3-(2-吡啶基)-1,2,4-三唑配体CoCu和Cd配合物的合成、结构及荧光性质
2017-09-12杜层层范建中李金萍王多志
杜层层范建中李金萍王多志
3-(2-吡啶基)-1,2,4-三唑配体CoCu和Cd配合物的合成、结构及荧光性质
杜层层范建中李金萍王多志*
(新疆大学化学化工学院,乌鲁木齐830046)
利用3-(2-吡啶基)-1,2,4-三唑配体(HL)和不同的金属盐设计合成了5个配合物[Co(HL)2(H2O)2](NO3)2(1)、[Cu2(L)2(NO3)2(H2O)4](2)、[Cu2(L)2(AcO)2(H2O)2]·6H2O(3)、[Cu2(L)2(HL)2(ClO4)2]·2CH3CN(4)和[Cd2(L)2(HL)2(NO3)2]·2H2O(5),并通过X射线单晶衍射、红外、元素分析、X射线粉末衍射和热重对配合物结构进行了表征。测试结果表明配合物1具有单核结构,并且可以通过氢键的相互作用形成二维超分子结构。配合物2~5为双核结构。配合物2和5可以通过氢键的相互作用形成二维超分子结构。配合物3通过氢键的相互作用形成三维超分子结构。研究了配合物中HL配体的配位模式。此外,研究了配体HL和配合物1和5的固态荧光性质及荧光寿命。
配合物;晶体结构;荧光性质
0 Introduction
During the past decades,many efforts have been devoted to coordination polymers(CPs)and metalorganic frameworks(MOFs)with interesting topological architectures,expanding functionalities and performances in many fields of catalysis[1],photoluminescence[2-3], magnetism[4-5],gas storage[6-7],electrochromism[8-9],drug delivery,and so on.However,it is still difficult to design and synthesis metal-organic frameworks with an intriguing diversity of architectures and properties[10-11]composed of simple inorganic salts and organic ligands due to the self-assembly progress highly influenced by several factors such as building units,temperature,pH value,reaction time,solvent,and metal-to-ligand ratio, etc[12-14].Among these,the design and selection of organic ligands with appropriate sites capable of coordinating with metal ions are quite critical in the construction of coordination polymers(CPs)and metalorganic frameworks(MOFs)[15-16],showing diversified structures and functional properties.
Great efforts have been focused on ligands based on azole heterocycle,which have both good coordination abilities and diverse coordination modes.As simple small molecular ligands,triazole and its derivatives are potential multi-topic ligands in constructions of coordination complexes[17-18].Ligand 1,2,4-Triazole is a very significant five-membered heterocyclic compound with good biological activity and widespread applications in medicament and pesticide such as weed removal,antimicrobials,decreasing blood pressure, antifungus,anti-convulsion,acariddisinfestations, insect disinfestations,plant growth regulator[19].In addition,3-(2-pyridyl)-1,2,4-triazole,as a polydentate ligand,can bind metal ions and provide a possible way to achieve more robust polymeric structure using nitrogen atoms with lone-pair electron,meanwhile the nitrogen atom existing in the form of N-H in the triazole ring can not only lose the hydrogen ion but also retain the hydrogen ion to coordinate with the metal.It is well known that metallic di-,tri-and tetranuclear complexes,so called small-size clusters, have attracted increasing attention owing to their potential to promote new catalytic reactions,which are no longer established by each mononuclear system andtobe minimal models for mixed-metal heterogeneous catalysts as well as photochemical devices[20-22].
Considering these aspects above mentioned,in this study,five transition metals complexes with mononuclear and dinuclear structure units were synthesized and structurally characterized by single crystal X-ray diffraction,infrared spectroscopy(IR),elemental analysis,powder X-ray diffraction(PXRD)and thermogravimetric analysis(TGA),using 3-(2-pyridyl)-1,2,4-triazole(HL)ligand as building blocks.We discussed the coordination modes of the ligand.Furthermore,the luminescent properties and fluorescence lifetimes of ligand HL,complexes 1 and 5 have been investigated.
1 Experimental
1.1 Materials and general methods
All the other reagents used for the syntheses were commercially available and employed without further purification.The ligand HL was prepared according to reported procedures[23].IR spectra were measured on a Brucker Equinox 55 FT-IR spectrometer with KBr Pellets in the range of 4 000~400 cm-1. Elemental analyses of C,H and N were performed on a Thermo Flash EA 1112-NCHS-O analyzer.Thermogravimetric analysis(TGA)measurements were on a Perkin-Elmer TG-7 analyzer heated from 25 to 800℃at the rate of 10℃·min-1under nitrogen atmosphere. The X-ray powder diffraction(PXRD)was recorded over the 2θ range of 5°~50°on a Bruker D8 Advance X-ray diffractometer operated at 40 kV and 40 mA using Cu Kα radiation(λ=0.154 8 nm).The fluorescence spectra of ligands and its complexes were obtained using a Hitachi F-4500 Fluorescence Spectrophotometer with a Xe arc lamp as the light source and bandwidths of 2.5 nm at room temperature.Fluorescence lifetime data were obtained on a Fluorolog-3 spectrofluorometer.
1.2 Synthesis of complex 1
A mixture of Co(NO3)2·6H2O(0.3 mmol),HL(0.3 mmol)and distilled water(5 mL)was sealed in a Teflon-lined reactor and heated at 130℃for 3 days,after the mixture was cooled to room temperature at a rate of 5℃·h-1.The pink block crystals were gained (ca.35%yield based on HL).Anal.Calcd.for C14H16CoN10O8(%):C,32.89;H,3.15;N,27.40.Found(%): C,32.60;H,3.48;N,27.95.IR(KBr,cm-1):3 377(s), 3 063(m),2 315(w),1 990(m),1 743(w),1 647(w), 1 568(m),1 514(m),1 472(s),1 447(s),1 412(m), 1 384(m),1 360(m),1 276(m),1 253(m),1 204(m), 1 147(m),1 116(m),1 093(m),1 030(m),1 010(m), 875(m),798(m),749(m),724(m),674(m),637(m), 495(m),412(m)(Fig.S1 of Supporting information).
