阿尔茨海默病的遗传学研究进展
2017-09-08李志鹏
高 珊 陈 超 李志鹏 杨 泽
※为通讯作者
阿尔茨海默病的遗传学研究进展
高 珊1,2陈 超2※李志鹏2杨 泽1※
阿尔茨海默病(Alzheimer’s disease,AD)是一种常见的中枢神经性退行性疾病。普遍认为AD的发生是遗传、环境、代谢等多种因素共同作用的结果。自20世纪末AD的研究进入到分子遗传学和基因组学的水平以来,陆续发现了一些AD的易感基因。本文将就2016年至今AD易感基因(ABCA7,APOE,CLU,ESR2,CR1,INPP5D,BIN1,CD2AP,PICALM,SLC242A,APP,MTHFR,PSEN1,PSEN2,SORL1,PTK2B,TOMM40,PTGS2,ZCWPW1,FERMT2)的遗传学研究进展作一综述。
阿尔茨海默病 易感基因
阿尔茨海默病(Alzheimer’s disease,AD)是一种常见的中枢神经性退行性疾病,包括进行性记忆衰退、认知功能下降、行为异常和社交功功能障碍。AD大多发生在65岁以上的患者,且世界范围内的发生趋势也成指数上涨,从60岁中段人群中的2%到80岁人群的30%至35%[1],严重威胁老年人的健康。AD的发病原因及机制尚不明确,普遍认为AD的发生是多种因素共同作用的结果,包括遗传、环境、代谢等多种因素,其中遗传因素起重要作用。
自20世纪末,Goate A等人发现了早发型阿尔茨海默病患者21号染色体上APP基因发生突变[2],这标志着AD的研究进入到分子遗传学和基因组学的水平。近些年随着全基因组关联研究(genome-wide association study,GWAS)和第二代基因测序技术(next generation sequencing,NGS)的蓬勃发展,又陆续发现了一些新的AD易感基因。目前已知与AD相关联的基因至少有50个[3],不同基因分别对应家族性早发、家族性晚发和散发性晚发三类AD。本文将就2016年至今AD易感基因的遗传学研究进展作一综述。
1.脂代谢相关基因
1.1 ABCA7基因 ABCA7基因编码ATP结合盒(ABC)转运蛋白的超家族成员,在单核细胞和巨噬细胞中高度表达,参与分子的跨膜转运,并受胆固醇的调节,这表明其在慢性炎症性疾病的作用。通过对人脑组织的研究[4,5],AD患者的ABCA7基因存在功能的潜在损失[6]。最近GWAS研究发现,ABCA7基因上的多肽位点rs4147929,rs200538373,rs142076058,rs376824416,rs78117248,rs3764650,rs3764605,rs7408475,rs3752246会增加AD的患病风险。
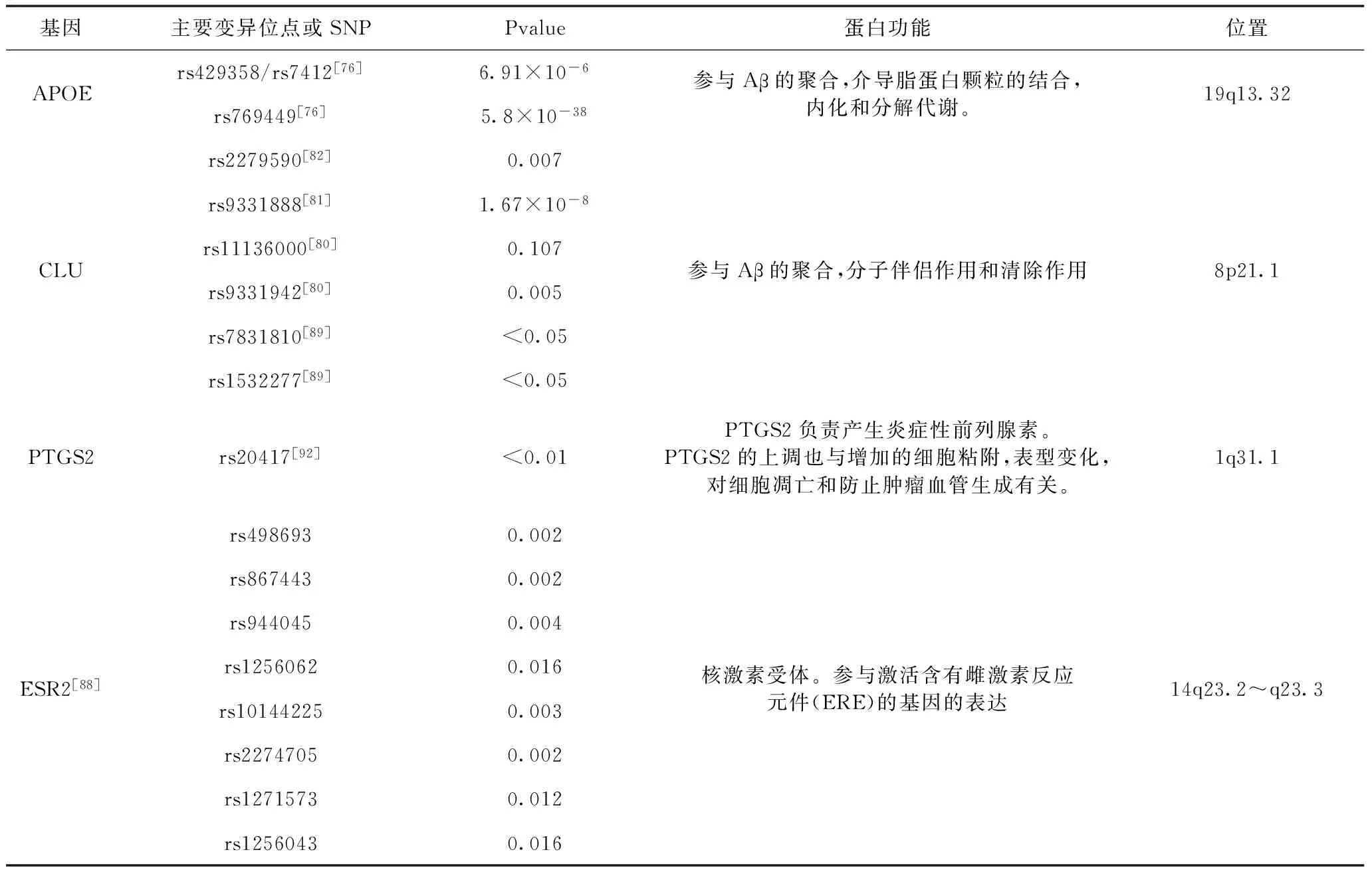
1.2 APOE基因 APOE基因是目前已被证实的散发性、迟发性(年龄>60)以及常染色体显性家族性AD的易感基因[7,8]。APOE基因的三个常见多态性,ε2,ε3和ε4导致了ApoE蛋白中的单个氨基酸变化且有强相关性改变发生阿尔茨海默病(AD)的可能性。AD的风险与APOE的等位基因相关(ε4>ε3>ε2),特别地,APOEε4与迟发性AD的风险增加相关约占患者的50%,APOEε2与迟发性AD的风险降低相关[7,9,10]。APOE不同的基因型对AD风险的影响很大程度上是ApoE蛋白对脑和脑血管中淀粉样蛋白β(Aβ)的积累产生的不同影响。但需要注意的是APOEε4对AD病程的发展不是充要条件,因此apoE的多态性不能单独用来诊断AD[11,12]。
