Complete mitochondrial genome of the leaf muntjac(Muntiacus putaoensis) and phylogenetics of the genus Muntiacus
2017-12-14GuoGangLiMingXiaZhangKyawSwaKyawWinMaungRuiChangQuanSoutheastAsiaBiodiversityResearchInstituteChineseAcademyofSciencesYezinNayPyiTaw058MyanmarCenterforIntegrativeConservationXishuangbannaTropicalBotanicalGardenChinese
Guo-Gang Li, Ming-Xia Zhang, Kyaw Swa, Kyaw-Win Maung, Rui-Chang Quan,*1 Southeast Asia Biodiversity Research Institute, Chinese Academy of Sciences, Yezin Nay Pyi Taw 058, Myanmar Center for Integrative Conservation, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Mengla Yunnan 6660,China Hponkan Razi Wildlife Sanctuary Offices, Putao Kachin 01051, Myanmar Forest Research Institute, Forest Department Ministry of Environmental Conservation and Forestry, Yezin Nay Pyi Taw 058, Myanmar
Complete mitochondrial genome of the leaf muntjac(Muntiacus putaoensis) and phylogenetics of the genus Muntiacus
Guo-Gang Li1,2, Ming-Xia Zhang1,2, Kyaw Swa3, Kyaw-Win Maung4, Rui-Chang Quan1,2,*1Southeast Asia Biodiversity Research Institute, Chinese Academy of Sciences, Yezin Nay Pyi Taw 05282, Myanmar2Center for Integrative Conservation, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Mengla Yunnan 666303,China3Hponkan Razi Wildlife Sanctuary Offices, Putao Kachin 01051, Myanmar4Forest Research Institute, Forest Department Ministry of Environmental Conservation and Forestry, Yezin Nay Pyi Taw 05282, Myanmar
The leaf muntjac (Muntiacus putaoensis) is an endemic deer species found in the east trans-Himalayan region. In recent years, population numbers have decreased due to heavy hunting and habitat loss, and little genetic data exists for this species, thus our knowledge of distribution rangs and population sizes likewise remain limited. We obtained mtDNA genes and the complete mitochondrial genome sequence of M. putaoensis using PCR, followed by direct sequencing. The complete mitogenome sequence was determined as a circular 16 349 bp mitochondrial genome,containing 13 protein-coding genes, two rRNA genes,22 tRNA genes, and one control region, the gene composition and order of which were similar to most other vertebrates so far reported. Most mitochondrial genes, except for ND6 and eight tRNAs, were encoded on the heavy strand. The overall base composition of the heavy strand was 33.1% A,29.3% T, 24.2% C, and 13.4% G, with a strong AT bias of 62.4%. There were seven regions of gene overlap totaling 95 bp and 11 intergenic spacer regions totaling 74 bp. Phylogenetic analyses (ML and BI) among the Muntiacus genus based on the sequenced of mitogenome and ND4L-ND4 supported M. putaoensis as a member of Muntiacus,most closely related to M. vuquangensis. However,when analyses based on cyt b included two more muntjacs, M. truongsonensis was most closely related to M. putaoensis rather than M.vuquangensis, and together with M. rooseveltorum,likely forming a M. rooseveltorum complex of the species. This study will help in the exploration of the evolutionary history and taxonomic status of the leafmuntjac, as well as its protection as a genetic resource.