1.3 Synthesis of complex 2
A mixture of Cu(NO3)2·3H2O(0.3 mmol)and HL (0.3 mmol)in methanol solution(15 mL)was refluxed for 3 h.After being cooled and filtrated,blue block crystals were obtained after several days with the evaporation of the filtrate(ca.45%yield based on HL). Anal.Calcd.for C14H16Cu2N10O10(%):C,27.49;H,2.64; N,22.90.Found(%):C,27.14;H,2.48;N,22.37.IR (KBr,cm-1):3 440(m),3 140(m),2 424(w),1 768(w), 1 658(m),1 613(m),1 529(w),1 485(m),1 452(m), 1 416(m),1 416(m),1 383(s),1 289(m),1 149(m), 1 097(m),1 047(m),1 020(m),952(w),894(w), 821(m),789(m),749(m),717(m),658(m),517(m), 415(m)(Fig.S1).
1.4 Synthesis of complex 3
A mixture of Cu(OAc)2·H2O(0.3 mmol)and HL (0.3 mmol)was dissolved in methanol solution(15 mL) and filtrated.Blue block crystals were formed after several days with the evaporation of the filtrate(ca. 34%yield based on HL).Anal.Calcd for C18H32Cu2N8O12(%):C,31.81;H,4.74;N,16.48.Found(%):C, 31.52;H,4.32;N,16.06.IR(KBr,cm-1):3 443(s), 3 135(m),2 495(w),2 202(w),1 956(w),1 878(w), 1 787(w),1 614(s),1 574(s),1 484(s),1 450(m), 1 403(m),1 340(m),1 286(m),1 260(m),1 213(w), 1 149(m),1 100(m),1 048(m),1 021(m),930(m), 899(m),801(m),758(m),720(m),686(m),645(m), 624(m),516(m),415(w)(Fig.S1).
1.5 Synthesis of complex 4
Complex 4 was obtained by a procedure similar to that for 3 except for using acetonitrile solution instead of methanol solution andCu(ClO4)2·6H2O instead of Cu(OAc)2·H2O.Blue block crystals were formed after several days with the evaporation of the filtrate.(ca.40%yield based on HL).Anal.Calcd.for C32H26Cl2Cu2N18O8(%):C,38.87;H,2.65;N,25.49. Found(%):C,38.54;H,2.22;N,25.08.IR(KBr,cm-1): 3 132(m),2 939(m),2 257(m),2 014(w),1 815(w), 1 732(w),1 615(m),1 571(m),1 544(w),1 480(m), 1 454(m),1 417(m),1 371(m),1 342(m),1 285(m), 1 264(m),1 214(w),1 178(m),1 146(m),1 105(s), 1 079(s),970(m),926(m),868(m),797(m),760(m), 747(m),720(m),663(m),620(m),515(m),483(m) (Fig.S1).
1.6 Synthesis of complex 5
Complex 5 was obtained via a similar method used for complex 3 except that Cu(OAc)2·H2O was replaced by Cd(NO3)2·4H2O,as well as methanol solution was replaced by deionized water.Colorless block crystals were formed after several days with the evaporation of the filtrate(ca.41%yield based on HL). Anal.Calcd.for C28H26Cd2N18O8(%):C,34.76;H,2.70; N,26.06.Found(%):C,34.49;H,2.31;N,25.83.IR (cm-1,KBr pellets):3 374(m),3 083(m),3 058(m), 3 022(m),2 978(m),2 947(m),2 714(w),2 621(w), 2 550(w),2 469(w),2 403(w),2 317(w),2 225(w), 2 142(w),1 974(w),1 903(w),1 864(w),1 737(w), 1 645(w),1 601(s),1 569(m),1 510(m),1 472(s), 1 446(m),1 415(s),1 349(s),1 273(s),1 251(m), 1 205(m),1 143(m),1 116(s),1 046(m),1 025(s), 1 006(m),874(m),802(m),751(m),725(m),671(m), 633(m),508(w),477(m)(Fig.S1).
1.7 Structure determination
X-ray single-crystal diffraction data for complex 1~5 were collected on a Bruker APEXⅡSmart CCD diffractometer at 293(2)K with Mo Kα radiation(λ= 0.071 073 nm)by ω-φ scan mode.The program SAINT[24]was used for integration of the diffraction profiles. Semi-empirical absorption corrections were applied using SADABS program.All the structures were solved by direct methods using the SHELXS program of the SHELXTL package and refined by full-matrix leastsquares methods with SHELXL[25].PLATON/SQUEEZE 55 was used to correct the data in the refinement process[26].Metal atoms in each complex were locatedfrom the E-maps and other non-hydrogen atoms were located in successive difference Fourier syntheses and refined with anisotropic thermal parameters on F2. Hydrogen atoms of carbon were included in calculated positions and refined with fixed thermal parametersriding on their parent atoms.Crystallographic data and experimentaldetailsforstructuralanalysesare summarized in Table 1.
