The association of inherited variation in the CLOCK gene with breast cancer tumor grade
2017-10-25NehaGuptaLiLiCherylThompson
Neha Gupta, Li Li, Cheryl L. Thompson,3
The association of inherited variation in the CLOCK gene with breast cancer tumor grade
Neha Gupta1, Li Li2,3, Cheryl L. Thompson1,3
Background:Sufficient sleep and maintenance of circadian rhythm are important to health.We have shown that short duration of sleep before diagnosis is associated with higher-grade tumors among breast cancer patients. Earlier studies suggest that genetic variation in the CLOCK gene is associated with risk of cancers, including breast cancer. Studies of the association of genetic variation, including in CLOCK, and tumor grade, a standard marker of tumor aggressiveness, are lacking.
Methods:We investigated the relationship between single nucleotide polymorphisms (SNPs)in the CLOCK gene and tumor grade and estrogen receptor, progesterone receptor, and human epidermal growth factor receptor 2 status in 293 breast cancer patients. Nine SNPs were determined by standard TaqMan assays. Tumor grade, receptor status, and other clinical variables were abstracted from medical records.
Results:Two SNPs were excluded because of poor genotyping performance. None of the remaining seven variants had a statistically significant association with breast cancer tumor grade or with receptor status.
Conclusion:As with all novel studies, further work is needed to examine the association of CLOCK and other genes in the circadian rhythm pathway with breast cancer tumor grade in other populations.
Breast cancer; SNP; tumor grade; CLOCK
Background
Breast cancer is the most common cancer in women worldwide, and is the second leading cause of death due to cancer among women[1]. Although the breast cancer mortality rates have been decreasing because of the improvement of detection technology, and the increase in prevention screenings [2], many women develop more aggressive forms of breast cancer that are much more likely to spread, causing the majority of the mortality due to breast cancer.
Our circadian rhythm is important to our health. This natural daily cycle influences many important body functions, such as sleep—wake cycles, hormone release,and body temperature, in addition to important normal biology functions such as DNA repair [3]. Long-term disruptions in circadian rhythms have been associated with obesity[4], diabetes [5], depression [6], and all-cause death [7]. Important to cancer, the circadian rhythm determines the cellular response to DNA repair and DNA stability [8].
The CLOCK gene is the master circadian regulator and key to the maintenance of our circadian rhythm. Variations in this gene affect the length of the rhythm period and change the circadian rhythm [3]. The CLOCK gene was one of the first genes in which a mutation caused an altered behavior instead of an altered physiological process [3]. Earlier studies showed an association between inherited variation in the CLOCK gene and the risk of breast cancer. These studies found significant associations between multiple common polymorphisms in CLOCK and breast cancer risk [9, 10].
In parallel, earlier studies by our group showed the association between sleep duration and tumor aggressiveness in breast cancer [11, 12]. These studies found significant associations with increased tumor grade as well as Oncotype DX recurrence score among breast cancer patients, with the effect found primarily among postmenopausal patients [11, 12].Recent mouse model studies have provided evidence corroborating our observed associations [13].
To date, no study has evaluated the association of CLOCK with breast cancer tumor aggressiveness. Here we examined the association between genetic variants in the CLOCK gene and tumor grade as well as tumor receptor status in breast cancer patients.
Materials and methods
Patient recruitment and data collection
From January 2007 to July 2012, patients at the University Hospitals Cleveland Medical Center with recently diagnosed breast cancer, including ductal carcinoma in situ (DCIS), were recruited as part of a larger case—control study. Participants were excluded because of prior nonsurgical treatment of any cancer, concurrent cancers, or known presence of a BRCA1 or BRCA2 mutation. All patients had to speak English to be eligible for the study, and the participants completed a phone survey on risk factors for breast cancer. All participants provided written informed consent, and they consented to their study data being linked to medical records and the donation of blood samples for genetic and biomarker studies. In addition, all participants provided informed consent for their responses to be linked to their medical record. The University Hospitals Cleveland Medical Center Institutional Review Board approved the study. Medical records were abstracted for diagnosis and tumor and clinical characteristics, including tumor grade, stage, and estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2(HER2) status.
We selected 496 breast cancer patients from this population to be genotypes. We then included only those who had received a diagnosis of invasive breast cancer (i.e., patients in whom DCIS only had been diagnosed were excluded) and for whom tumor grade was available.
