth ability at various pH values and temperatures. c. Acidification capability in sterile rice extract (SRE). Overall, 41 yeast and 60 LAB isolates were isolated from the spontaneous fermentation of rice. The technological characteristics of these isolates were then tested. Of the 41 yeast isolates (Table 2), 40 possessed catalase activity, with activity being strong (++) in 36 isolates and weak (+) in 4 isolates.β-glucosidase activity was detected in 12 isolates: 10 isolates with weak and 2 with strong (CSY 10 and 17) activity. Of the remainder, 22 isolates exhibited lipase activity (17 with weak and 5 with strong activity), 10 isolates showed α-amylase activity (9 with weak and 1 with strong activity), and 9 isolates showed proteolytic activity (4 with weak and 5 with strong activity). The growth ability of the isolates was defined by their growth value. An incubation temperature of 10 ℃ could inhibit yeast growth, indicated byGAvalues below 0.1 for CSY 38 and 41. However, there was no significant difference between the growth of yeast isolates at 25, 40 ℃. Regarding pH, growth was generally inhibited at pH 3 and 7, but eight isolates (CSY 01, 07, 13, 14, 15, 27, 30, and 40) showed highGAvalues at pH 3, 4, 5, 6, and 7. Only 2 isolates (CSY 07 and 19) reduced the pH of SRE below 5.0 after 36 h. After 48 h, 6 isolates (CSY 01, 03, 08, 10, 17, and 19) acidified the broth to a pH below 4.0. Finally, the technological traits of the yeast isolates were analyzed using PCA. A biplot of principal component 1 (variability of 21.6%) and principal component 2 (variability of 16.3%) was constructed [Figure 1(a)]. Two important groups, A1 and A2, could be distinguished according to the position of the variables on the PCA map. The group A1 included the isolates CSY 01, 13, 15, and 40, showing high growth at pH 3, 5, and 6; the group A2 included the isolates CSY 07, 14, 27, and 30 showing high growth at pH 4 and 7. Apart from these two groups, some interesting isolates were observed (Table 2): CSY 16 had the trait of strong lipase activity, CSY 17 the traits of strongα-amylase, proteolytic, andβ-glucosidase activities, and CSY 41 the trait of strong proteolytic enzyme activity. Therefore, the isolates, CSY 01, 07, 13, 14, 15, 16, 17, 27, 30, 40, and 41, were selected for the next phase of analysis. The results of the technological characterization of LAB are shown in Table 3. Regarding extracellular enzymes, 23 isolates exhibited no catalase activity, with activity being strong (++) for 30 isolates and weak (+) for 7 isolates. Proteolytic andα-amylase activities were only detected in 3 isolates (CSL 13, 29,and 31);β-glucosidase activity was detected in 13 isolates (3 with weak and 10 with strong activity, with the strongest values exhibited by isolates CSL 06 and 23). Otherwise, 36 isolates possessed lipase activity (25 with weak and 11 with strong activity). Regarding growth ability at different temperatures, 31 isolates showed strong growth ability (GA> 0.7) at 40 ℃ and 10 isolates at 25 ℃. At 10 ℃, all isolates exhibited low growth ability (GA<0.4). Regarding the effect of pH, 49 isolates showed high growth at pH 4, 5, 6, and 7 but at pH 3, the growth of all isolates was inhibited. The SRE pH fell below 5.0 in 8 isolates after 4 h and in 21 isolates after 12 h. After 48 h, all isolates had acidified the broth to a pH below 4.0 and 31 isolates to a pH below 3.0. 