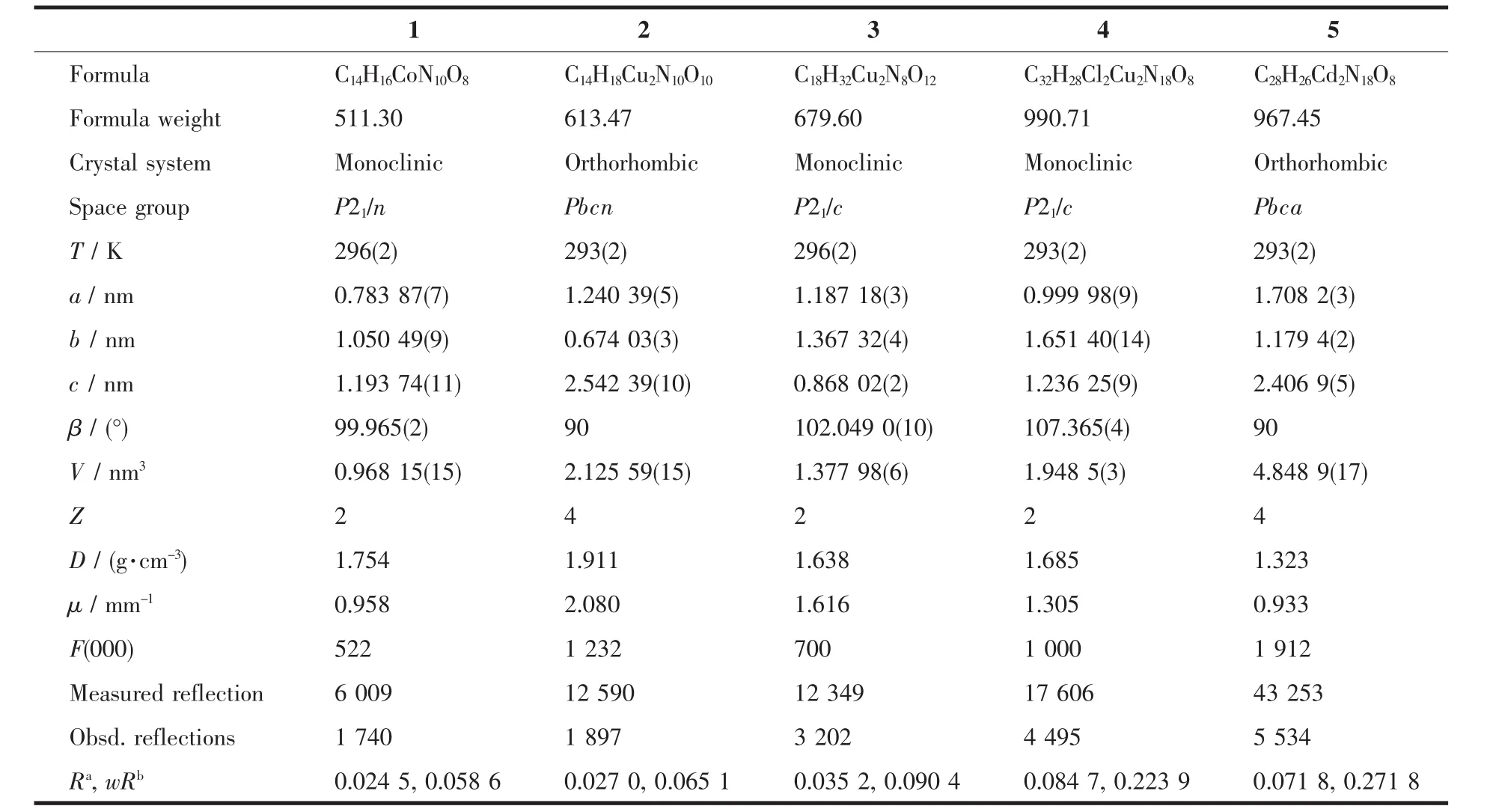
Table1 Crystallographic data for complexes 1~5
CCDC:1054520,1;1054521,2;1054522,3; 1054523,4;1054524,5.
2 Results and discussion
2.1 Structure description of 1
Complex 1 crystallizes in the monoclinic symmetry space group P21/n.Crystallographic data and experimental details for structural analyses were summarized in Table 1,and the selected bond distances and angles were listed in Table 2.The X-ray diffraction analysis shows that 1 has a mononuclear structure. The centrosymmetric unit contains a half of Cocenter,one HL ligand,one coordinated water molecule and one NO3-anion.The center Coion is coordinated to four N atoms from two distinct chelating HL ligands in the equatorial plane and two O atoms from two distinct water molecules at the axial positions to completeitsdistortedoctahedroncoordination geometry(Fig.1a).Ligand HL,a typical chelating ligand,adopts a coordination mode(modeⅠin Scheme 1)to link the Coion with the Co-N bond distances of 0.215 15(12)and 0.216 12(12)nm,respectively,which are in the normal range.The N(2)#1-Co(1)-N(3)and N(2)-Co(1)-N(3)angels are 77.62(5)° and 102.38(5)°,respectively(Table 2).It should be pointed out that O(4)-H(5)…O(3)#1 and N(5)-H(8)…O(1)intermolecular hydrogen-bonding interactions are observed between adjacent mononuclear structures. Thus the adjacent mononuclear structures are arranged into a one-dimensional chain by the intermolecular hydrogen-bonding interactions(Fig.1b)(O(4)…O(3)#1 andN(5)…O(1)separations are 0.284 03(19)and 0.292 5(2)nm,respectively).The chains are further expandedbytheO(4)-H(6)…O(2)#2hydrogenbonding interactions to generate a two-dimensionalsupramolecular sheet with the distance of 0.280 33(19) nm between O(4)and O(2)#2(Fig.1c).In complex 1, the uncoordinated NO3-anions serving as counteranions locate at the cavities of the sheets and form O-H…O hydrogen-bondingswiththecoordinatedwater molecules(Table 3).