Genotyping
Nine SNPs in the CLOCK gene (rs7698022, rs6850524,rs11133391, rs11133389, rs13102385, rs11932595, rs1801260,rs3749474, rs1048004) were chosen for inclusion in this study on the basis of previous investigation and association with risk of breast cancer [9]. DNA was extracted from buffy coats by standard methods. ABI TaqMan assays were used to determine genotype according to the manufacturer’s protocol. For quality control, any sample or SNP with a less than 90% call rate was excluded from further analysis.
Statistical analysis
Differences in the distribution of SNPs and categorical variables between different tumor grades and tumor receptor status was determined by a chi-square test. If any cell count was less than five, Fisher’s exact test was used instead. Differences in continuous measures were determined by analysis of variance. Multivariate modeling using stepwise multinomial regression with tumor grade as the outcome was used to test the association of each SNP, with adjustment for age and race. In all cases, SNPs were treated as categorical variables.Because such a large portion of our population self-reported as being white, to minimize possible confounding due to population stratification and loss of power due to adjustment for race, all analyses were repeated with white samples only, and regressions were done with adjustment for age only. Statistical analysis was performed with SAS version 9.4 (SAS Institute,Cary, NC, USA).
Results
The clinical characteristics of the patients in our final study population are shown in Table 1. Most of the patients hadearly-stage cancer, with about half (50.2%) presenting with stage 1 cancer, and many others (36.9%) presenting with stage 2 cancer (Table 1). Most of the patients were ER positive(78.5%), and most were PR positive (68.6%). Most patients were HER2 negative (78.2%). In our sample, 20.5% of the patients had a grade 1 tumor, 46.8% had a grade 2 tumor and 32.8% had a grade 3 tumor.

Table 1. Patient population clinical characteristics
Table 2 describes the demographic and lifestyle characteristics of the patient population by tumor grade. Most (91.8%)of our population self-reported as being white, and more than half (67.6%) did not have a family history of breast cancer.Overall, the average age of our patients was 58.1 years (standard deviation 10.8 years) and the mean BMI was 28.0 kg/m2(standard deviation 6.4 kg/m2). Patients with higher-grade tumors tended to be younger than patients with lower-grade tumors (P=2.7×10−3; Table 2). Otherwise, there were no statistically significant differences between patients by tumor grades (Table 2).
Two SNPs (rs11133389 and rs3749474) were excluded as a result of poor genotyping performance. None of the remaining SNPs were statistically significantly associated with tumor grade in our population by univariate analyses (Table 3), with all P values less than 0.3. Multivariate analyses adjusted for patient age showed similar findings (Table 3), with no statistically significant results identified. Analyses limited to self-reporting whites displayed very similar results (data not shown).
While investigating the association of these SNPs with tumor receptor status, we found a statistically significant association of the rs11133391 SNP with ER status (Table 4). In our sample, patients with the CC genotype were more likely to have an ER-positive tumor, whereas the tumors of those with the TT genotype were more likely to be ER negative (P=0.020).However, after adjustment for age, this was no longer statistically significant (P=0.091). None of the other six SNPs were statistically significantly associated with ER, PR, or HER2 status in either univariate or multivariate analyses (P>0.1; Table 4).
Discussion
We did not find evidence for risk association of CLOCK gene with breast cancer grade. To our knowledge, this is the first study to explore the association of inherited variation in the CLOCK gene with breast cancer tumor grade. Other studies have suggested associations with breast cancer risk.Motivated by our earlier findings suggesting an inverse association between sleep duration and tumor aggressiveness [11,12], we hypothesized that inherited variation in the CLOCK gene, a known regulator of sleep, would be associated with tumor aggressiveness. Even though the population represents the patient demographic at University Hospitals Cleveland Medical Center, most of our patients self-reported as being white. Further studies need to be done in other sample populations to generalize this finding to other patient populations.
We found a statistically significant association of the rs11133391 SNP with ER status. However, this did not remain significant in multivariate analyses. Further, since these P values were not adjusted for multiple testing, we hesitate to conclude there is an association. However, it is an intriguing finding and encourages the analysis of this association in other independent populations, as well as larger populations with more statistical power.