Muntiacus; Muntiacus putaoensis;Mitogenome; Phylogenetics
INTRODUCTION
Muntjacs (Muntiacus spp., Muntiacinae, Cervidae) are a group of small solitary deer occurring in the forests of Asia. They are of great interest to evolutionary biologists and mammalogists due to their considerable chromosome variations (from 2n=6(Muntiacus reevesi) to 2n=46 (M. muntjak)) (Fontana & Rubini,1990) and discovery of new species in the last decades of the twentieth century, including M. vuquangensis (Tuoc et al., 1994),M. truongsonensis (Giao et al., 1998), and M. putaoensis(Amato et al., 1999; Rabinowitz et al., 1999).1
Muntiacus putaoensis is the most recently discovered species in this muntjac genus. It is named after the town of Putao, located in the most northern part of Myanmar, which is also its most recognizable reference point (Amato et al., 1999).Compared with other species, it is the smallest muntjac (mean adult body mass 12 kg), and is only half the size of the sympatric M. muntjak (22–29 kg) (Rabinowitz et al., 1999).Local people call it the ‘leaf deer’ because it is so small it can be wrapped in a single leaf of Phrynium capitatum (Rabinowitz &Khaing, 1998; Rabinowitz et al., 1999). Until recently,information on the distribution range of the species was limitedto northern Myanmar. However, it has since been found in the adjoining hill forests of southeastern Tibet, with the local name similar in meaning to its Myanmar name (Choudhury, 2007;Choudhury, 2009; Datta et al., 2003; James et al., 2008).Muntiacus putaoensis is endemic to the east trans-Himalayan region (including northern Myanmar and southeastern Tibet,China), the population of the leaf muntjac has decreased dramatically in recent years due to considerable degradation of its natural habitat and heavy hunting pressure (Rabinowitz &Khaing, 1998; Rao et al., 2010; Schaller & Rabinowitz, 2004).This species is listed as data deficient (DD) in the IUCN Red List of Threatened species as there is a lack of certainty about its taxonomy, distribution, population, natural history, and threats (Timmins & Duckworth, 2016). Previously the species was only known to occur along a 70 km east to west stretch in northern Myanmar, the new discovery means a two-fold increase in the total east-west range of this species (James et al., 2008). It also indicates that the extent of its geographic range is still being determined.
The leaf muntjac was characterized and confirmed primarily by its diagnostic mitochondrial DNA (mtDNA) with several partial fragments (Amato et al., 1999). However, its taxonomic status remains controversial as it falls within a group of closely related ‘little’ muntjacs with strong morphological similarities,including M. rooseveltorum and M. truongsonensis (James, et al., 2008; Timmins & Duckworth, 2016). This species is characterized by short thin pedicles, small unbranched antlers,and relatively large preorbital fossa, with both males and females possessing canines (Rabinowitz et al., 1999). Because hunters usually cut off the lower jaw and part of the upper jaw, it is difficult to obtain complete skulls to compare with other muntjacs. In addition, the similarities in morphology also create difficulties when marking comparisons, thus requiring molecular genetic analyses. To date however, genetic data remain scarce,with only partial segments of mtDNA currently sequenced(Amato et al., 1999; James et al., 2008), which are insufficient for confirming the identity of this species or clarifying its phylogenetic relationship relative to other muntjacs.
Here, we sequenced and analyzed the complete mitochondrial genome and two mtDNA segments of M. putaoensis, the aim of this study was to: 1) provide fundamental genetic data for further conservation genetic studies for this near cryptic mammal; 2) discuss the possible relationships among the genus of Muntiacus, especially that of M. putaoensis, M.rooseveltorum, and M. truongsonensis; and, 3) provide further discussion on the evolutionary history of this species.
MATERIALS AND METHODS
Sampling and laboratory procedures
Seventeen M. putaoensis specimens were collected from Putao,Kachin state, northern Myanmar in three field trips (December 2015, December 2016 to January 2017, and May 2017) (Figure 1; Supplementary Table S1). Three skin samples were preserved in 95% ethanol, and 14 dry meat samples were preserved using silica gel for subsequent analyses. Voucher specimens were deposited in the Southeast Asia Biodiversity Research Institute, Chinese Academy of Sciences, in Nay Pyi Taw, Myanmar (Supplementary Table S1).