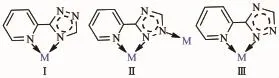
Scheme 1Coordination modes of the ligand
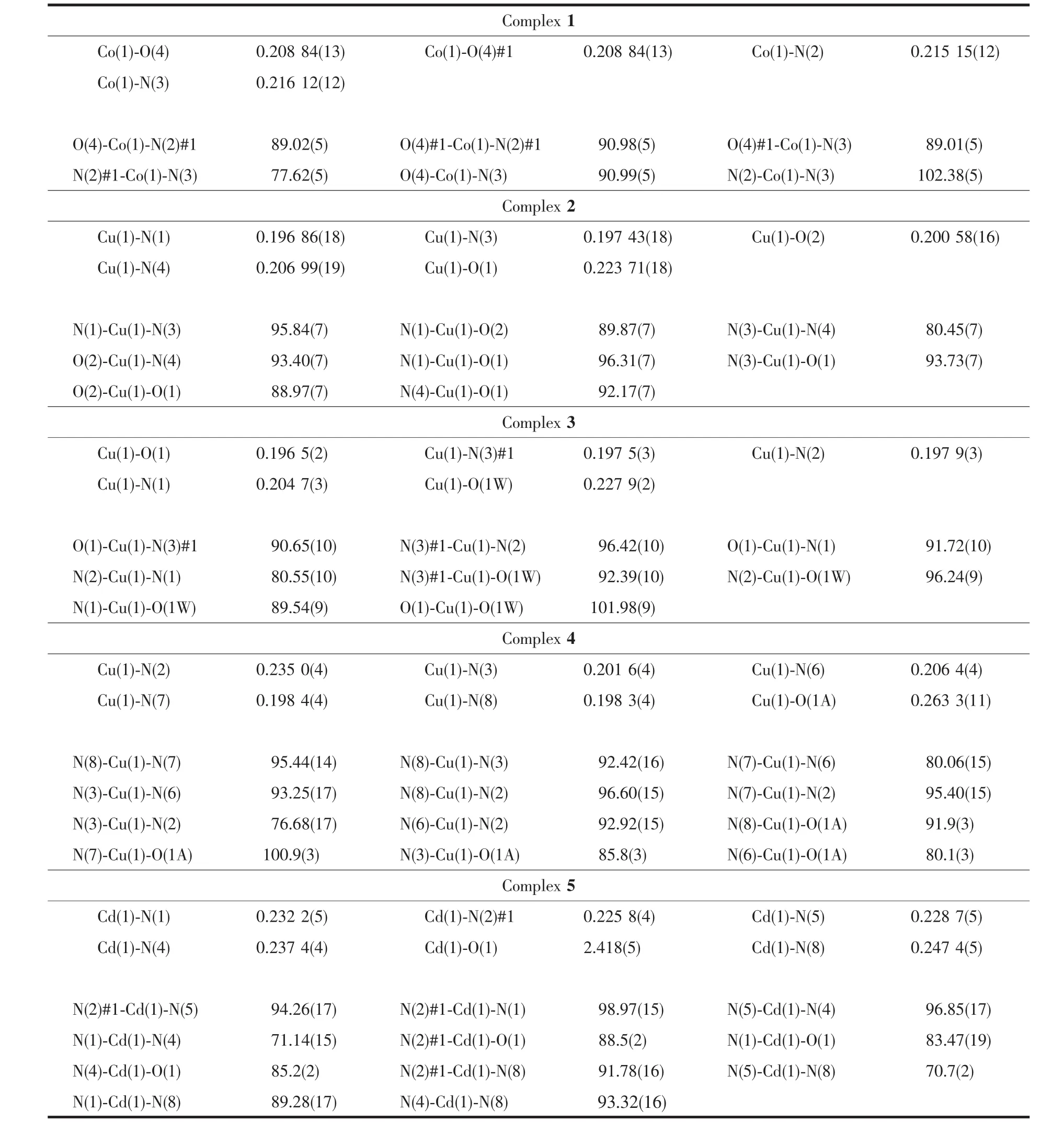
Table2 Selected bond distances(nm)and angles(°)for complexes 1~5
2.2 Structure description of 2
Complex2hasacentrosymmetricdinuclear structure.Single-crystalX-raydiffractionanalysis reveals that complex 2 crystallizes in the orthorhombic system,with space group Pbcn.The asymmetric unit of complex 2 contains one Cucation,one L ligand, two coordinated water molecules and one NO3-anion. The center Cuion is six-coordinated by three nitrogen atoms from two distinct L-ligands(Cu(1)-N(1) 0.196 86(18)nm,Cu(1)-N(3)0.197 43(18)nm),two Oatoms from four distinct water molecules(Cu(1)-O(1) 0.223 71(18)nm,Cu(1)-O(2)0.200 58(16)nm),one oxygen atom from one nitrate ion(Cu(1)-O(3)0.276 8(18) nm,Fig.2a).The equatorial plane of slightly distorted octahedral coordination geometry is occupied by atoms N1,N3,N4 and O2,while O1 and O3 occupy axial positions withthe O(1)-Cu-O(3)bond angle of 164.82(3)°.The ligand adopts one single coordination mode(modeⅡin Scheme 1)acting as a tridentate linker to connect two Cucations with the nonbonding Cu…Cu distance being 0.399 3 nm.It is worth noting that O(4)and O(5)atoms of NO3-which adopts monodentate coordination mode are not coordinated to Cuions,which present intermolecular hydrogenbonding interactions with O(2)atoms of water molecules (the distances of O(2)…O(4)#2 and O(2)…O(5)are 0.281 3(3)and 0.274 9(3)nm,respectively.O(2)-H(3w)…O(4)#2 and O(2)-H(4w)…O(5)angles are 173(3)°and 165(3)°,respectively).Therefore,such molecules are interconnected via the O-H…O hydrogen bonding interactions to give an infinite 1D chain along the b-axis as shown in Fig.2b.Additionally, the nitrogen atoms of the triazole rings and coordinated water molecules are involved in the strong and directional O(1)-H(1W)…N(2)#1 hydrogen bonds, which connect the adjacent dimer units to generate a 2D supramolecular network(Fig.2c,Table 3).