图1 酵母菌和乳酸菌在大米提取物培养基(SRE)中的培养时的pH值变化Figure 1 Kinetics of acidification in sterile rice extracts (SRE) of isolated yeasts and lactic acid bacteria The technological traits of the LAB isolates were also analyzed using PCA. A biplot of principal component 1 (variability of 34.1%) and principal component 2 (variability of 16.2%) was constructed[Figure 1(b)]. According to the position of the variables on the PCA map, the B1 group, consisting of CSL 07, 09, 15, 20, 27, 30, 36, 37, 41, 46, 49, 54, 55, and 58, was distinguished by showing a strong growth ability at pH values of 3, 4, 5, 6, and 7 and temperatures of 10, 25, and 40 ℃. Other interesting isolates were also identified: CSL 13, 29, and 31 possessed strongα-amylase and proteolytic enzyme activities and CSL 06 and 23 had strong lipase andβ-glucosidase activities. Thus, these technologically relevant isolates, CSL 06, 07, 09, 13, 15, 20, 23, 27, 29, 30, 31, 36, 37, 41, 46, 49, 54, 55, and 58, were selected for further analysis. The genotype and phylogenetic trees of the technologi-cally relevant isolates were identified and are shown in Figure 2(a) and Figure 2(b). For yeasts, the isolates, CSY 01, 15, 17, and 40, were classified asLachanceaspp with one isolate, CSY 13, identified asS.cerevisiae. Otherwise, the isolates CSY 07, 14, 16, 27, 30, and 41, showed a high degree of similarity and could be classified asTrichos-poronspp. 图2 酵母菌和乳酸菌发酵性能主成分分析图Figure 2 Principal component analysis (PCA) of the technological traits of yeasts and lactic acid bacteria表2 自然发酵大米分离的酵母菌发酵性能参数†Table 2 Technological characteristics of the yeast isolates from spontaneously fermented rice IsolatesActivityofdifferentenzymesaCβ-GLAPGrowthatdifferenttemperaturesb10℃25℃40℃GrowthatdifferentpHvaluesc34567Acidificationcapability(r)dCSY01++-++--0.1~0.4>0.70.4~0.70.4~0.70.4~0.7>0.7>0.70.4~0.70.04~0.05CSY02+++---0.1~0.40.4~0.70.4~0.70.4~0.7>0.7>0.70.4~0.70.4~0.7<0.03CSY03++++-+0.1~0.40.4~0.70.4~0.70.4~0.70.4~0.70.4~0.70.4~0.70.4~0.70.04~0.05CSY04++----0.1~0.40.4~0.70.4~0.70.4~0.70.4~0.70.4~0.7>0.70.4~0.70.03~0.04CSY05++-++--0.1~0.40.4~0.70.4~0.70.4~0.70.4~0.70.4~0.70.4~0.70.4~0.7>0.05CSY06+----0.1~0.40.4~0.70.1~0.40.4~0.70.4~0.70.4~0.70.4~0.70.4~0.7<0.03CSY07++++-+0.1~0.4>0.70.1~0.40.1~0.4>0.7>0.70.4~0.7>0.7>0.05CSY08++----0.1~0.40.4~0.70.4~0.70.4~0.70.4~0.70.4~0.70.4~0.70.4~0.70.04~0.05CSY09++-+--0.1~0.40.4~0.70.4~0.70.4~0.70.4~0.7>0.7>0.70.4~0.70.03~0.04CSY10++++--0.1~0.40.4~0.70.1~0.40.1~0.40.4~0.70.1~0.40.4~0.70.4~0.70.04~0.05CSY11++-+--0.1~0.40.4~0.70.4~0.70.4~0.70.4~0.7>0.70.4~0.70.4~0.7<0.03CSY12++--+-0.1~0.40