Total genomic DNA was extracted from tissue using a DNeasy Blood & Tissue Kit (Qiagen, Shanghai, China). The master mixture for polymerase chain reaction (PCR) contained approximately 100 ng of template DNA, 1 μL (10 pmol) of each primer, 5 μL of 10× reaction buffer, 2 μL of dNTPs (2.5 mmol/L of each), and 2.0 U of Taq DNA polymerase, in a total volume of 50 μL. Reactions were carried out on a Veriti Thermal Cycler(Applied Biosystems, Carlsbad, CA, USA) and always included a negative control. The M. putaoensis mtDNA segments (ND4LND4 and cyt b) were amplified using PCR with universal primers from previous studies (Irwin et al., 1991; Wang & Lan,2000). After visualization of the fragments using 1% agarose gel,the PCR products were sequenced from both ends using an ABI PRISM 3700 sequencing system with using the same primers as for PCR (Beijing Tianyi Huiyuan Bioscience and Technology Incorporation, Beijing, China).
To obtain the whole mitogenomic sequence of M. putaoensis,we designed 16 pairs of primers based on previous studies(Hassanin et al., 2012; Larkin et al., 2007; Martins et al., 2017;Shi et al., 2003; Wu & Fang, 2005; Zhang et al., 2004a, b). The primer sequences and approximate lengths of the amplified fragments from the whole genome are listed in Supplementary Table S2. All the primers were synthesized by Beijing Tianyi Huiyuan Bioscience and Technology Incorporation (Beijing, China).
Sequence analyses
Two mitochondrial DNA segments sequences were edited using the program DNASTAR 5.0 (DNASTAR Inc.), and were aligned using the CLUSTALW algorithm implemented in MEGA 5.05 with default parameters (Larkin et al., 2007; Tamura et al., 2011).Positions of ambiguous alignment and those from variable length intergenic regions were excluded from further analyses.Identical haplotypes were collapsed using DNASP 5.1 (Librado &Rozas, 2009). The base composition of the mitogenomic sequence was analyzed using MEGA 5.05 (Tamura et al., 2011).We annotated the genome sequence using DOGMA (Wyman et al., 2004).
Phylogenetic analyses
Phylogenies of the two mtDNA segments and mitogenome were constructed using maximum likelihood (ML) implemented in PHYML 3.0 (Guindon et al., 2010). Bayesian inference (BI)was implemented in MRBAYES 3.2.1 (Ronquist et al., 2012)using different parameter estimates for the two mtDNA segments and mitogenome. The most appropriate nucleotide substitution models for the two segments and genome were selected using the Akaike Information Criterion in JMODELTEST 2.1.4 (Darriba et al., 2012). To assess the statistical significance of the hypothesized lineages, bootstrap analysis with 200 replicates was used for the ML analyses, with other settings set to default. The posterior distributions were obtained by Markov Chain Monte Carlo (MCMC) analysis with one cold chain and three heated chains. Samples of the trees and parameters were drawn every 100 steps from a total of one million MCMC generations. Three additional runs were conducted, beginning with random trees. A 50% majority rule consensus of the postburn (using a burn-in of 25%) for all generations was computed for all four runs. Elaphodus cephalophus and Tragulus kanchil were chosen as outgroups to root the tree.

Figure 1 Maximum likelihood (ML) and Bayesian inference (BI) phylogenetic trees (GTR+G model) for Muntiacus based on 20 complete genomes from mitochondrial DNA
RESULTS AND DISCUSSION
Sequence data and genome content
Cyt b sequences with a total length 1 140 bp were obtained from all 17 M. putaoensis individuals with four variable sites.We obtained 1 844 bp of unambiguous sequences (ND4LND4) from 13 individuals, which contained the complete sequences of NADH dehydrogenase subunit 4L (ND4L),NADH dehydrogenase subunit 4 (ND4), tRNASer, and tRNAHisgenes, and a partial sequence of the tRNALeugene, with 15 variable sites and six parsimony-informative sites. We detected three cyt b haplotypes and eight ND4L-ND4 haplotypes separately (GenBank accession Nos. MF737179–MF737190, Supplementary Table S1).