Table3 Hydrogen bonding geometry for 1~3 and 5

Fig.1 View of the coordination environment of Coion in 1(a);(b)One dimensional chain formed by hydrogen bonding interactions;(c)Two dimensional network formed by hydrogen bonding interactions
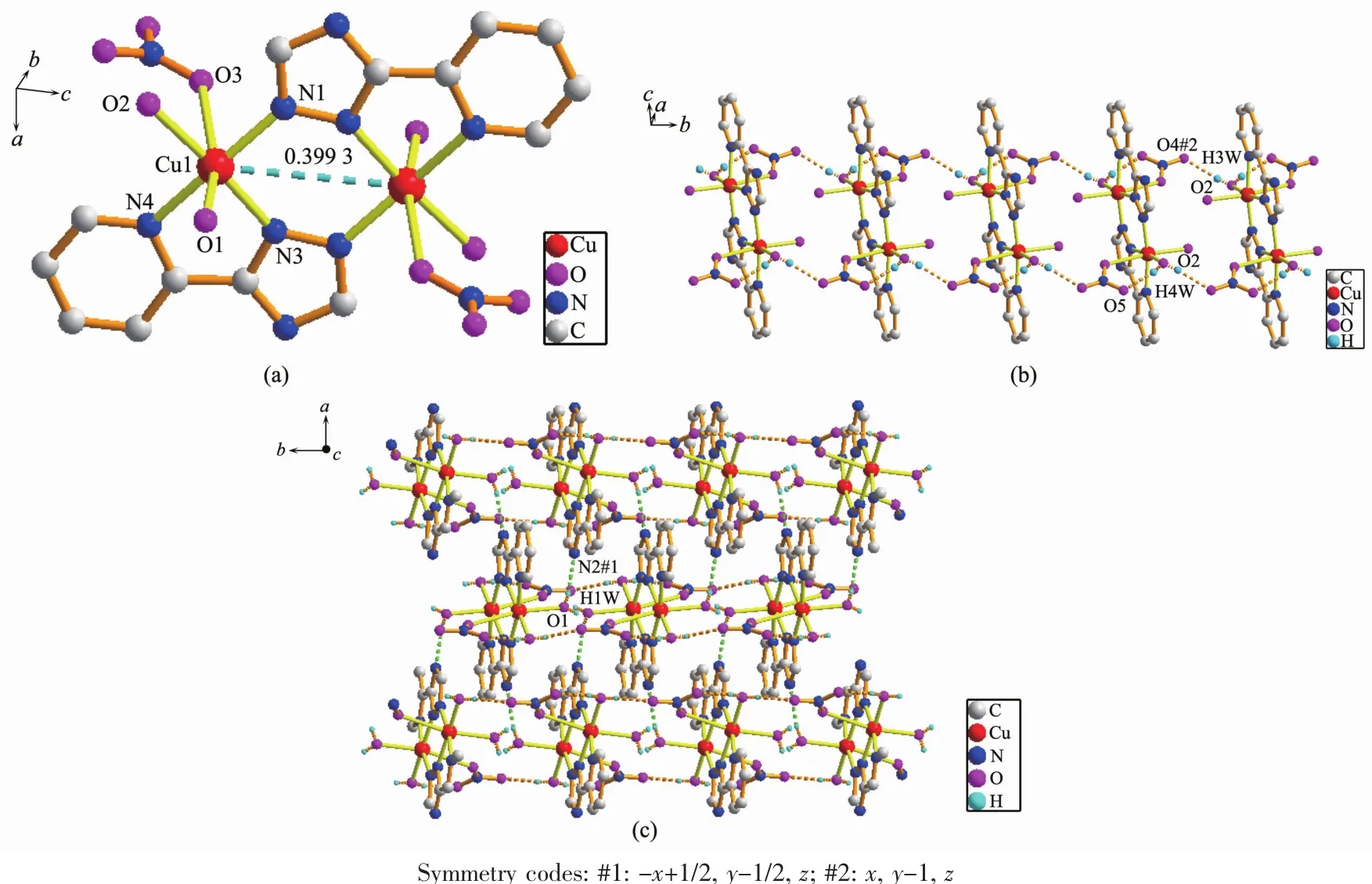
Fig.2 (a)View of the coordination environment of Cuion in 2;(b)One dimensional chain formed by hydrogen bonding interactions;(c)Two dimensional network formed by hydrogen bonding interactions
2.3 Structure description of 3
The molecular structure of 3 is shown in Fig.3a. The X-ray diffraction analysis shows that 3 crystallizes in the monoclinic space group P21/c.There are one Cucation,one L-ligand,one acetate ion,one coordinated water molecule and three lattice water molecules in the asymmetric unit of complex 3.The center Cuion is six-coordinated by three N atoms from two distinct L-ligands,one O atom from a water molecule and two O atoms from one acetate ion.Acetate ion acts as a typical chelating ligand coordinated to the Cuion with the Cu-O bond distances being 0.196 5(2) and 0.273 9(2)nm,the O-Cu-O angel being 53.31°. The ligand is tridentate linker to concatenate Cuions(modeⅡin Scheme 1),two nitrogen atoms(N1 and N2)of which chelate and coordinate one center cation to form a five-membered ring,at the same time, two central copper atoms and four nitrogen atoms make a six-membered ring improving the stability of the molecule to a large extent.Additionally,the hydrogenbonding interactions are observed between the adjacent structures with the O(1W)…O(2W),O(1W)…N(4)#2, O(2W)…O(2)#1,O(3W)…O(2),O(3W)…O(4W),O(4W)…O(1)and O(4W)…O(2W)separation of 0.277 7(3), 0.2756(3),0.2874(3),0.287 2(3),0.274 8(3),0.280 1(3) and0.2752(4)nm,respectively.Therefore,thedinuclear structure are expanded by O(1W)-H(1W)…O(2W), O(2W)-H(8W)…O(2)#1,O(3W)-H(5W)…O(2),O(3W) -H(6W)…O(4W),O(4W)-H(3W)…O(1)and O(4W)-H(4W)…O(2W)to form a two-dimensional network (Fig.3b),then further expanded by the O(1W)-H(2W)…N(4)#2 hydrogen-bonding interactions to form a three-dimensional framework(Fig.3c).The hydrogenbonding parameters are listed in Table 3.

Fig.3 (a)Coordination environment of Cuion in 3;(b)Two dimensional network formed by hydrogen bonding interactions;(c)Three dimensional framework formed by hydrogen bonding interactions
2.4 Structure description of 4
X-ray single crystal diffraction reveals that 4 indicates a dinuclear structure crystallizing in the monoclinic space group P21/c.Each asymmetric unit of 4 contains one Cucation,one L-ligand,one HL ligand,one ClO4-anion and two lattice CH3CN molecules.Similar to 2 and 3,complex 4 also has a dinuclear structure which consists of two Cuions, two ClO4-anions,four ligands and two CH3CN molecules(Fig.4).The Cuion is linked to one oxygen atom from one ClO4-anion and five nitrogen atoms from three different ligands,demonstrating a distorted octahedral coordination environment.The best equatorial plane is formed by four nitrogen atoms(N3,N6, N7 and N8),and the axial positions are occupied by N2 and O1A.The Cu-N bond distances range from 0.198 3(4)to 0.235 0(4)nm,and the Cu-O length is0.263 3(11)nm,which are in the normal range of those observed in copper complexes.Interestingly,there are two types of coordination modes of the ligands in the structure(modeⅡandⅢin Scheme 1).One acts as a bidentate ligand to collaborate one Cuion,while the other adopts a tridentate chelating-bridging coordination mode to connect two crystallography equivalent Cuions with a Cu…Cu distance of 0.401 5 nm,slightly longer than that of 2(0.399 3 nm)and that of 3(0.398 8 nm).Moreover,two-coordinated ClO4-anions serve as counter-anions synchronously adopting a monodentate coordination mode.