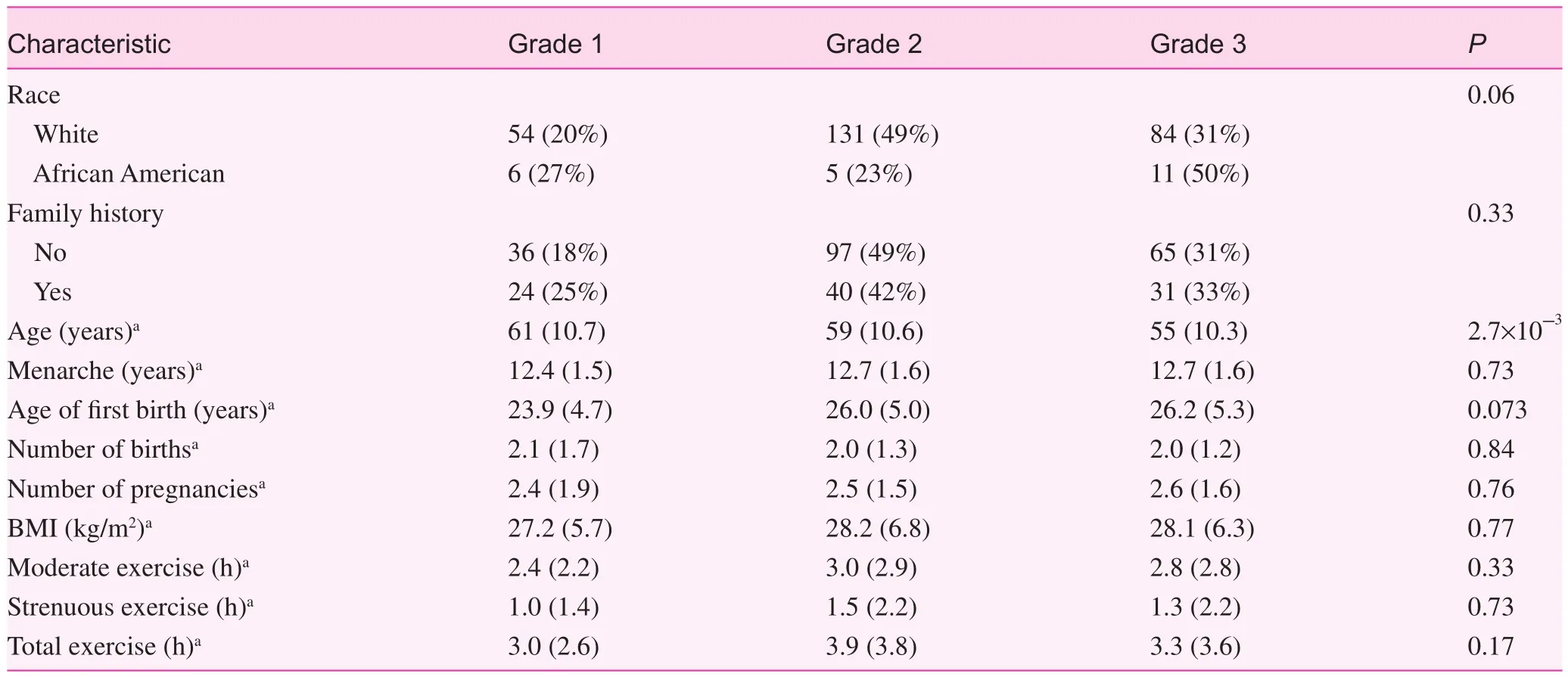
Table 2. Population characteristics and distribution by tumor grade

Table 3. SNPs in the CLOCK gene and tumor grade

Table 4. SNPs in the CLOCK gene and tumor receptor status
There were technical problems that diminished the results from the study. From a total of 496 patients, 55 patients had a SNP call rate of less than 90%, and two SNPs exhibited a poor genotyping performance. An additional 148 patients were not included in this study because of diagnosis of DCIS or missing data on tumor grade. This resulted in a total of 293 patients available for analysis. Thus the low sample size is a limitation and may be the cause of our null findings. While we were sufficiently powered (>80% power at α=0.05) to detect moderate to large effects (differences in frequency of 18% or greater),we were underpowered to detect smaller effects, particularly for the SNPs with lower minor allele frequencies. Thus there is potential that the SNPs we studied here have smaller associations with breast cancer tumor grade than we were able to detect. Importantly, our study was limited by the study of only seven SNPs in a single gene in the circadian rhythm pathway and breast cancer tumor grade. Although these were chosen to capture most of the variation in the CLOCK gene, and because of previous investigations of their association with breast cancer risk, we cannot rule out the association of other variations in this gene with breast cancer grade.