1.3 CLU基因 CLU基因也为ApoJ基因,是晚发性AD易感基因之一[13]。CLU基因高度保守,并且广泛表达,其参与细胞周期、炎症反应、脂质运输、膜循环和细胞凋亡、激活神经元营养因子并促进神经元存货[14]。CLU表达的丛集素与Aβ结合并阻止Aβ聚集[15,16],并且丛集素能增强溶酶体对Aβ的降解[17,18],并通过血脑屏障清除Aβ[17]。通过GWAS分析研究,CLU基因上的多肽位点rs9331896,rs2279590,rs9331888,rs11136000,rs9331942,rs7831810,rs1532277与AD相关,而对CLU及其相关基因的研究对AD进程的研究有重要意义。
1.4 ESR2基因 雌激素是不仅对生殖系统,而且对中枢神经系统有影响的多效激素。目前,有人认为星形胶质细胞合成和分泌的雌激素可以调节突触形成和突触传递[19]。在过去几十年中,已经发表了关于雌激素在中枢神经系统中的有益作用的一些研究[20,21]。此外,雌激素与氧化应激保护、神经递质调节、情绪障碍和基因表达有关[22~24]。另一方面,女性绝经后雌激素水平降低是AD的重要危险因素[25,26]。然而,雌激素与AD相关的主要机制仍然未知。雌激素代谢物结合雌激素受体(ERs),其属于作为配体激活转录因子的核激素受体的超家族[27,28]。ESR2基因编码ERs的亚型ERβ。ERβ参与情感、情绪、行为和认知的调节,这都对AD产生影响[29,30]。此外,现在已经认识到雌激素通过ER亚型的不同激活影响ApoE水平[31,32]。ESR2基因中的一些在以前的研究中已经与AD相关,并且它们似乎参与基因增强子活性或基因调控[33,34]。研究表明,ESR2基因多态性与AD发病晚期有关[35],该基因的变异可能改变疾病易感性。
1.5 PTGS2基因 编码PTGS2的基因位于1q31.1在两个与阿尔茨海默病相关的区域之间[69]。PTGS2基因也称为环加氧酶2基因(COX2),已被证明在中枢神经系统的炎性细胞中被普遍表达[70]。
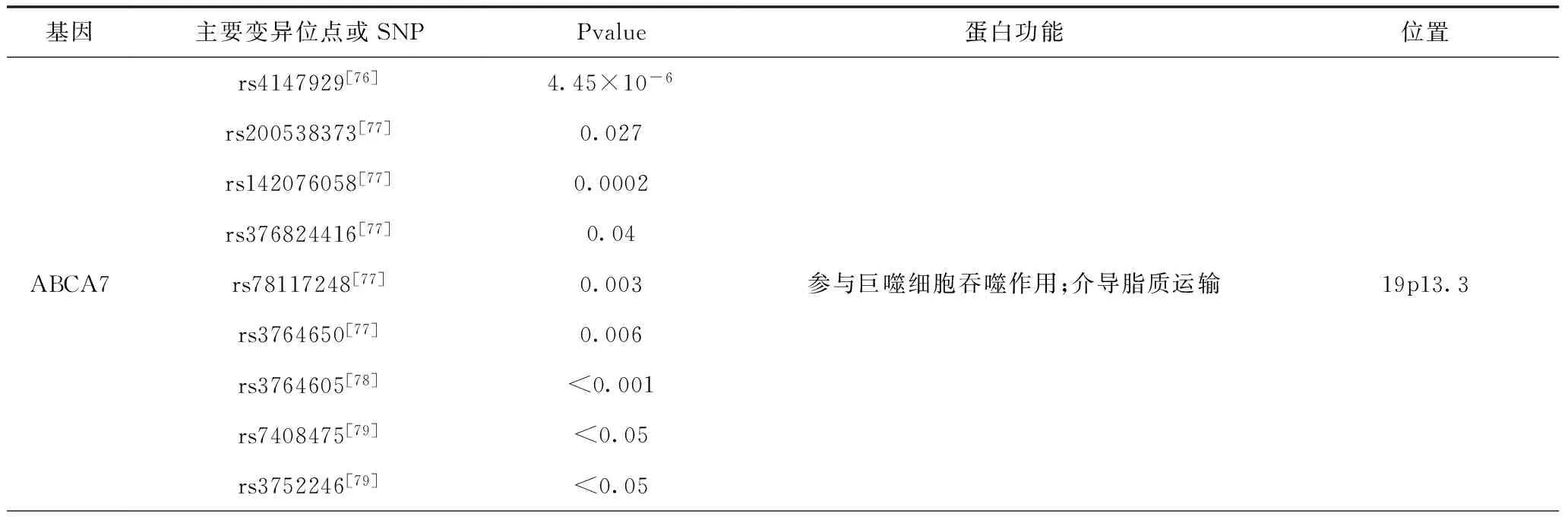
表1 AD脂代谢相关易感基因
续表1
2.免疫调节相关基因——CR1基因
CR1基因是补体激活(RCA)家族的受体成员,位于1号染色体的“簇RCA”区域。在GWAS研究中,CR1基因被认为是AD的风险基因[36]。CR1表达的凝集素蛋白在聚集和沉积中,直接影响Aβ。凝集素的存在会影响Aβ的形态和毒性[37,38]。凝集素可调节可溶性Aβ向不溶性的形式转化,如低聚物的转化,从而抑制Aβ的毒性和沉积。凝集素参与Aβ交叉形成复杂的血脑屏障,其可调节Aβ的清除[39]。本文提到的rs6656401,rs4266886,rs61822977是AD的易感基因,可增加AD患病的风险。
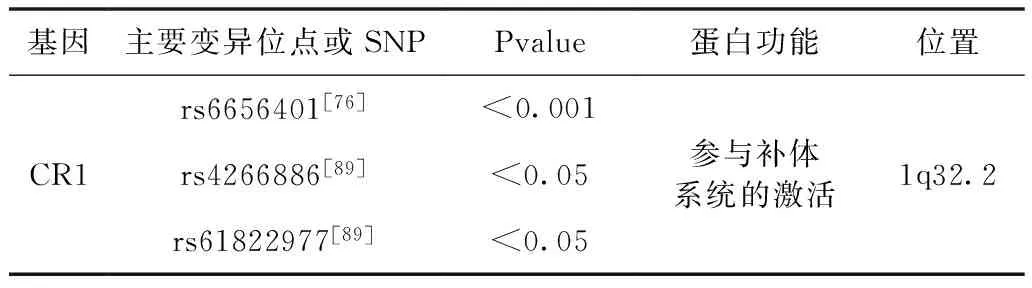
表2 AD免疫调节相关易感基因
3.内吞作用相关基因
3.1 BIN1基因 BIN1基因为APOE后最明显的LOAD易感基因座,其编码的桥联整合蛋白1具有普遍表达性和组织特异性,包括脑中特异性同型(iso1)和普遍存在的较小同型(iso9)[40~42]。经研究发现在AD患者的大脑中,iso1的水平显著降低,而iso9的水平提高[41]。已知BIN1的功能包括调节膜曲率、介导网格蛋白内吞作用和膜泡运输[41,12~18]。BIN1基因的多态性,一些会破坏BIN1接到的网格蛋白内吞作用,降低APP的加工作用[43],从而对LOAD起保护作用。相反一些破坏了对网格蛋白内吞作用的抑制作用,就会增加其对LOAD的风险。本文中提到的rs6733839,rs12989701,rs744373为LOAD的易感基因。