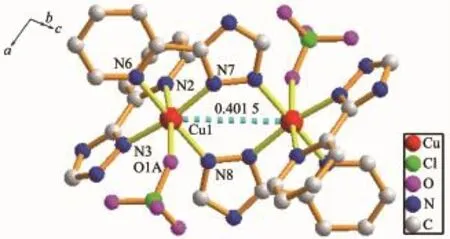
Fig.4 View of the coordination environment of Cuion in 4
2.5 Structure description of 5
The asymmetric unit of 5 consists of one Cdcation,one L-ligand,one HL ligand and one NO3-anion,which crystallizes in the orthorhombic space group Pbca.Structure determination indicates that complex 5 possesses a dinuclear structure,similar to 2,3 and 4,constructed from the ligands,NO3-anions and the center Cdions,which are seven-coordinated by five N atoms from three distinct chelating ligands and two O atoms from nitrate ions(Cd(1)-O(1)0.241 8(5) nm,Cd(1)-O(3)0.272 8(5)nm,Fig.5a).The Cd-N bond lengths are in the range of 0.225 8(4)~0.247 4(5) nm.The CdO2N5seven-coordination fashion can be treated as a distorted pentagonal bipyramidal geometry, where O(1),O(3),N(1),N(5)and N(8)atoms comprise the equatorial plane,while N(2)and N(4)atoms occupy the axial positions.In complex 5,the coordination modes of the ligands are the same as that of complex 4.In the dinuclear unit,the non-bonding Cd…Cd length is 0.434 8 nm.The two-coordinated nitrate ions serve as counter-anions synchronously.The hydrogen bonds are formed between uncoordinated nitrate atom and nitrate oxygen atom,and the distance of N(6)and O(2)is 0.291 5 nm(Table 3).Finally,intermolecular hydrogen-bonding(N(6)-H(6A)…O(2))interactions assembleneighboringstructureunitstoforma two-dimensional(2D)sheet(Fig.5b).The SQUEEZE/ SQUEEZE 55 program was used to remove the contributions of the lattice water molecules for complex 5,so part of the hydrogen-bonding parameters are listed in Table 3.
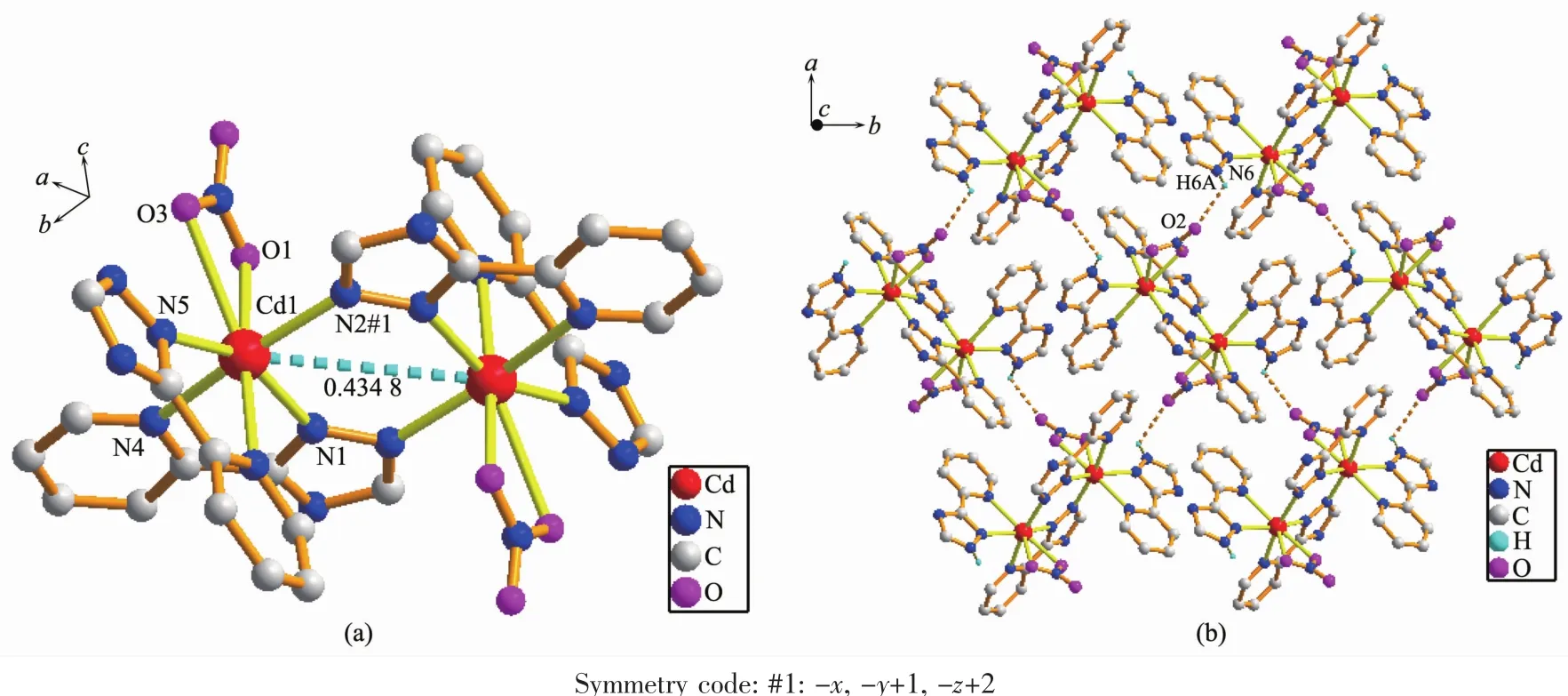
Fig.5 (a)Coordination environment of Cdion in 5;(b)Two dimensional network formed by hydrogen bonding interactions
2.6 PXRD patterns
To confirm the mass identity and phase purity of 1~5,X-ray powder diffraction(XRPD)experiments were carried out for complexes 1~5.The XRPD experimental and simulated patterns of the corresponding complexes are shown in Fig.6.The PXRD patterns of as-synthesizedcomplexes1~5areapproximately coincident with the simulated ones from the singlecrystal data,indicating high purity and homogeneity of them.