.4~0.70.4~0.70.4~0.70.4~0.7>0.7>0.70.4~0.7<0.03CSY13++-+--0.1~0.4>0.7>0.70.4~0.7>0.7>0.7>0.7>0.7<0.03CSY14++++++0.1~0.40.4~0.70.4~0.70.1~0.4>0.70.4~0.7>0.7>0.70.03~0.04CSY15++-+++-0.1~0.40.4~0.70.4~0.70.4~0.70.4~0.7>0.7>0.70.4~0.70.03~0.04CSY16+++++--0.1~0.40.4~0.7>0.70.1~0.40.1~0.40.1~0.40.1~0.40.1~0.40.03~0.04CSY17++++-+++0.1~0.40.4~0.70.4~0.70.1~0.40.4~0.70.4~0.70.4~0.70.4~0.70.03~0.04CSY18++-++-0.1~0.40.4~0.70.4~0.70.4~0.70.4~0.7>0.7>0.70.4~0.70.03~0.04CSY19++-+--0.1~0.40.4~0.70.4~0.70.4~0.70.4~0.7>0.7>0.70.4~0.7>0.05CSY20++----0.1~0.4>0.70.4~0.70.4~0.70.4~0.7>0.7>0.70.4~0.7<0.03CSY21++----0.1~0.40.4~0.70.4~0.70.4~0.70.4~0.70.4~0.7>0.70.4~0.7<0.03CSY22++----0.1~0.4>0.70.4~0.70.4~0.70.4~0.7>0.7>0.70.4~0.7<0.03CSY23+----0.1~0.40.4~0.70.4~0.70.1~0.40.4~0.70.4~0.70.4~0.70.4~0.7<0.03CSY24++++--0.1~0.40.4~0.70.4~0.70.1~0.40.4~0.70.4~0.70.4~0.70.4~0.7<0.03CSY25++-+--0.1~0.40.1~0.40.1~0.40.4~0.70.4~0.70.4~0.70.4~0.70.4~0.70.04~0.05CSY26++----0.1~0.40.4~0.70.4~0.70.4~0.70.4~0.70.4~0.70.4~0.70.4~0.7<0.03CSY27++++++++0.1~0.40.4~0.70.1~0.40.1~0.4>0.7>0.70.4~0.70.4~0.7<0.03CSY28++----0.1~0.40.4~0.70.4~0.70.4~0.70.4~0.7>0.70.4~0.70.1~0.4<0.03CSY29++----0.1~0.40.4~0.70.4~0.70.4~0.70.4~0.7>0.70.4~0.70.4~0.7<0.03CSY30+++++-0.1~0.40.4~0.70.4~0.70.1~0.4>0.7>0.70.4~0.7>0.7<0.03CSY31+++---0.1~0.40.4~0.70.4~0.70.1~0.4>0.70.4~0.70.1~0.4>0.7<0.03CSY32++-+--0.1~0.4>0.7>0.70.4~0.70.4~0.7>0.7>0.70.4~0.7<0.03CSY33++--++0.1~0.40.4~0.7>0.70.4~0.70.4~0.7>0.7>0.70.4~0.7<0.03CSY34+----0.1~0.40.4~0.70.4~0.70.4~0.7>0.70.4~0.70.4~0.70.4~0.7<0.03CSY35++-+-++0.1~0.40.4~0.70.4~0.70.1~0.40.4~0.70.4~0.70.4~0.70.4~0.7<0.03CSY36-+--++0.1~0.40.4~0.70.4~0.70.1~0.40.4~0.70.4~0.70.4~0.70.4~0.7<0.03CSY37+-+--0.1~0.40.4~0.70.4~0.70.4~0.70.4~0.70.4~0.70.4~0.7>0.7<0.03CSY38++----<0.10.4~0.70.4~0.70.4~0.70.4~0.70.4~0.70.4~0.70.4~0.7<0.03CSY39++-+--0.1~0.40.4~0.70.4~0.70.4~0.70.4~0.70.4~0.70.4~0.70.4~0.70.03~0.04CSY40++-+++-0.1~0.40.4~0.7>0.70.1~0.40.4~0.7>0.7>0.70.4~0.7<0.03CSY41++--+++<0.10.4~0.70.4~0.70.1~0.40.1~0.40.1~0.40.4~0.70.4~0.7<0.03 † a. Catalase (C),β-glucosidase (β-G), lipase (L),α-amylase (A) and proteolytic enzyme (P) activities; b. Growth of yeasts incubated at different pH values; c. Growth of yeasts incubated at different temperatures; d. Acidification capability of yeasts in sterile rice extract (SRE). 表3 自然发酵大米分离的乳酸菌发酵性能参数†Table 3 Technological characteristics of LAB isolates from spontaneously fermented rice 续表3 IsolatesEnzymesaCβ-GLAPTemperatureb10℃25℃40℃pHc34567Acidificationcapability(r)dCSL51++-+--<0.10.1~0.40.1~0.4<0.1>0.70.4~0.7>0.70.4~0.7>0.05CSL52-----0.1~0.40.4~0.7>0.70.4~0.7>0.7>0.7>0.7>0.70.04~0.05CSL53-+++--<0.1>0.7>0.70.4~0.7>0.7>0.7>0.7>0.70.04~0.05CSL54+----0.1~0.40.4~0.7>0.70.4~0.7>0.7>0.7>0.7>0.70.04~0.05CSL55-----0.1~0.40.4~0.7>0.70.4~0.7>0.7>0.7>0.7>0.70.04~0.05CSL56-++++--<0.1>0.7>0.70.4~0.7>0.7>0.7>0.7>0.70.04~0.05CSL57++-+--<0.10.1~0.4>0.7<0.1>0.7<0.1>0.7<0.10.04~0.05CSL58-----0.1~0.40.4~0.7>0.70.1~0.4>0.7>0.7>0.7>0.70.04~0.05CSL59++-+--<0.10.1~0.4>0.70.1~0.4>0.70.4~0.7>0.7>0.70.04~0.05CSL60++-+--<0.10.1~0.40.1~0.4<0.1>0.7>0.7>0.70.1~0.40.04~0.05 † a. Catalase (C),β-glucosidase (β-G), lipase (L),α-amylase (A) and proteolytic enzyme (P) activities; b. Growth of lactic acid bacteria incubated at different pH values; c. Growth of lactic acid bacteria incubated at different temperatures; d. Acidification capability of lactic acid bacteria in sterile rice extract (SRE). Regarding LAB(Figure 3), two species could be classified: the isolates CSL 06 and 23 were identified as from theL.plantarumgroup and the isolates CSL 07, 09, 13, 15, 20, 27, 29, 30, 31, 36, 37, 41, 46, 49, 54, 55, and 58 asL.fermentum, but CSL 13 and 15 were a relatively long evolutionary distance from the others. 图3 采用最大似然法(ML)构建的酵母菌ITS-5.8s-ITS序列的系统发育树和乳酸菌16S RNA gene序列的系统发育树Figure 3 Phylogenetic tree based on the ITS-5.8s-ITS sequences of isolated yeasts and the complete 16S rRNA gene sequences of isolated LAB using the maximum likelihood (ML) method Thus, isolates with interesting technological traits and antimicrobial activity were selected: CSY 01 possessed a strong acidification capability compared with the others from theLachanceaspp. group; CSY 07 had stronger extracellular enzymes activity, higher growth at different pH values and temperatures, and a stronger acidification capacity than others from theTrichosporonspp. group; in theL.fermentumgroup, CSL 30 showed higher quality technological traits and more significant antimicrobial activities, but CSL 13 and 15 may be subspecies, and may thus be selected for further tests as particular isolates; CSL 23 possessed a stronger antimicrobial activity than CSL 06 inLactobacillusplantarum. Thus, the isolates CSY 01, 07, and 13 as well as CSL 13, 15, 23, and 30, were selected for fermenting the rice noodles. The textural and cooking qualities of rice noodles fermented using the different isolates were significantly different (P<0.05) (Table 4). The tensile strength of cooked rice noodles fermented by CSL 23 and 30 was significantly higher than those fermented by the other isolates. The hardness of noodles fermented by CSL 15, 23,and 30 was significantly higher than that of other samples. The cooked noodles fermented by CSL 15 and 23 exhibited lower adhesiveness values than the naturally fermented noodles. The noodles fermented by the yeast isolates (CSY 01, 07, and 13) were less cohesive than the naturally fermented noodles, which were less cohesive than noodles fermented by the LAB isolates. Regarding cooking quality, cooking loss was generally negatively correlated with cooked weight. The noodles fermented by CSY 13 and CLS 23 and 30 had significantly lower cooking losses and higher cooked weight than noodles fermented using the other isolates. 