3.2 CD2AP基因 CD2AP编码CD2相关蛋白,这种支架蛋白参与内吞作用和膜运输作用和动态肌动蛋白重塑作用。CD2AP在大脑的神经元与毛细血管中有表达。在AD的果蝇模型中,发现果蝇的cindr为人类CD2AP直系同源物,也被认为是介导Tau调节的疾病的机制[44]。研究发现CD2AP可影响体外的Aβ水平和Aβ42/Aβ40的比例,但对体内的Aβ代谢影响甚微[45]。近期[46]研究显示,神经元的神经突长度、复杂度、生成的锥形伪足数与CD2AP的表达相符。而在发散性AD患者外周血淋巴细胞(PBL)中检测到CD2AP表达显著降低,即可能CD2AP在全身参与了发散性AD。
3.3 PICALM基因 PICALM基因编码参与网格蛋白介导的内吞作用,其为细胞内运输蛋白质、脂质、生长因子和神经递质的重要步骤。PICALM蛋白对于突触前膜的神经递质释放不可或缺,对记忆形成和神经元功能十分重要[47]。PICALM的缺陷减少了Aβ的清除和加速Aβ的病理学进程,这对Aβ在脑内的稳态和清除的治疗有影响[48]。PICALM水平也与磷酸化、自噬相关的蛋白水平相关,也与AD中的tau包涵体相关。文中提到的SNP rs3851179位于PICALM上游约90kb,保护性变体A与微血管系统中适度增加的PICALM表达相关,这可能有助于Aβ的清除,并可降低AD风险。此外,PICALM rs3851179 A等位基因与92~93岁的患者相比,具有更好的认知能力,rs3851179 GA或AA基因型显示出显着的效应与GG携带者相比,在药物遗传学测定过程中防止AD快速进展[49]。
3.4 SLC242A基因GWAS 对于与AD相关的SLC24A4的功能知之甚少。SLC24A4可能参与神经发育[50]。另外,SLC24A4基因位于RIN3基因旁边,其与早期内吞途径中的BIN1有相互作用。
3.5 SORL1基因 遗传和生物学证据表明,分拣蛋白相关受体1(SORL1)的紊乱功能与AD相关。作为AD的神经病理学标志,功能性SORL1降低脑中的淀粉样蛋白β水平,从而降低神经毒性淀粉样蛋白β斑块的负荷[59]。SORL1通过结合淀粉样蛋白前体蛋白(APP)降低淀粉样蛋白-β水平,防止其加工成淀粉样蛋白-β,通过结合淀粉样蛋白-β并将其引导至溶酶体降解[60]。对于SORL1,有趣的是,基因中常见和罕见的变体都能提高LOAD的风险。分子研究表明,SORL1的蛋白质编码区中的变体影响编码蛋白的功能,即Sortilin相关受体,导致淀粉样蛋白前体蛋白加工的改变,以及作为淀粉样蛋白斑块成分的淀粉样β肽的下游分泌[61]。
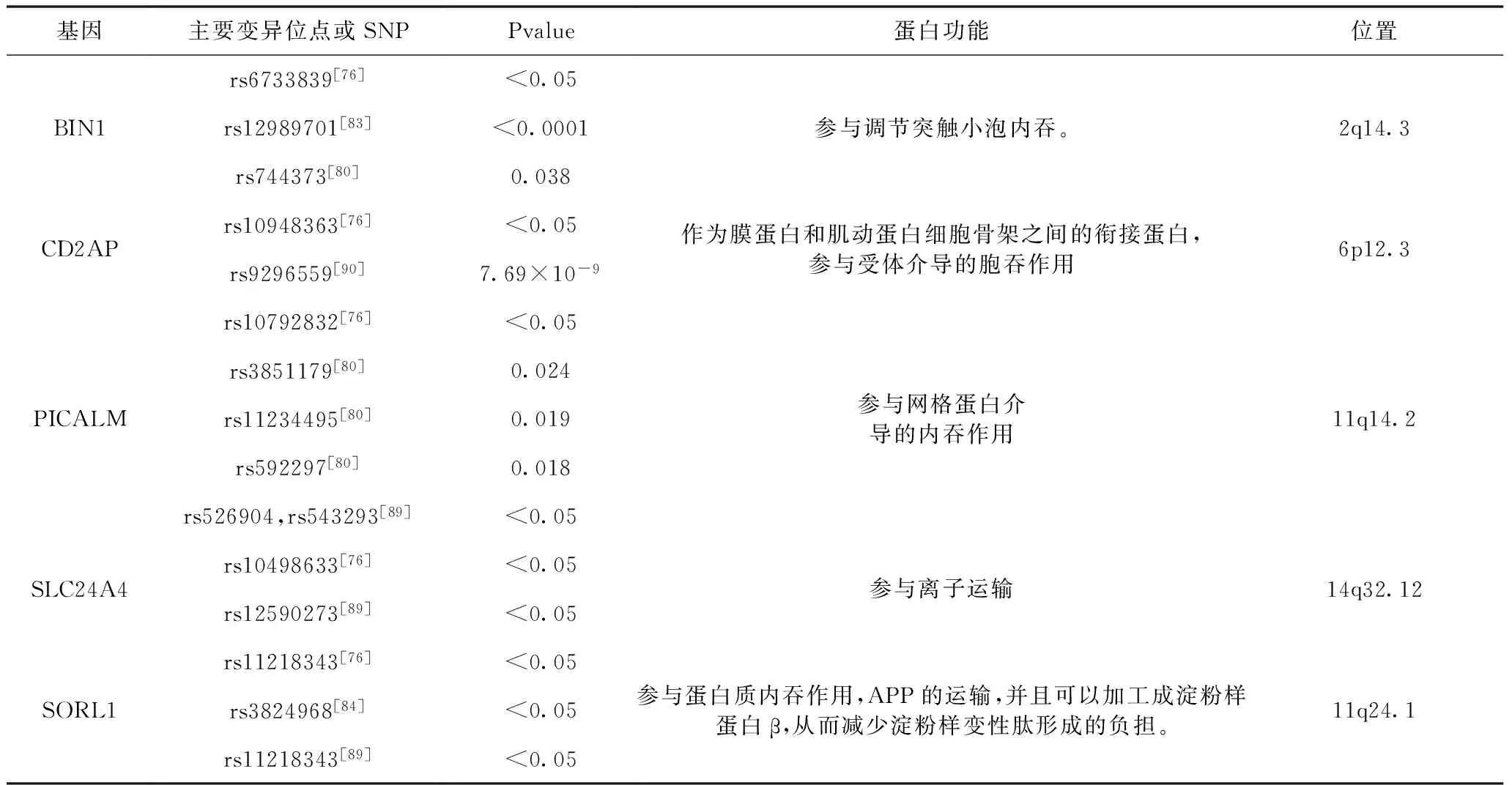
表3 AD内吞作用相关易感基因
4.神经递质相关基因
4.1 APP基因 APP基因与AD高度相关,AD的特征在于淀粉样蛋白斑,突触损失和细胞死亡。由APP基因编码的细胞表面受体和跨膜前体蛋白,经β-分泌酶和γ-分泌酶水解产生的Aβ片段是阿尔茨海默病患者闹钟淀粉样蛋白斑的蛋白质基础。Aβ是不同因素对AD的共同通路,也是AD发展的重要因素[51]。Aβ在正常人脑中经分泌酶释放到胞外区,此过程中Aβ将分裂成两半,阻止Aβ的合成。若APP突变,涉及常染色体显性阿尔茨海默病的产生。这是因为游离的Aβ变长,经聚集生成不可逆具神经毒性的沉淀,导致生成淀粉样蛋白斑进而引发AD[52]。