2.7 Thermogravimetric analysis
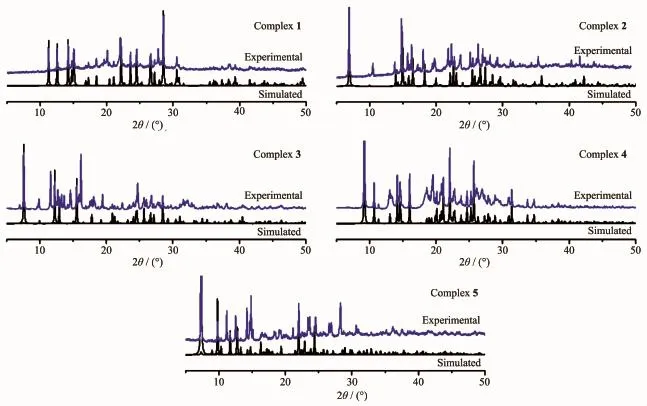
Fig.6 PXRD patterns of complexes 1~5
To examine the thermal stability of complexes 1~5,the TGA analyses of complexes 1~5 were carried out(Fig.7).The TGA curves indicate the weight loss of 1 can mainly be divided into two steps.The first step shows a weight loss from 130 to 260℃corresponding to the loss of two lattice NO3-anions and two coordinated water molecules(Obsd.31.93%;Calcd. 31.3%),then the removal of ligands occurs within the range of 260~650℃.The residue,estimated as CoO, has an observed mass of 16.51%compared with the calculated value of 14.60%.The TG analysis of 2 demonstrates a curve gradient phase about the weight loss of 11.10%in the temperature range of 130~240℃corresponding to the loss of four coordinated water molecules(Calcd.11.46%).After that,a weight loss of 23.20%rapidly occurs from 280 to 450℃where the NO3-anions are destroyed(Calcd.20.27%).Then, complex 2 undergoes a weight loss of L-ligands.The percentage of residual weight obtained from the graph (26.88%)is in well agreement with the calculated value of 26.01%indicating that the final product obtained is copper oxide(CuO).The first stage of weight loss of complex 3 from 130 to 260℃corresponds to the release of six lattice water molecules and two coordinated water molecules(Obsd.19.25%;Calcd. 21.21%).The second stage of weight loss from 300 to 650℃is ascribed to the gradual loss of CH3COO-and the decomposition of L-ligands.The residue is calculated to be Cu(Obsd.16.20%;Calcd.18.70%).For 4, the first step of consecutive weight loss of 7.85% taking place in the temperature range of 100~180℃is attributed to the evacuation of two lattice CH3CN molecules(Calcd.8.29%).The dehydration product experiences almost one-step weight loss from 250 to 650℃,which can be attributed to the thermal decomposition of the ligands and ClO4-.The remaining fraction is 18.70%of the initial one,which is well consistent with the expected value for the formation of a stoichiometric quantity of CuO(Calcd.16.05%).TheTG curve of 5 shows a weight loss from 100 to 150℃, and the further decomposition occurs in the range of 230~650℃,which is attributed to the elimination of NO3-anions,L-and HL ligands,whose losses have no definite boundary.The final residual may be CdO (Obsd.26.30%;Calcd.26.55%).
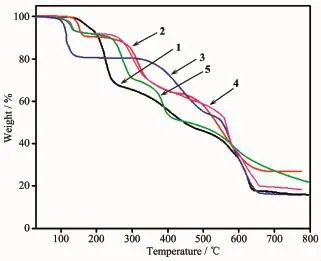
Fig.7 TGA curves of complexes 1~5
2.8 Luminescent properties
The solid-state photoluminescence properties of complexes 1 and 5 as well as the free nitrogencontainingligandHLwereinvestigatedatroom temperature,as shown in Fig.8.The free ligand HL exhibits blue fluorescent emission band at 405 nm upon excitation at 245 nm.Upon the same excitation at 245 nm,the emission spectra of complexes 1 and 5 exhibit similar shapes and maximums at 413 and 405 nm,respectively.Since the d10metal ions are difficult to oxidize or reduce due to their d10configuration,the emission band of 1 is neither metal-to-ligand charge transfer(MLCT)nor ligand-to-metal charge transfer (LMCT)in nature.It can probably be assigned to the intraligand π→π*transitions[27].In spite of Coions probablyquenchingtheluminescenceemissions, fortunately,the complex 1 also shows emission.The slight red-shift of complex 1 compared to that of the ligand HL may be ascribed to the fact that the coordination of Coion increases the conformation rigidity of the ligand and reduces the loss of energy[28]. Their emission bands in the blue area suggest that two complexes may be potential candidates as blue-light emitting materials.Furthermore,the emission decay lifetimes of HL ligand and complexes 1 and 5 were measuredundercorrespondingexcitation/emission maxima at room temperature as shown in Fig.9.Luminescent lifetimes of HL ligand and complexes 1 and 5 are fitted by triexponential curves with τ1=2.41 ns (15.19%),τ2=16.71ns(51.39%),τ3=0.54ns(33.41%)for HL,τ1=2.25 ns(9.52%),τ2=17.58 ns(46.17%),τ3=0.57 ns(44.31%)for 1 and τ1=3.27 ns(64.13%),τ2=15.71 ns(19.17%)τ3=1.06 ns(16.71%)for 5,respectively.

Fig.8 Emission spectra of 1,5 and HL in the solid state at room temperature

Fig.9 Luminescence decays of HL and 1,5 under corresponding excitation/emission maxima at room temperature
3 Conclusions
In summary,Five new complexes[Co(HL)2(H2O)2] (NO3)2(1),[Cu2(L)2(NO3)2(H2O)4](2),[Cu2(L)2(AcO)2(H2O)2]·6H2O(3),[Cu2(L)2(HL)2(ClO4)2]·2CH3CN(4) and[Cd2(L)2(HL)2(NO3)2]·2H2O(5)(HL=3-(2-pyridyl)-1,2,4-triazole)have been synthesized and structurally characterizedbysinglecrystalX-raydiffraction, infrared spectroscopy(IR),elemental analysis,powder X-ray diffraction(PXRD)and thermogravimetric analysis(TGA).Complex 1 was prepared under the hydrothermal condition,complex 2 was produced via reflux, complexes 3~5 were synthesized by the evaporation of the solvent at room temperature.Complex 1 has a mononuclear structure and complexes 2~5 have dinuclear structures.It is interesting that 1,2 and 5 are further assembled to form two dimensional(2D)supramolecular structures as well as 3 is further assembled to form three dimensional(3D)supramolecular structure by hydrogen-bonding interactions.This result indicates that hydrogen-bonding interactions play an important role in the formation of supramolecular networks. Moreover,the ligand HL and corresponding complexes 1 and 5 display strong blue fluorescent emission at room temperature and shorter fluorescence lifetimes, which shows that they may be potential candidates as blue-light emitting materials.