The complete mitochondrial genome features of M. putaoensis were identical to those of most other muntjac deer (e.g.Hassanin et al., 2012; Shi et al., 2003; Wu & Fang, 2005;Zhang et al., 2004a, b). The complete mitogenome sequence was a circular molecule 16 349 bp in length, and included 13 protein-coding genes (ATP6, ATP8, COI-III, cyt b, ND1-6, and ND4L), two rRNA genes (12S and 16S rRNA), 22 tRNA genes,and one non-coding control region (D-loop), most of which were encoded on the heavy strand, except for ND6 and eight tRNAs(tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer, tRNAGlu,and tRNAPro). Notably, gene overlap and separation, common characteristics found in other vertebrate mitochondrial genomes,were also observed in the M. putaoensis mitogenome. There were seven regions of gene overlap totaling 95 bp (varying from 1 to 40 bp) and 11 intergenic spacer regions totaling 74 bp(varying from 1 to 32 bp) (Table 1). The overall base composition of the heavy strand was 33.1% A, 29.3% T, 24.2%C, and 13.4% G with a strong AT bias of 62.4%. Nearly all 13 protein-coding genes started with the common initiation codon ATG, whereas ND2, ND3, and ND5 started with ATA. In addition,10 protein-coding genes shared the complete stop codons TAA or TAG (TAA for ND1, COI, COII, ATP6, ND4L, ND5, and ND6,and TAG for ND2, ATP8, and ND3), whereas the remaining three protein-coding genes (ND4, COIII and cyt b) possessed incomplete stop codons with a terminal T or TA. The 22 tRNA genes were interspersed along the genome, with lengths varying from 60 to 75 bp. The lengths of 12S rRNA and 16S rRNA were 958 and 1 568 bp, respectively, located between tRNAPheand tRNALeu(UUR)and separated by tRNAVal. The D-loop region was located between tRNAProand tRNAPhe, and was 914 bp in length. The base composition of the D-loop was A-29.9%,T-31.9%, C-23.1%, and G-15.1%, re fl ecting a strong feature in A and T (A+T=61.8%).