4.2 PSEN1和PSEN2基因 PSEN1和PSEN2是γ-分泌酶复合物的重要元素,其在APP的蛋白水解切割中具有关键作用。在早老素基因中鉴定的大多数突变聚集在跨膜结构域(TMD)中,并且它们通过APP处理导致淀粉样蛋白生成肽Aβ1~42的过量产生[56]。与PSEN2和APP基因相比,PSEN1基因突变更为频繁。迄今为止,在全球范围内已经确定了超过216个人有突变,PSEN1基因影响了476个家庭。PSEN1突变的AD患者的临床照片通常显示非常早的发病年龄和快速的疾病过程。然而,根据皮质区域病变的形态,确定了AD在遗传学上与PSEN1相关,即使在同一家族中也可能表现出广泛的表型变异性[57]。近来,焦点关注在小脑区域体积的损失与退行性皮层区域特异性互联,尤其突出了小脑在AD相关神经中的参与程度[58]。
4.3 PTK2B基因 PTK2B属于FAK蛋白质,已知其在细胞粘附途径中直接相互作用[62]。PTK2B已被报道与整合素下游的CAS家族成员物理相互作用[63]。经研究观察得到苍蝇,小鼠和人AD大脑中Tau与粘着斑基因PTK2B之间的强相互作用,共同表明PTK2B是AD中Tau病理学的重要早期标记物和调节剂。Fak(PTK2B)作为Tau毒性调节剂。其中,粘着斑激酶Fak在眼睛和独立的粘连相关性翼状起泡分析中表现为强的Tau毒性抑制剂。因此,人Tau和PTK2B蛋白在体外生物化学相互作用,PTK2B与AD患者和转基因Tau小鼠脑中进行性病理分期的超磷酸化和低聚Tau共定位。这些数据表明PTK2B作为Tau毒性的早期标记物和体内调节剂[64]。
4.4 ZCWPW1基因 ZCWPW1是组蛋白修饰读者,涉及表观遗传调控[71]。ZCWPW1作为GATS,PILRB和TRIM4的eQTL(表达数量性状位点),并显示影响RFX3和CTCF结合的指示[72]。此外,在一个大的LD区域,ZCWPW1区域中称为NYAP1的候选基因可激活神经元中的PI3K信号通路,从而抑制LOAD发生[73,74]。

表4 AD神经递质相关易感基因
5.线粒体相关基因——TOMM40基因
TOMM40(外部线粒体膜转移酶40)基因是位于19q13.2,其与APOE连锁不平衡(LD)[65]。TOMM40编码与Tom22,Tom7,Tom5和Tom6形成复合物的线粒体蛋白(Tom40),并参与外线粒体膜的蛋白质前体的整合[66]。此外,已经描述了TOMM40与APOE,APP和Aβ的相互作用,促进线粒体毒性和降低线粒体迁移率[67]。TOMM40,rs2075650中的单核苷酸多态性(SNPs)是APOE基因座周围与AD密切相关的第二个标记,已被证明是参与海马完整性,杏仁核体积和情景记忆[68]。
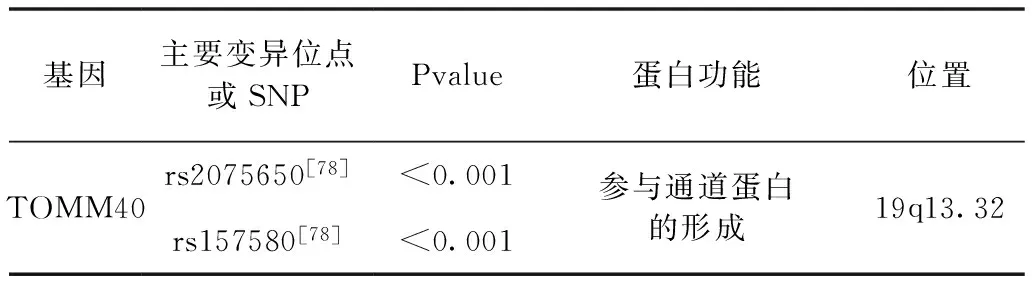
表5 AD线粒体相关易感基因
6.其他与AD相关的易感基因
6.1 MTHFR基因 高血清同型半胱氨酸水平(Hcy)的升高与多种血管疾病和AD相关。Hcy可以通过几种机制引起脑损伤,如糖尿病肾病A修复的损伤,β-淀粉样蛋白肽生成的增强和神经元对淀粉样蛋白的敏感化[53,54]。已有一些机制的提出来说明Hcy导致AD:①Hcy可以诱导钙流入和氧化应激,产生游离氧自由基,加速糖尿病肾病A损伤并最终导致神经元凋亡,②Hcy参与AD的病理生理学中的淀粉样蛋白途径参,③Hcy损害血管内皮功能,刺激血管平滑肌增生,打破凝血和出血通路之间的平衡,并介导血栓形成。最终,这些副作用减少了对大脑的供血,加速了神经细胞的凋亡[55]。
6.2 INPP5D、FERMT2基因 通过GWAS分析,得到了AD的易感基因,INPP5D和FERMT2[75]。这两种基因在AD中的作用尚不清楚,但是这两个基因改变了AD中的一直的途径。INPP5D参与了免疫反应,FERMT2还参与了tau蛋白代谢[75]。这些基因出现在基因密集区,因此目前仍不清楚它们具体和哪些基因相关联。