表4 不同菌种发酵鲜湿米粉的质构和蒸煮品质†Table 4 Textural and cooking quality of rice noodles fermented with different starter cultures † Mean values in the same column bearing superscripts with the same letters are not significantly different (P>0.05); n. Natural fermentation without starter culture. A promising strain of microorganism for starter cultures in rice noodle manufacturing must have some desirable traits: functional extracellular enzyme activity, high growth over a wide range of pH values and temperatures, a strong acidification capability and a strong antimicrobial ability. Extracellular enzymatic activity is a critical property for a strain to be used as a starter because it can modify the quality of raw material through producing some metabolites and secondary compounds (Bevilacqua, Corbo, & Sinigaglia, 2012).β-glucosidase can help release secondary compounds which can potentially enhance the flavor of foods (Palmeri & Spagna 2007). Catalase can prevent the oxidation reaction to help improve the quality of the food material (Bevilacqua, Perricone, Cannarsi, Corbo, & Sinigaglia, 2009). Previous studies have shown that changes in rice granules during the fermentation process, such as slight etching of the starch crystal and reduced contents of lipid and protein, can improve the quality of rice noodles (Lu, Li, Min, Wang, & Tatsumi, 2005). Thus, it can be concluded thatα-amylase, lipase, and proteolytic enzymes were active in the isolates examined in the present study. To evaluate the effectiveness of a starter at different pH values and temperatures, the growth ability of isolates was calculated at incubation temperatures of 10, 25,and 40 ℃ and at pH values of 3, 4, 5, 6, and 7. From an industrial point of view, starters with high growth ability under ordinary conditions are easy to culture in large quantities (Bevilacqua, Beneduce, Sinigaglia, & Corbo, 2013). To be relevant to industrial conditions, the assay of growth ability in the present study was conducted under aerobic conditions. As the results detailing the technological characteristics are multifactorial, selecting the most promising isolates cannot be achieved through simple analysis techniques. Principal component analysis can be used to reduce the dimensionality of the data. Based on this multivariate approach, as detailed earlier, 11 yeast isolates and 19 LAB isolates were selected as candidates for preparing improved starters. Determining the sequence of the 16S rRNA gene is an important technique for differentiating prokaryotic microorganisms (Nagpal, Fox, & Fox, 1998). For the fungal kingdom, the ITS sequence has generally been applied for species discrimination and has a more clearly defined barcode gap for fungi (Schoch, Seifert, Huhndorf, Robert, Spouge, Levesque, & Chen, 2012). Based on these methods, the technologically relevant isolates were identified and classified. By combining the technological traits with antimicrobial activity, the promising species were selected for producing fermented rice noodles. Rice noodles of high quality require several special textural and cooking traits: a higher tensile strength, hardness, cohesiveness, and cooked weight, and a lower adhesiveness and cooking