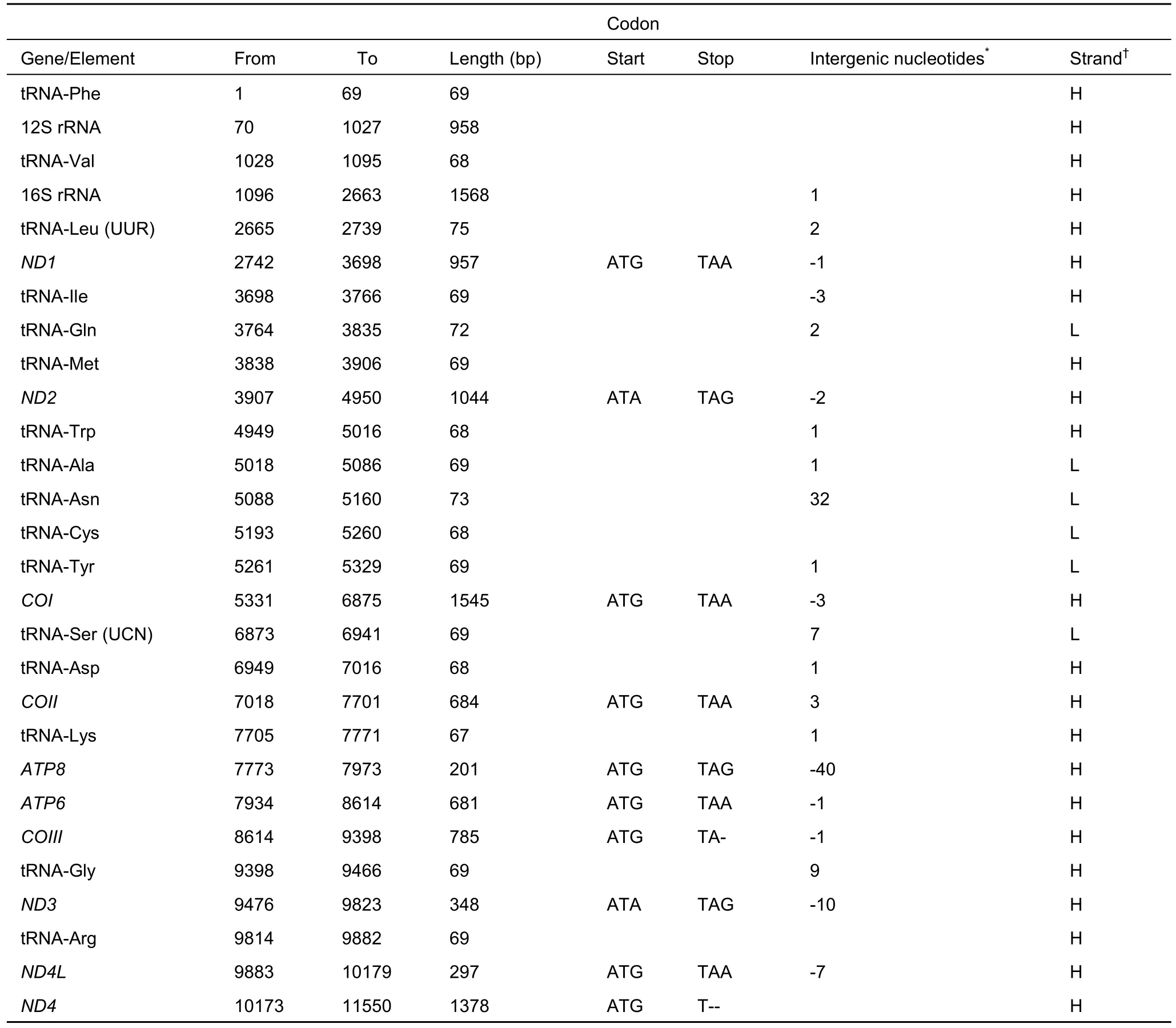
Table 1 Characteristics of the mitochondrial genome of Muntiacus putaoensis

Continued
Phylogenetic analysis
The phylogeny of the genus Muntiacus has long been debated(James et al., 2008; Timmins & Duckworth, 2016). Here, we conducted a phylogenetic analysis using complete mitochondrial genomes from five muntjac species (others have not yet been sequenced for mitochondrial genomes) (Figure 1). Additional sequences and species were included in phylogenetic analysis based on ND4L-ND4 (Figure 2A) and cyt b (Figure 2B). The ML and BI analyses recovered the same tree topologies (Figure 1 and 2).
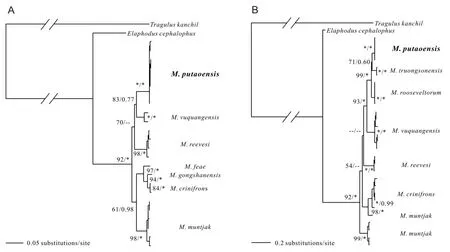
Figure 2 Maximum likelihood (ML) and Bayesian inference (BI) phylogenetic trees (GTR + G model) for Muntiacus based on ND4L-ND4 genes (A) and cyt b gene (B)
The three different level sequences obtained in this study (i.e.,complete mitogenome, ND4L-ND4, and cyt b) provide insight into the taxonomic status of Muntiacus.
Based on the complete genomes (Figure 1), the data showed two major clades within the genus of Muntiacus, one of which included M. putaoensis, M. vuquangensis and M. reevesi, and the other M. crinifrons and M. muntjak. In the latter clade, M.crinifrons was positioned on a distinct branch as sister to M.muntjak. Within M. muntjak, three phylogenetically and geographically distinct groups were previously identified by Martins et al. (2017). We determined the leaf muntjac (M.putaoensis) to be a member of genus Muntiacus, most closely related to M. vuquangensis, with M. reevesi as their sister species. However, as genomic data for other muntjacs is still unavailable, further genomic research on more species may illuminate different relationships within this taxon.