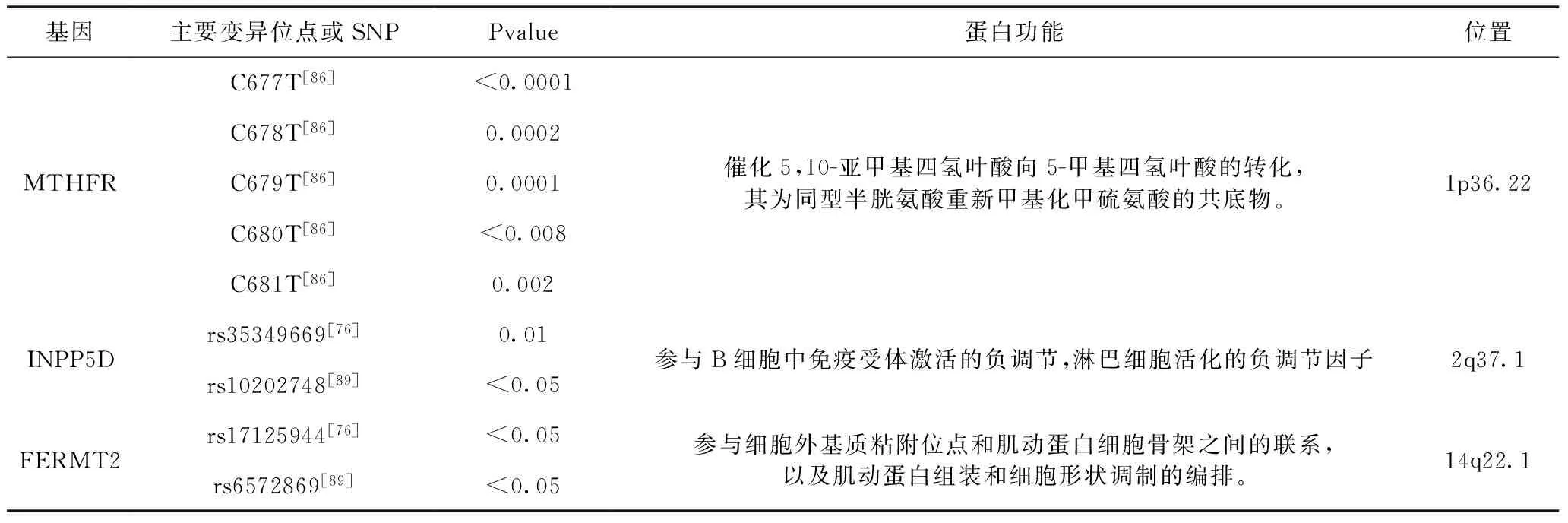
表6 其他与AD相关易感基因
AD是一种多基因多种因素共同影响的老年疾病,而AD给家庭和社会带了诸多的压力,这也致使探明AD形成机制并确定合理的治疗方案尤为重要。发现和识别新的易感基因,有助于为了解AD的形成机制及对AD的防治提供新的方向。随着全基因组关联分析和第二代测序的发展,将有越来越多的易感基因添加到已知的队列中,一些待发现的相关通路途径和细胞机制也将被识别鉴定。
1 Alagiakrishnan K, Gill SS,Fagarasanu A.Genetics and epigenetics of Alzheimer’s disease[J].Postgrad Med J,2012,88(1043): 522-529.
2 Ferri CP,Prince M,Brayne C,et al.Global prevalence of dementia: a Delphi consensus study[J].Lancet,2005,366(9503): 2112-2117.
3 Querfurth H W,LaFerla F M.Alzheimer’s Disease[J].New England Journal of Medicine,2010,362(4): 329-344.
4 Vasquez JB,Fardo DW,Estus S.ABCA7 expression is associated with Alzheimer’s disease polymorphism and disease status[J].Neurosci Lett,2013,556: 58-62.
5 Allen M,Zou F,Chai HS,et al.Novel late-onset Alzheimer disease loci variants associate with brain gene expression[J].Neurology,2012,79(3): 221-228.
6 Steinberg S,Stefansson H,Jonsson T,et al.Loss-of-function variants in ABCA7 confer risk of Alzheimer’s disease[J].Nat Genet,2015,47(5): 445-447.
7 Corder EH,Saunders AM,Strittmatter WJ,et al.Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families[J].Science,1993,261(5123): 921-923.
8 Shaw P,Lerch JP,Pruessner JC,et al.Cortical morphology in children and adolescents with different apolipoprotein E gene polymorphisms: an observational study[J].Lancet Neurol,2007,6(6): 494-500.
9 Saunders AM,Strittmatter WJ,Schmechel D,et al.Association of apolipoprotein E allele epsilon 4 with late-onset familial and sporadic Alzheimer’s disease[J].Neurology,1993,43(8): 1467-1472.