loss. Tensile testing assesses the breaking strength of noodles to act as an indicator of their quality (Fari, Rajapaksa, & Ranaweera, 2011). Hardness has been shown to be a dominant factor in the quality perception of rice noodles, being negatively correlated with cooking loss, but positively correlated with cooking weight (Wang, Warkentin, Vandenberg, & Bing, 2014). As rice noodles have low adhesiveness, they are easy to separate from each other. This lower adhesiveness as well as high cohesiveness can improve aspects of mouthfeel, such as increasing the slipperiness of the noodles (Chen, Sagis, Legger, Linssen, Schols, & Voragen, 2002). In the present study, rice noodles fermented by the LAB isolate, CSL 23, exhibited the required qualities of higher tensile strength, higher hardness, lower adhesiveness, higher cohesiveness, lower cooking loss, and higher cooked weight. In the ecological system of fermented foods, the actions of yeasts and LAB cannot be completely separated, as they can interact through molecular, nutritional, and metabolic factors (Frey-Klett, Burlinson, Deveau, Barret, Tarkka, & Sarniguet, 2011). These symbiotic interactions include mutualism, commensalism, and parasitism. Stadie, Anna, Ehrmann, & Vogel (2013) have described the interaction between yeasts and LAB in a water/kefir medium and demonstrated that they both significantly increased cell yield in a co-culture system because of molecular interaction, trophic interaction, and metabolite exchange. Tada, Katakura, Ninomiya, & Shioya (2007) used the symbiotic interactions betweenLactobacilluskefiranofaciensandSaccharomycescerevisiaeto enhance the production of kefiran. In the present study, rice noodles fermented with the yeast isolate, CSY 13, exhibited better qualities of higher tensile strength, higher hardness, lower adhesiveness, and lower cooking loss than noodles fermented by the other yeast isolates. In conclusion, two potential isolates,Saccharomycescerevisiae(CSY 13) andLactobacillusplantarum(CSL 23), have been shown to be the most suitable starter cultures for producing fermented rice noodles. [1] AIDOO K, NOUT M, SARKAR P. Occurrence and function of yeasts in Asian indigenous fermented foods[J]. FEMS Yeast Res, 2006, 6: 30-39. [2] ALFONZO A, VENTIMIGLIA G, CORONA O, et al. Diver-sity and technological potential of lactic acid bacteria of wheat flours[J]. Food Microbiol, 2013, 36(2): 343-354. [3] BELLEMAIN E, CARLSEN T, BROCHMANN C, et al. ITS as an environmental DNA barcode for fungi: an in silico appr-oach reveals potential PCR biases[J]. Bmc Microbiology, 2010, 10(1): 189. [4] BEVILACQUA A, BENEDUCE L, SINIGAGLIA M, et al. Selection of yeasts as starter cultures for table olives[J]. Journal of Food Science, 2013, 78(5): M742-M751. [5] BEVILACQUA A, CORBO M , SINIGAGLIA M. Selection of yeasts as starter cultures for table olives: A step-by-step procedure[J]. Frontiers in Microbiology, 2012, 3: Article 194. [6] BEVILACQUA A, PERRICONE M, CANNARSI M, et