Supporting information is available at http://www.wjhxxb.cn
[1]Zhou Z,He C,Xiu J H,et al.J.Am.Chem.Soc.,2015,137: 15066-15069
[2]JI Qi(吉沁),CHEN Li-Zhuang(陈立庄).Chinese J.Inorg. Chem.(无机化学学报),2017,33(5):874-880
[3]WANG Xin-PING(王新萍),LI Ying-Ying(李莹莹),LIU Yong(刘勇),et al.Chinese J.Inorg.Chem.(无机化学学报), 2017,33(5):823-829
[4]Xia B,Wang K,Wang Q L,et al.CrystEngComm,2017,19: 811-816
[5]Armentano D,Martínez-Lillo J.Cryst.Growth Des.,2016,16: 1812-1816
[6]Chen Q,Chang Z,Song W C,et al.Angew.Chem.Int.Ed., 2013,52:11550-11553
[7]Gao Q,Xu J,Cao D P,et al.Angew.Chem.Int.Ed.,2016, 55:15027-15030
[8]Procopio E Q,Bonometti V,Panigati M,et al.Inorg.Chem., 2014,53:11242-11251
[9]Choi T L,Lee K H,Joo W J,et al.J.Am.Chem.Soc.,2007, 129:9842-9843
[10]Tranchemontagne D J,Mendoza-Cortés J L,OKeeffe M,et al. Chem.Soc.Rev.,2009,38:1257-1283
[11]Jiang H L,Tatsu Y,Lu Z H,et al.J.Am.Chem.Soc.,2010, 132:5586-5587
[12]Ni J,Wei K J,Liu Y Z,et al.Cryst.Growth Des.,2010,10: 3964-3976
[13]Ou G C,Feng X L,Lu T B.Cryst.Growth Des.,2011,11: 851-856
[14]Stephenson A,Argent S P,Riis-Johannessen T,et al.J.Am. Chem.Soc.,2011,133:858-870
[15]Wang D Z.Polyhedron,2012,35:142-148
[16]Chu Q,Su Z,Fan J,et al.Cryst.Growth Des.,2011,11:3885 -3894
[17]Wang D Z,Zhang Q,Zhang J B,et al.Polyhedron,2012,42: 216-226
[18]Sun C Y,Zheng X J,Gao S,et al.Eur.J.Inorg.Chem.,2005: 4150-4159
[19]Sahin O,Kantar C,Sasmaz S,et al.J.Struct.Chem.,2014, 1067:83-87
[20]Tanase T,Toda H,Yamamoto Y.Inorg.Chem.,1997,36: 1571-1577
[21]Zhao M,Xue S S,Jiang X Q,et al.J.Mol.Catal.A.Chem., 2015,396:346-352
[22]Das M,Harms K,Ghosh B N,et al.Polyhedron,2015,87: 286-292
[23]Zhang J J,Zhang C H,Yang N F,et al.Chem.Res.Appl., 2010,22:749-753
[24]SAINT Software Reference Manual,Madison,WI,1998.
[25]Sheldrick G M.SHELXS-97,Program for Solution and Refinement of Crystal Structures,University of Göttingen, Germany,1997.
[26]Spek A L.Acta Crystallogr.Sect.A,1990.
[27]LI Jin-Ping(李金萍),FAN Jian-Zhong(范建中),WANG Duo-Zhi(王多志).Chinese J.Inorg.Chem.(无机化学学报), 2016,32(5):753-761
[28]Wang X W,Chen J Z,Liu J H.Cryst.Growth Des.,2007,7: 1227-1229
Co,Cuand CdComplexes Based on 3-(2-Pyridyl)-1,2,4-triazole: Syntheses,Structures,and Fluorescent Properties
DU Ceng-CengFAN Jian-ZhongLI Jin-PingWANG Duo-Zhi*
(College of Chemistry and Chemical Engineering,Xinjiang University,Urumqi 830046,China)
Five complexes[Co(HL)2(H2O)2](NO3)2(1),[Cu2(L)2(NO3)2(H2O)4](2),[Cu2(L)2(AcO)2(H2O)2]·6H2O(3), [Cu2(L)2(HL)2(ClO4)2]·2CH3CN(4)and[Cd2(L)2(HL)2(NO3)2]·2H2O(5)utilizing 3-(2-pyridyl)-1,2,4-triazole ligand (HL)as well as different metal salts have been synthesized and structurally characterized by single crystal X-ray diffraction,infrared spectroscopy(IR),elemental analysis,powder X-ray diffraction(PXRD)and thermogravimetric analysis(TGA).The structural analyses reveal that complex 1 has a mononuclear structure and forms a 2D supramolecular structure via hydrogen bonding interactions.Complexes 2~5 have dinuclear structures.Complexes 2 and 5 are arranged into 2D supramolecular structurs by the corresponding hydrogen bonding interactions,and complex 3 is also arranged into a 3D supramolecular structure through hydrogen bonding interactions.The coordination modes of HL ligand in the complexes were studied.Moreover,the fluorescent properties and fluorescence lifetimes of ligand HL,complexes 1 and 5 were investigated in the solid at room temperature. CCDC:1054520,1;1054521,2;1054522,3;1054523,4;1054524,5.
complex;crystal structure;luminescent property
O614.81+2;O614.121;O614.24+2
A
1001-4861(2017)09-1685-12
10.11862/CJIC.2017.194
2017-04-22。收修改稿日期:2017-06-01。
新疆维吾尔自治区自然科学基金(No.2015211C266)资助项目。*
。E-mail:wangdz@xju.edu.cn