10 Saunders AM.Apolipoprotein E and Alzheimer disease: an update on genetic and functional analyses[J].J Neuropathol Exp Neurol,2000,59(9): 751-758.
11 Chiang GC,Insel PS,Tosun D,et al.Hippocampal atrophy rates and CSF biomarkers in elderly APOE2 normal subjects[J].Neurology,2010,75(22): 1976-1981.
12 Caselli RJ,Dueck AC,Osborne D,et al.Longitudinal modeling of age-related memory decline and the APOE epsilon4 effect[J].N Engl J Med,2009,361(3): 255-263.
13 Harold D,Abraham R,Hollingworth P,et al.Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease[J].Nature genetics,2009,41(10): 1088-1093.
14 Nuutinen T,Suuronen T,Kauppinen A,et al.Clusterin: a forgotten player in Alzheimer’s disease[J].Brain Res Rev,2009,61(2): 89-104.
15 Oda T,Wals P,Osterburg HH,et al.Clusterin (apoJ) alters the aggregation of amyloid beta-peptide (A beta 1-42) and forms slowly sedimenting A beta complexes that cause oxidative stress[J].Exp Neurol,1995,136(1): 22-31.
16 Matsubara E,Frangione B,Ghiso J.Characterization of apolipoprotein J-Alzheimer’s A beta interaction[J].J Biol Chem,1995,270(13): 7563-7567.
17 Bell RD,Sagare AP,Friedman AE,et al.Transport pathways for clearance of human Alzheimer’s amyloid beta-peptide and apolipoproteins E and J in the mouse central nervous system[J].J Cereb Blood Flow Metab,2007,27(5): 909-918.
18 DeMattos RB,Cirrito JR,Parsadanian M,et al.ApoE and clusterin cooperatively suppress Abeta levels and deposition: evidence that ApoE regulates extracellular Abeta metabolism in vivo[J].Neuron,2004,41(2): 193-202.
19 Hu R,Cai W Q,Wu X G,et al.Astrocyte-derived estrogen enhances synapse formation and synaptic transmission between cultured neonatal rat cortical neurons[J].Neuroscience,2007,144(4): 1229-1240.
20 Gillies G E,McArthur S.Estrogen Actions in the Brain and the Basis for Differential Action in Men and Women: A Case for Sex-Specific Medicines[J].Pharmacological Reviews,2010,62(2): 155-198.
21 McEwen BS.Invited review: Estrogens effects on the brain: multiple sites and molecular mechanisms[J].J Appl Physiol,2001,91(6): 2785-2801.
22 Douma SL,Husband C,O’Donnell ME,et al.Estrogen-related mood disorders: reproductive life cycle factors[J].ANS Adv Nurs Sci,2005,28(4): 364-375.
23 Giddabasappa A,Bauler M,Yepuru M,et al.17-β estradiol protects ARPE-19 cells from oxidative stress through estrogen receptor-β[J].Invest Ophthalmol Vis Sci,2010,51(10): 5278-5287.
24 Liu F,Day M,Mu?iz LC,et al.Activation of estrogen receptor-beta regulates hippocampal synaptic plasticity and improves memory[J].Nat Neurosci,2008,11(3): 334-343.
25 Fillit H.Estrogens in the pathogenesis and treatment of Alzheimer’s disease in postmenopausal women[J].Ann N Y Acad Sci,1994,743: 233-238.
26 Paganini-Hill A,Henderson VW.Estrogen deficiency and risk of Alzheimer’s disease in women[J].Am J Epidemiol,1994,140(3): 256-261.
27 Ascenzi P,Bocedi A,Marino M.Structure-function relationship of estrogen receptor alpha and beta: impact on human health[J].Mol Aspects Med,2006,27(4): 299-402.
28 Parker MG.Mortyn Jones Memorial Lecture.Structure and function of the oestrogen receptor[J].J Neuroendocrinol,1993,5(3): 223-228.
29 Sundermann EE,Maki PM,Bishop JR.A review of estrogen receptor alpha gene (ESR1) polymorphisms,mood,and cognition[J].Menopause,2010,17(4): 874-886.
30 ter Horst GJ.Estrogen in the limbic system[J].Vitam Horm,2010,82: 319-338.
31 Srivastava RA,Srivastava N,Averna M,et al.Estrogen up-regulates apolipoprotein E (ApoE) gene expression by increasing ApoE mRNA in the translating pool via the estrogen receptor alpha-mediated pathway[J].J Biol Chem,1997,272(52): 33360-33366.
32 Wang JM,Irwin RW,Brinton RD.Activation of estrogen receptor alpha increases and estrogen receptor beta decreases apolipoprotein E expression in hippocampus in vitro and in vivo[J].Proc Natl Acad Sci USA,2006,103(45): 16983-16988.
33 Albagha OM,McGuigan FE,Reid DM,et al.Estrogen receptor alpha gene polymorphisms and bone mineral density: haplotype analysis in women from the United Kingdom[J].J Bone Miner Res,2001,16(1): 128-134.
34 Maruyama H,Toji H,Harrington CR,et al.Lack of an association of estrogen receptor alpha gene polymorphisms and transcriptional activity with Alzheimer disease[J].Arch Neurol,2000,57(2): 236-240.
35 Pirskanen M,Hiltunen M,Mannermaa A,et al.Estrogen receptor beta gene variants are associated with increased risk of Alzheimer’s disease in women[J].Eur J Hum Genet,2005,13(9): 1000-1006.
36 Lambert JC,Heath S,Even G,et al.Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease[J].Nat Genet,2009,41(10): 1094-1099.
37 Ma J Y,Yee A,Bryan Brewer H,et al.Amyloid-associated proteins α1-antichymotrypsin and apolipoprotein E promote assembly of Alzheimer β-protein into filaments[J].Nature,1994(6501): 92-94.DOI:10.1038/372092a0.
38 DeMattos RB,O’dell MA,Parsadanian M,et al.Clusterin promotes amyloid plaque formation and is critical for neuritic toxicity in a mouse model of Alzheimer’s disease[J].Proc Natl Acad Sci USA,2002,99(16): 10843-10848.
39 Bell RD,Sagare AP,Friedman AE,et al.Transport pathways for clearance of human Alzheimer’s amyloid beta-peptide and apolipoproteins E and J in the mouse central nervous system[J].J Cereb Blood Flow Metab,2007,27(5): 909-918.