al. Technological and spoiling characteristics of the yeast microflora isolated from Bella Di Cerignola table olives[J]. International Journal of Food Science & Technology, 2010, 44(11): 2 198-2 207. [7] CARIDI A, PULVIRENTI A, RESTUCCIA C, et al. Screening for yeasts able to hydrolyse arbutin in the presence of glucose or ethanol[J]. Annals of Microbiology, 2005, 55(55): 43-46. [8] CHAM S, SUWANNAPORN P. Effect of hydrothermal treatment of rice flour on various rice noodles quality[J]. Journal of Cereal Science, 2010, 51(3): 284-291. [9] CHEN Z, SAGIS L, LEGGER A, et al. Evaluation of Starch Noodles Made from Three Typical Chinese Sweet-potato Starches[J]. Journal of Food Science, 2010, 67(9): 3 342-3 347. [10] FARI M J M, RAJAPAKSA D, RANAWEERA K K D S. Quality characteristics of noodles made from selected varieties of Sri Lankan rice with different physicochemical characteristics[J]. Journal of the National Science Foundation of Sri Lanka, 2011, 39(1): 53-60. [11] FREY Klett P, BURLINSON P, DEVEAU A, et al. Bacterial-fungal interactions: hyphens between agricultural, clinical, environmental, and food microbiologists[J]. Microbiology and Molecular Biology Reviews : MMBR, 2011, 75(4): 583-609. [12] FU Bin-xiao. Asian noodles: History, classification, raw materials, and processing[J]. Food Research International, 2008, 41(9): 888-902. [13] GAMERO-Sandemetrio E, GMEZ-Pastor R, MATALLANA E. Zymogram profiling of superoxide dismutase and catalase activities allows Saccharomyces and non-Saccharomyces species differentiation and correlates to their fermentation performance[J]. Appl Microbiol Biotechnol, 2013, 97(10): 4 563-4 576. [14] HEILIG H, ZOETENDAL E G, VAUGHAN E E, et al. Molecular Diversity of Lactobacillus spp. and Other Lactic Acid Bacteria in the Human Intestine as Determined by Specific Amplification of 16S Ribosomal DNA[J]. Appl Environ Microbiol, 2002, 68(1): 114-123. [15] HONG X, CHEN J, LIU L, et al. Metagenomic sequencing reveals the relationship between microbiota composition and quality of Chinese Rice Wine[J]. Scientific Reports, 2016, 6: 26 621. [16] KOUKER G, JAEGER K E. Specific and sensitive plate assay for bacterial lipases[J]. Applied & Environmental Microbiology, 1987, 53(1): 211-213. [17] LEROY F, VUYST L D. Lactic acid bacteria as functional starter cultures for the food fermentation industry[J]. China Dairy Industry, 2004, 15(2): 67-78. [18] LU Z H, PENG H H, CAO W, et al. Isolation, characterization and identification of lactic acid bacteria and yeasts from sour Mifen, a traditional fermented rice noodle from China[J]. Journal of Applied Microbiology, 2008, 105(3): 893-903. [19] LU Zhan-hui, LI Li-te, MIN Wei-hong, et al. The effects of natural fermentation on the physical properties of rice flour and the rheological characteristics of rice noodles[J]. International Journal of Food Science & Technology, 2005, 40(9): 985-992. [20] NAGPAL M L, FOX K F, FOX A. Utility of 16S-23S rRNA spacer region methodology: how similar are interspace regions within a genome and between strains for closely related organisms[J]. Journal of Microbiological Methods, 1998, 33(3): 211-219. [21] PALMERI R, SPAGNA G.