Previous phylogenetic information has been provided for the genus Muntiacus from mtDNA genes (Amato et al., 1999;Wang & Lan, 2000). Based on eight haplotypes of M.putaoensis obtained in this study, combined with the ND4LND4 sequences of six other muntjacs downloaded from GenBank, our phylogenetic results showed M. putaoensis as a member of genus Muntiacus (Figure 2A), consistent with the above mitochondrial genome findings.
However, when M. truongsonensis and M. rooseveltorm were included in analyses based on cyt b (Figure 2B), we found that M. truongsonensis, rather than M. vuquangensis, was most closely related to M. putaoensis. Furthermore, we observed only weak support (bootstrap supports for ML/posterior probability in BI=71/0.60; Figure 2B) among these three species,and that they all likely belonged to the M. rooseveltorum complex of species. This result is consistent with previous morphological studies (James et al., 2008; Timmins & Duckworth,2016).
Complete mitogenome data of M. putaoensis would be fundamental to research phylogentic relationship and conservation genetics of muntjac genus. As the current conclusion is based on analysis of only a few mtDNA fragments, comparison of longer sequences or genomes will be more convincing. More studies based on genome are required to better illuminate wholesome view of phylogenetics and evolution in this extraordinary taxon.
ACKNOWLEDGEMENTS
The authors sincerely thank Prof. Shu-Qiang Li and Dr. Yu Song for their support with the laboratory work. We also thank Mr. Kyaw Kyaw Han and Shuyin Huang for sample collection, thank Ms. Nan Sun and Alice Hughes for proofreading the manuscript. This work was possible because of the support from the Forest Research Institute, Hponkan Razi Wildlife Sanctuary and Hkakaborazi National Park Offices, Myanmar.
Amato G, Egan MG, Rabinowitz A. 1999. A new species of muntjac,Muntiacus putaoensis (Artiodactyla: Cervidae) from northern Myanmar.Animal Conservation, 2(1): 1-7.
Choudhury A. 2007. Discovery of leaf deer Muntiacus putaoensis in Nagaland with a new northerly record from Arunachal Pradesh. Journal of the Bombay Natural History Society, 104: 205-208.
Choudhury A. 2009. Records and distribution of Gongshan and leaf muntjacs in India. Deer Specialist Group News, 23: 2-7.
Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods, 9(8): 772.Datta A, Pansa J, Madhusudan MD, Mishra C. 2003. Discovery of the leaf deer Muntiacus putaoensis in Arunachal Pradesh: An addition to the large mammals of India. Current Science, 84(3): 454-458.
Fontana F, Rubini M. 1990. Chromosomal evolution in Cervidae.Biosystems, 24(2): 157-174.
Giao PM, Tuoc D, Dung VV, Wikramanayake ED, Amato G, Arctander P,MacKinnon JR. 1998. Description of Muntiacus truongsonensis, a new species of muntjac (Artiodactyla: Muntiacidae) from central Vietnam, and implications for conservation. Animal Conservation, 1(1): 61-68.
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O.2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology,59(3): 307-321.
Hassanin A, Delsuc F, Ropiquet A, Hammer C, Van Vuuren BJ, Matthee C,Ruiz-Garcia M, Catzeflis F, Areskoug V, Nguyen TT. 2012. Pattern and timing of diversification of Cetartiodactyla (Mammalia, Laurasiatheria), as revealed by a comprehensive analysis of mitochondrial genomes. Comptes Rendus Biologies, 335(1): 32-50.
Irwin DM, Kocher TD, Wilson AC. 1991. Evolution of the cytochrome b gene of mammals. Journal of Molecular Evolution, 32(2): 128-144.
James J, Ramakrishnan U, Datta A. 2008. Molecular evidence for the occurrence of the leaf deer Muntiacus putaoensis in Arunachal Pradesh,north-east India. Conservation Genetics, 9(4): 927-931.
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA,McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD,Gibson TJ, Higgins DG. 2007. Clustal W and clustal X version 2.0.Bioinformatics, 23(21): 2947-2948.
Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 25(11): 1451-1452.
Martins RF, Fickel J, Le M, Van Nguyen T, Nguyen HM, Timmins R, Gan HM, Rovie-Ryan JJ, Lenz D, Forster DW, Wilting A. 2017. Phylogeography of red muntjacs reveals three distinct mitochondrial lineages. BMC Evolutionary Biology, 17: 34.
Rabinowitz A, Khaing ST. 1998. Status of selected mammal species in North Myanmar. Oryx, 32(3): 201-208.
Rabinowitz A, Myint T, Khaing ST, Rabinowitz S. 1999. Description of the leaf deer (Muntiacus putaoensis), a new species of muntjac from northern Myanmar. Journal of Zoology, 249(4): 427-435.
Rao M, Htun S, Zaw T, Myint T. 2010. Hunting, livelihoods and declining wildlife in the Hponkanrazi Wildlife Sanctuary, North Myanmar.Environmental Management, 46(2): 143-153.
Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Hohna S,Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, 61(3): 539-542.
Schaller GB, Rabinowitz A. 2004. Species of barking deer (genus Muntiacus) in the eastern Himalayan region. The Journal of the Bombay Natural History Society, 101(3): 442-444.
Shi YF, Shan XN, Li J, Zhang XM, Zhang HJ. 2003. Sequence and organization of the complete mitochondrial genome of the Indian muntjac(Muntiacus muntjak). Acta Zoologica Sinica, 49(5): 629-636. (in Chinese)
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011.MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods.Molecular Biology and Evolution, 28(10): 2731-2739.
Timmins R J, Duckworth JW. 2016. Muntiacus putaoensis. The IUCN Red List of Threatened Species 2016: e.T136479A22159478.http://dx.doi.org/10.2305/IUCN.UK.2016-1.RLTS.T136479A22159478.en
Tuoc D, Dung VV, Dawson S, Arctander P, MacKinnon J. 1994. Introduction of a new large mammal species in Vietnam. Science and Technology News,3: 4-12.
Wang W, Lan H. 2000. Rapid and parallel chromosomal number reductions in muntjac deer inferred from mitochondrial DNA phylogeny. Molecular Biology and Evolution, 17(9): 1326-1333.
Wu HL, Fang SG. 2005. Mitochondrial DNA genetic diversity of black muntjac (Muntiacus crinifrons), an endangered species endemic to China.Biochemical Genetics, 43(7-8): 407-416.
Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics, 20(17): 3252-3255.
Zhang HJ, Li J, Shi YF, Zhang XM, Xu CH, Shan XN. 2004a. Complete sequence of black muntjac (Muntiacus crinifrons) mitochondrial genome.Chinese Journal of Biochemistry and Molecular Biology, 20(4): 513-518. (in Chinese)
Zhang XM, Shan XN, Shi YF, Zhang HJ, Li J, Zheng AL. 2004b. Sequence and organization of Muntiacus reevesi mitochondrial genome. Hereditas(Beijing), 26(6): 849-853.
10 August 2017; Accepted: 07 September 2017
s: This work was supported by a grant from the Southeast Asia Biodiversity Research Institute, Chinese Academy of Sciences (Y4ZK111B01)
*Corresponding author, E-mail: quanrc@xtbg.ac.cn
10.24272/j.issn.2095-8137.2017.058
杂志排行
Zoological Research的其它文章
- Nine new species of the spider genus Stedocys(Araneae, Scytodidae) from China and Thailand
- A new species of rain-pool frog (Dicroglossidae:Fejervarya) from western Thailand
- A new species of the Southeast Asian genus Opisthotropis (Serpentes: Colubridae: Natricinae) from western Hunan, China
- Bird diversity in northern Myanmar and conservation implications
- Myanmarorchestia victoria sp. nov. (Crustacea,Amphipoda, Talitridae), a new species of landhopper from the high altitude forests in Myanmar
- A new species of sisorid catfish of the genus Exostoma from the Salween drainage, Yunnan, China