40 DuHadaway JB,Lynch FJ,Brisbay S,et al.Immunohistochemical analysis of Bin1/Amphiphysin II in human tissues: diverse sites of nuclear expression and losses in prostate cancer[J].J Cell Biochem,2003,88(3): 635-642.
41 Tan MS,Yu JT,Tan L.Bridging integrator 1 (BIN1): form,function,and Alzheimer’s disease[J].Trends Mol Med,2013,19(10): 594-603.
42 Wechsler-Reya R,Sakamuro D,Zhang J,et al.Structural analysis of the human BIN1 gene.Evidence for tissue-specific transcriptional regulation and alternate RNA splicing[J].J Biol Chem,1997,272(50): 31453-31458.
43 Prokic I,Cowling BS,Laporte J.Amphiphysin 2 (BIN1) in physiology and diseases[J].J Mol Med,2014,92(5): 453-463.
44 Shulman JM,Imboywa S,Giagtzoglou N,et al.Functional screening in Drosophila identifies Alzheimer’s disease susceptibility genes and implicates Tau-mediated mechanisms[J].Hum Mol Genet,2014,23(4): 870-877.
45 Liao F,Jiang H,Srivatsan S,et al.Effects of CD2-associated protein deficiency on amyloid-β in neuroblastoma cells and in an APP transgenic mouse model[J].Molecular Neurodegeneration,2015,10(1).
46 Harrison BJ,Venkat G,Lamb JL,et al.The Adaptor Protein CD2AP Is a Coordinator of Neurotrophin Signaling-Mediated Axon Arbor Plasticity[J].J Neurosci,2016,36(15): 4259-4275.
47 Bushlin I,Petralia RS,Wu F,et al.Clathrin assembly protein AP180 and CALM differentially control axogenesis and dendrite outgrowth in embryonic hippocampal neurons[J].J Neurosci,2008,28(41): 10257-10271.
48 Zhao Z,Sagare AP,Ma Q,et al.Central role for PICALM in amyloid-β blood-brain barrier transcytosis and clearance[J].Nat Neurosci,2015,18(7): 978-987.
49 Ando K,Tomimura K,Sazdovitch V,et al.Level of PICALM,a key component of clathrin-mediated endocytosis,is correlated with levels of phosphotau and autophagy-related proteins and is associated with tau inclusions in AD,PSP and Pick disease[J].Neurobiol Dis,2016,94: 32-43.
50 Larsson M,Duffy DL,Zhu G,et al.GWAS findings for human iris patterns: associations with variants in genes that influence normal neuronal pattern development[J].Am J Hum Genet,2011,89(2): 334-343.
51 Saunders AM.Apolipoprotein E and Alzheimer disease: an update on genetic and functional analyses[J].J Neuropathol Exp Neurol,2000,59(9): 751-758.
52 Hardy J,Selkoe DJ.The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics[J].Science,2002,297(5580): 353-356.
53 Kruman II,Culmsee C,Chan SL,et al.Homocysteine elicits a DNA damage response in neurons that promotes apoptosis and hypersensitivity to excitotoxicity[J].J Neurosci,2000,20(18): 6920-6926.
54 Kruman II,Kumaravel TS,Lohani A,et al.Folic acid deficiency and homocysteine impair DNA repair in hippocampal neurons and sensitize them to amyloid toxicity in experimental models of Alzheimer’s disease[J].J Neurosci,2002,22(5): 1752-1762.
55 Kimura M,Umegaki K,Higuchi M,et al.Methylenetetrahydrofolate reductase C677T polymorphism,folic acid and riboflavin are important determinants of genome stability in cultured human lymphocytes[J].J Nutr,2004,134(1): 48-56.
56 Campion D,Dumanchin C,Hannequin D,et al.Early-onset autosomal dominant Alzheimer disease:prevalence,genetic heterogeneity,and mutation spectrum[J].American Journal of Human Genetics,1999,65(3):664.
57 Bruni AC,Bernardi L,Colao R,et al.Worldwide distribution of PSEN1 Met146Leu mutation: a large variability for a founder mutation[J].Neurology,2010,74(10): 798-806.
58 Guo CC,Tan R,Hodges JR,et al.Network-selective vulnerability of the human cerebellum to Alzheimer’s disease and frontotemporal dementia[J].Brain,2016,139(Pt 5): 1527-1538.
59 Cuccaro ML,Carney RM,Zhang Y,et al.SORL1 mutations in early-and late-onset Alzheimer disease[J].Neurol Genet,2016,2(6): e116.
60 Holstege H,van der Lee SJ,Hulsman M,et al.Characterization of pathogenic SORL1 genetic variants for association with Alzheimer’s disease: a clinical interpretation strategy[J].Eur J Hum Genet,2017,25(8): 973-981.
61 Vardarajan BN,Zhang Y,Lee JH,et al.Coding mutations in SORL1 and Alzheimer disease[J].Ann Neurol,2015,77(2): 215-227.
62 Tikhmyanova N,Little JL,Golemis EA.CAS proteins in normal and pathological cell growth control[J].Cell Mol Life Sci,2010,67(7): 1025-1048.
63 Beck TN,Nicolas E,Kopp MC,et al.Adaptors for disorders of the brain? The cancer signaling proteins NEDD9,CASS4,and PTK2B in Alzheimer’s disease[J].Oncoscience,2014,1(7): 486-503.
64 Dourlen P,Fernandez-Gomez FJ,Dupont C,et al.Functional screening of Alzheimer risk loci identifies PTK2B as an in vivo modulator and early marker of Tau pathology[J].Mol Psychiatry,2017,22(6): 874-883.
65 Roses AD,Lutz MW,Amrine-Madsen H,et al.A TOMM40 variable-length polymorphism predicts the age of late-onset Alzheimer’s disease[J].Pharmacogenomics J,2010,10(5): 375-384.
66 Rapaport D.How does the TOM complex mediate insertion of precursor proteins into the mitochondrial outer membrane[J].J Cell Biol,2005,171(3): 419-423.
67 Devi L.Accumulation of Amyloid Precursor Protein in the Mitochondrial Import Channels of Human Alzheimer’s Disease Brain Is Associated with Mitochondrial Dysfunction[J].Journal of Neuroscience,2006,26(35): 9057-9068.
68 Shen L,Kim S,Risacher SL,et al.Whole genome association study of brain-wide imaging phenotypes for identifying quantitative trait loci in MCI and AD: A study of the ADNI cohort[J].Neuroimage,2010,53(3): 1051-1063.
69 Blacker D,Bertram L,Saunders AJ,et al.Results of a high-resolution genome screen of 437 Alzheimer’s disease families[J].Hum Mol Genet,2003,12(1): 23-32.