β-Glucosidase in cellular and acellular form for winemaking application[J]. Enzyme & Microbial Technology, 2007, 40(3): 382-389. [22] RUBIO R, JOFRÉ A, MARTN B, et al. Characterization of lactic acid bacteria isolated from infant faeces as potential probiotic starter cultures for fermented sausages[J]. Food Microbiology, 2014, 38(4): 303-311. [23] SARAN S, ISAR J, SAXENA R K. A modified method for the detection of microbial proteases on agar plates using tannic acid[J]. Journal of Biochemical & Biophysical Methods, 2007, 70(4): 697-699. [24] SCHOCH CL, SEIFERT KA, HUHNDORF S, et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi[J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(16): 6 241-6 246. [25] LUCA Settanni, GIUSI Ventimiglia, ANTONIO Alfonzo, et al. An integrated technological approach to the selection of lactic acid bacteria of flour origin for sourdough production[J]. Food Research International, 2013, 54(2): 1 569-1 578. [26] SANDHU K S, KAUR M. Studies on noodle quality of potato and rice starches and their blends in relation to their physicochemical, pasting and gel textural properties[J]. LWT-Food Science and Technology, 2010, 43(8): 1 289-1 293. [27] STADIE J, GULITZ A, EHRMANN M A, et al. Metabolic activity and symbiotic interactions of lactic acid bacteria and yeasts isolated from water kefir[J]. Food Microbiology, 2013, 35(2): 92-98. [28] STRAUSS M L, JOLLY N P, LAMBRECHTS M G, et al. Screening for the production of extracellular hydrolytic enzymes by non-Saccharomyces wine yeasts[J]. Journal of Applied Microbiology, 2001, 91(1): 182-190. [29] TADA S, KATAKURA Y, NINOMIYA K, et al. Fed-batch coculture of Lactobacillus kefiranofaciens with Saccharomyces cerevisiae for effective production of kefiran[J]. Journal of Bioscience & Bioengineering, 2007, 103(6): 557-562. [30] TAN Hong-zhuo, LI Zai-gui, TAN Bin. Starch noodles: History, classification, materials, processing, structure, nutri-tion, quality evaluating and improving[J]. Food Research International, 2009, 42(5): 551-576. [31] WANG N, WARKENTIN T D, VANDENBERG B, et al. Physicochemical properties of starches from various pea and lentil varieties, and characteristics of their noodles prepared by high temperature extrusion[J]. Food Research International, 2014, 55(55): 119-127. [32] ZANIRATI D F, JR A M, SANDES S H, et al. Selection of lactic acid bacteria from Brazilian kefir grains for potential use as starter or probiotic cultures[J]. Anaerobe, 2015, 32: 70-76.1.3 Genotypic identification
1.4 Application in the production of rice noodles
1.5 Statistical analysis
2 Results
2.1 Technology characterization
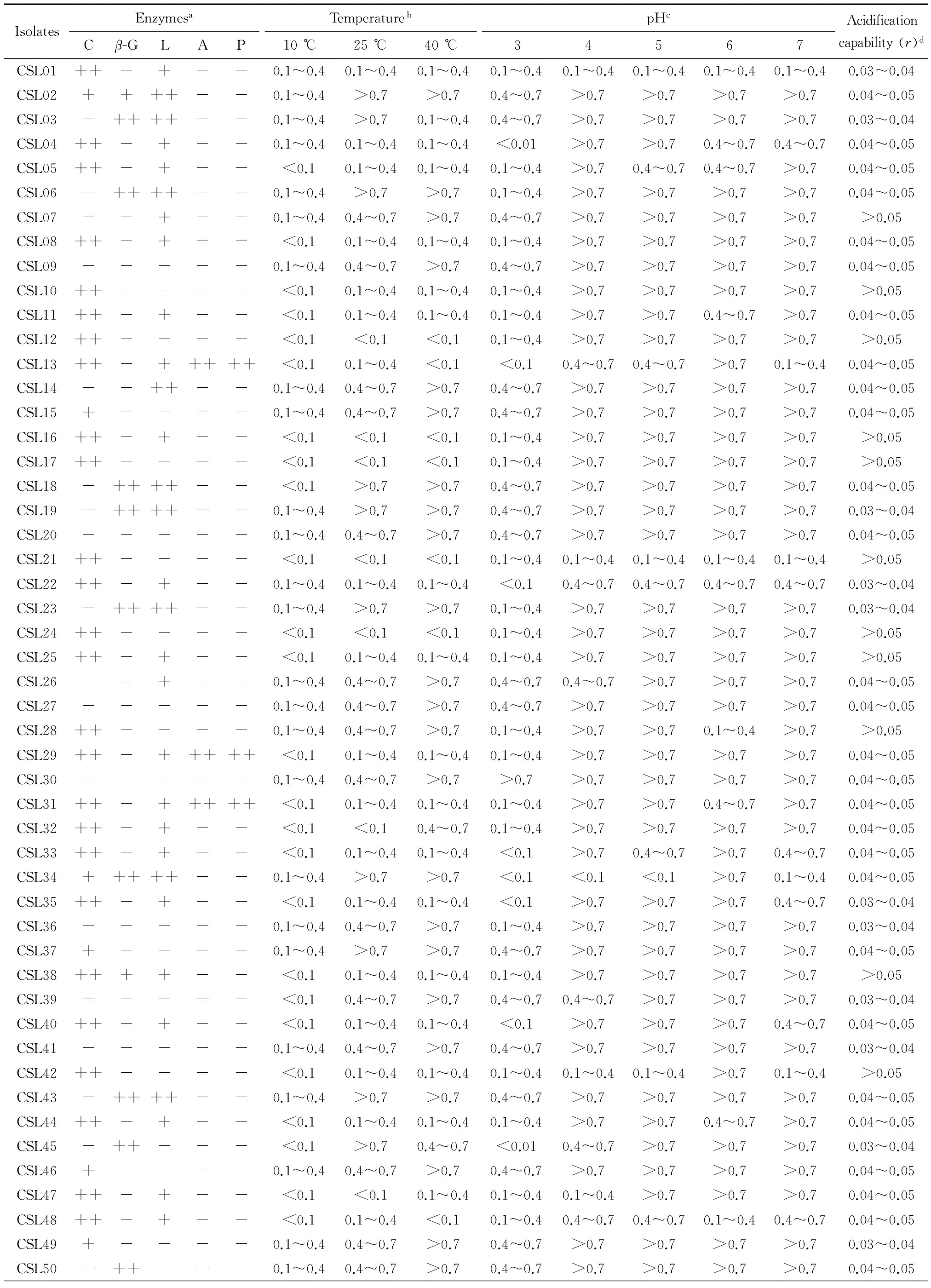
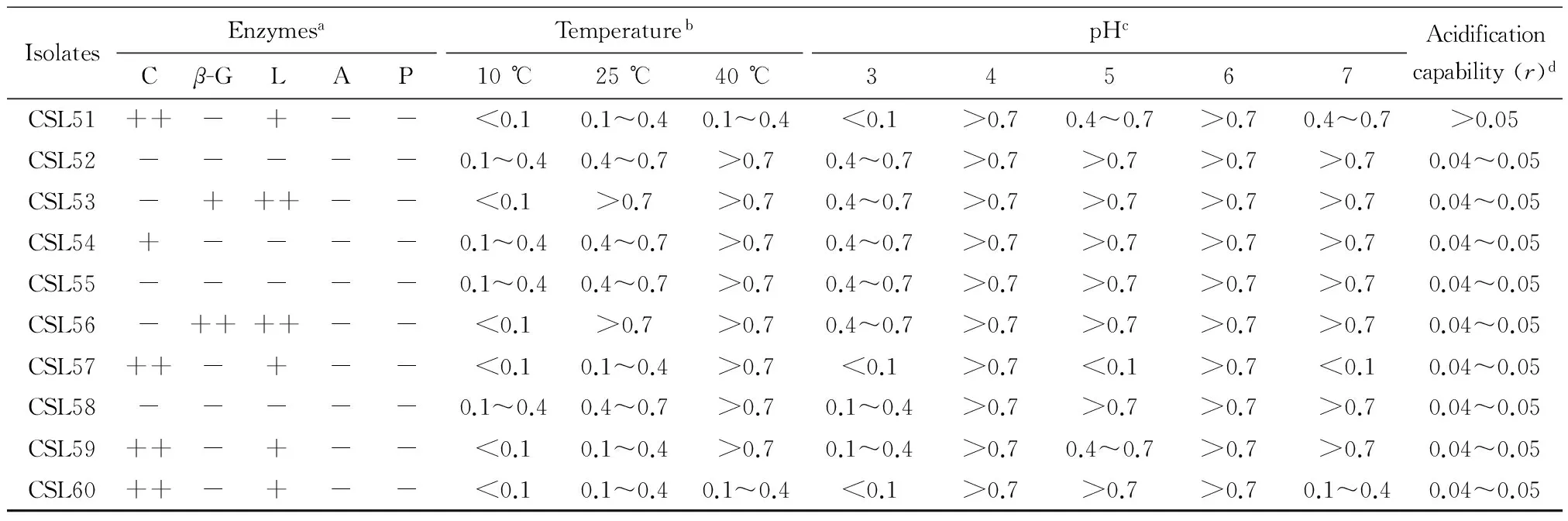
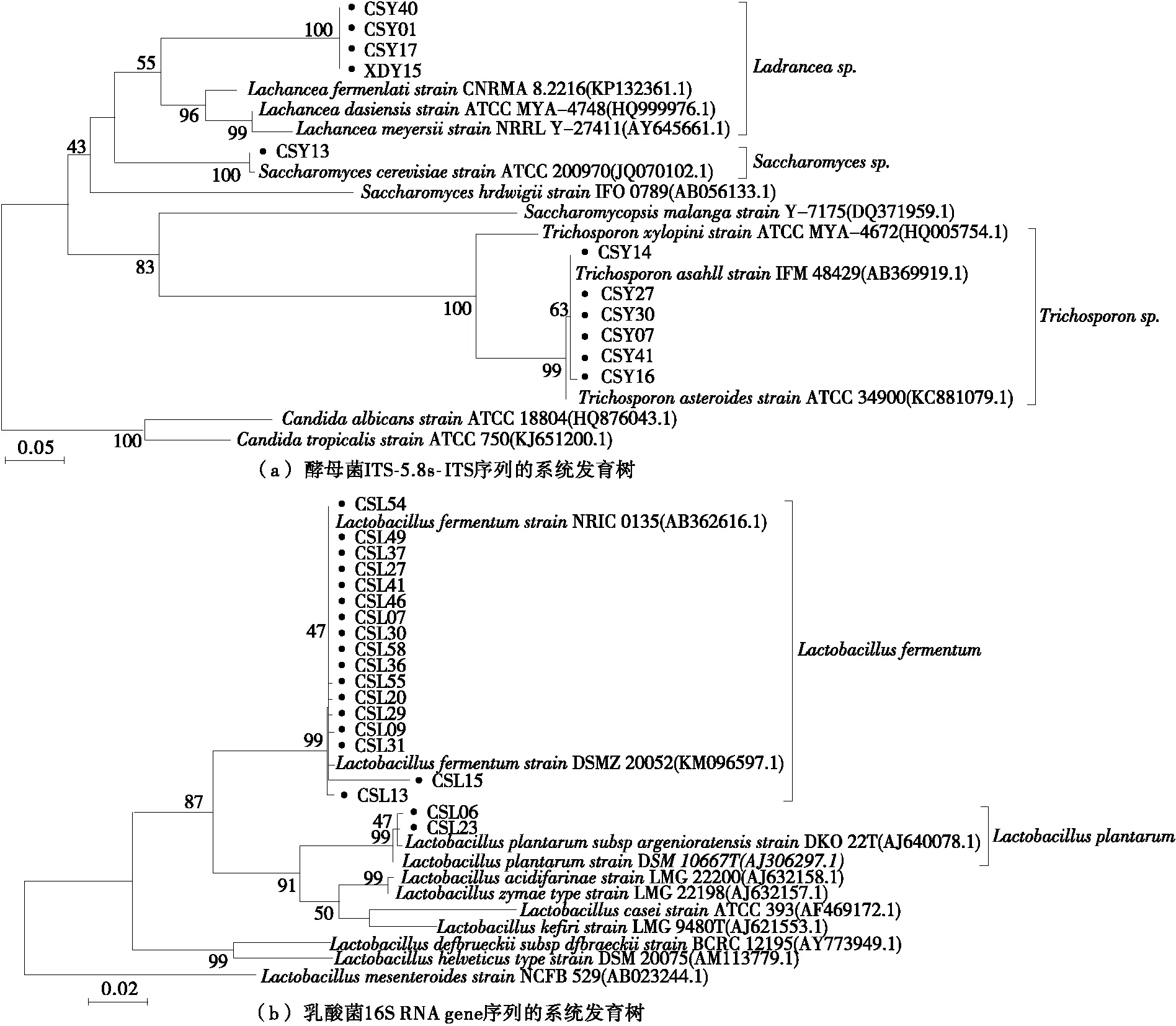
2.2 Genotypic identification
2.3 Quality of rice noodles

3 Discussion