In conclusion, this study is the first to examine the association between circadian rhythm SNPs and breast cancer tumor grade and receptor status. Although we did not observe large or moderate effects of individual variations in the CLOCK gene with tumor aggressiveness, sample size limitations exclude our ability to draw conclusions with regard to smaller effect. Since this area is largely completely unexplored, more studies will need to be done to further this line of inquiry.Further studies should consider additional genes in the circadian rhythm pathway and should also investigate the association with aggressiveness of other cancers.
Acknowledgments
This publication was made possible by the Clinical and Translational Science Collaborative of Cleveland,UL1TR000439 from the National Center for Advancing Translational Sciences component of the National Institutes of Health (NIH) and NIH Roadmap for Medical Research.Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.We also thank the study participants for their contributions to this study.
Conflict of interest
The authors declare no conflict of interest.
Funding
Funding for this study was provided by the National Cancer Institute (grant number K07CA136758).
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016;66:7—30.
2. Moss SM, Wale C, Smith R, Evans A, Cuckle H, Duffy SW.Effect of mammographic screening from age 40 years on breast cancer mortality in the UK Age trial at 17 years’ follow-up: a randomised controlled trial. Lancet Oncol 2015;16:1123—32.
3. Vitaterna MH, Takahashi JS, Turek FW. Overview of circadian rhythms. Alcohol Res Health 2001;25:85—93.
4. Cappuccio FP, Taggart FM, Kandala NB, Currie A, Peile E,Stranges S, et al. Meta-analysis of short sleep duration and obesity in children and adults. Sleep 2008;31:619—26.
5. Bopparaju S, Surani S. Sleep and diabetes. Int J Endocrinol 2010;2010:759509.
6. Adrien J. Neurobiological bases for the relation between sleep and depression. Sleep Med Rev 2002;6:341—51.
7. Cappuccio FP, D’Elia L, Strazzullo P, Miller MA. Sleep duration and all-cause mortality: a systematic review and meta-analysis of prospective studies. Sleep 2010;33:585—92.
8. Savvidis C, Koutsilieris M. Circadian rhythm disruption in cancer biology. Mol Med 2012;18:1249—60.
9. Hoffman AE, Yi CH, Zheng T, Stevens RG, Leaderer D, Zhang Y, et al. CLOCK in breast tumorigenesis: genetic, epigenetic, and transcriptional prof i ling analyses. Cancer Res 2010;70:1459—68.
10. Li J, Humphreys K, Heikkinen T, Aittomäki K, Blomqvist C,Pharoah P, et al. A combined analysis of genome-wide association studies in breast cancer. Breast Cancer Res Treat 2011;126:717—27.
11. Khawaja A, Rao S, Li L, Thompson CL. Sleep duration and breast cancer phenotype. J Cancer Epidemiol 2013;2013:467927.
12. Thompson CL, Li L. Association of sleep duration and breast cancer OncotypeDX recurrence score. Breast Cancer Res Treat 2012;134:1291—5.
13. Hakim F, Wang Y, Zhang SX, Zheng J, Yolcu ES, Carreras A,et al. Fragmented sleep accelerates tumor growth and progression through recruitment of tumor-associated macrophages and TLR4 signaling. Cancer Res 2014;74:1329—37.
1. Department of Nutrition, Case Western Reserve University,Cleveland, OH, USA
2. Department of Family Medicine and Community Health,Case Western Reserve University, Cleveland, OH, USA
3. Case Comprehensive Cancer Center, Case Western Reserve University, Cleveland, OH, USA
Cheryl L. Thompson, PhD
Department of Nutrition, Case Western Reserve University,10900 Euclid Ave, Cleveland,OH 44106, USA
Tel.: +1-216-3683956
E-mail:
cheryl.l.thompson@case.edu
21 February 2017;
Accepted 8 June 2017
杂志排行
Family Medicine and Community Health的其它文章
- Primary care and cancer
- Use of prostate-specific antigen testing in Medicare beneficiaries:Association with previous evaluation
- Symptoms predicting health-related quality of life in prostate cancer patients treated with localized radiation therapy
- Complex multimorbidity and health outcomes in older adult cancer survivorsa
- The relationship between anxiety about prostate cancer among patients with biochemical cancer recurrence and the use of complementary and alternative medicines, diet, and exercise
- Short sleep duration as a contributor to racial disparities in breast cancer tumor grade