70 Chen Q,Liang B,Wang Z,et al.Influence of four polymorphisms in ABCA1 and PTGS2 genes on risk of Alzheimer’s disease: a meta-analysis[J].Neurol Sci,2016,37(8): 1209-1220.
71 He F,Umehara T,Saito K,et al.Structural insight into the zinc finger CW domain as a histone modification reader[J].Structure,2010,18(9): 1127-1139.
72 Rosenthal SL,Barmada MM,Wang X,et al.Connecting the dots: potential of data integration to identify regulatory SNPs in late-onset Alzheimer’s disease GWAS findings[J].PLoS ONE,2014,9(4): e95152.
73 Lambert JC,Ibrahim-Verbaas CA,Harold D,et al.Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease[J].Nat Genet,2013,45(12): 1452-1458.
74 Yokoyama K,Tezuka T,Kotani M,et al.NYAP: a phosphoprotein family that links PI3K to WAVE1 signalling in neurons[J].EMBO J,2011,30(23): 4739-4754.
75 Lambert JC,Ibrahim-Verbaas CA,Harold D,et al.Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease[J].Nat Genet,2013,45(12): 1452-1458.
76 Jakobsdottir J,Van der Lee SJ,Bis JC,et al.Rare Functional Variant in TM2D3 is Associated with Late-onset Alzheimer’s Disease[J].PLOS genetics,2016,12(10):e1006327.
77 Kunkle BW,Carney RM,Kohli MA,et al.Targeted sequencing of ABCA7 identifies splicing,stop-gain and intronic risk variants for Alzheimer disease[J].Neurosci Lett,2017,649: 124-129.
78 Bao J,Wang X J,Mao Z F.Associations Between Genetic Variants in 19p13 and 19q13 Regions and Susceptibility to Alzheimer Disease: A Meta-Analysis[J].Medical Science Monitor,2016,22: 234-243.
79 Desikan RS,Fan CC,Wang Y,et al.Genetic assessment of age-associated Alzheimer disease risk: Development and validation of a polygenic hazard score[J].PLoS Med,2017,14(3): e1002258.
80 Wang HZ,Bi R,Hu QX,et al.Validating GWAS-Identified Risk Loci for Alzheimer’s Disease in Han Chinese Populations[J].Mol Neurobiol,2016,53(1): 379-390.
81 Zhang S,Li X,Ma G,et al.CLU rs9331888 Polymorphism Contributes to Alzheimer’s Disease Susceptibility in Caucasian But Not East Asian Populations[J].Mol Neurobiol,2016,53(3): 1446-1451.
82 Huang F,Shang Y,Luo Y,et al.Lower Prevalence of Alzheimer’s Disease among Tibetans: Association with Religious and Genetic Factors[J].J Alzheimers Dis,2016,50(3): 659-667.
83 Dong X,Zhang L,Meng Q,et al.Association Between Interleukin-1A,Interleukin-1B,and Bridging integrator 1 Polymorphisms and Alzheimer’s Disease: a standard and Cumulative Meta-analysis[J].Mol Neurobiol,2017,54(1): 736-747.
84 Huang CC,Liu ME,Kao HW,et al.Effect of Alzheimer’s Disease Risk Variant rs3824968 at SORL1 on Regional Gray Matter Volume and Age-Related Interaction in Adult Lifespan[J].Sci Rep,2016,6: 23362.
85 Li YQ,Tan MS,Wang HF,et al.Common variant in PTK2B is associated with late-onset Alzheimer’s disease: A replication study and meta-analyses[J].Neurosci Lett,2016,621: 83-87.
86 Rai V.Folate Pathway Gene Methylenetetrahydrofolate Reductase C677T Polymorphism and Alzheimer Disease Risk in Asian Population[J].Indian J Clin Biochem,2016,31(3): 245-252.
87 Bagyinszky E,Youn Y C,An S S A,et al.Mutations,associated with early-onset Alzheimer’s disease,discovered in Asian countries[J].Clinical Interventions in Aging,2016,11: 1467-1488.
88 Chen LH,Fan YH,Kao PY,et al.Genetic Polymorphisms in Estrogen Metabolic Pathway Associated with Risks of Alzheimer’s Disease: Evidence from a Southern Chinese Population[J].J Am Geriatr Soc,2017,65(2): 332-339.
89 Desikan RS,Fan CC,Wang Y,et al.Genetic assessment of age-associated Alzheimer disease risk: Development and validation of a polygenic hazard score[J].PLoS Med,2017,14(3): e1002258.
90 Tao QQ,Liu ZJ,Sun YM,et al.Decreased gene expression of CD2AP in Chinese patients with sporadic Alzheimer’s disease[J].Neurobiol Aging,2017,56: 212-215.
91 Lee JH,Lee AJ,Dang LH,et al.Candidate gene analysis for Alzheimer’s disease in adults with Down syndrome[J].Neurobiol Aging,2017,56: 150-158.
92 Chen Q,Liang B,Wang Z,et al.Influence of four polymorphisms in ABCA1 and PTGS2 genes on risk of Alzheimer’s disease: a meta-analysis[J].Neurol Sci,2016,37(8): 1209-1220.
Advances in genetics of Alzheimer’s disease
(GAOShan1,2,CHENChao2,LIZhipeng2,YANGZe1.1.InstituteofGeriatrics,MinistryofHealth,BeijingHospital,Beijing100730.2.BeijingNormalUniversity(Zhuhai),Zhuhai518000,China.)
Alzheimer’s disease (AD) is a common central neurodegenerative disease.It is generally believed that the occurrence of AD is the result of the interaction among genetic,environmental,metabolic and other factors.The AD research reach level of molecular genetics and genomics since the end of the 20th century have gradually revealed some genes implicated in AD.Here we review the AD risk genes (ABCA7,APOE,CLU,ESR2,CR1,INPP5D,BIN1,CD2AP,PICALM,SLC242A,APP,MTHFR,PSEN1,PSEN2,SORL1,PTK2B,TOMM40,PTGS2,ZCWPW1,FERMT2 ) for AD since 2016.
Alzheimer’s disease, Risk gene
1.北京医院 北京老年医学研究所,100730 2.北京师范大学珠海分校 518000
国家自然科学基金(81460203,8321120527,81370265,8147108,81400710),卫生部公益性研究基金(201192008)和国家科技部十二五支撑计划(2012BAI10B01),北京市科技新星计划(Z12133002512058)。
10.3969/j.issn.1672-4860.2017.04.006
2017-7-15
※Correspondingauthor:ChenChao,YangZe.
