含吡咯环的缩氨基硫脲席夫碱镍、铜配合物的晶体结构及与DNA的相互作用
2017-07-05李晓静毛盼东吴伟娜寇凯刘树阳王元
李晓静 毛盼东 吴伟娜 寇凯 刘树阳 王元
(河南理工大学化学化工学院,焦作454000)
含吡咯环的缩氨基硫脲席夫碱镍、铜配合物的晶体结构及与DNA的相互作用
李晓静 毛盼东 吴伟娜*寇凯 刘树阳 王元*
(河南理工大学化学化工学院,焦作454000)
合成并通过单晶衍射、元素分析及红外光谱表征了配合物[Ni(L1)2]·2DMF(1),[Cu(L1)2]·THF·0.25MeOH·2.25H2O(2),[Ni(L2)2] ·2MeOH(3)和[Cu(L2)2]·2EtOH(4)的结构(HL1:5-甲酰基-3,4-二甲基-吡咯-2-甲酸乙酯缩硫代氨基脲,HL2:5-甲酰基-2,4-二甲基-吡咯-3-甲酸乙酯缩4-异丙基氨基硫脲)。单晶衍射结果表明,除溶剂分子不同外,配合物1~4的结构相似。每个配合物的中心金属离子分别与来自2个阴离子L-配体的N2S2电子供体配位,采取扭曲的平面正方形配位构型。荧光光谱结果表明,配合物与DNA的相互作用强于其配体。
吡咯;缩氨基硫脲;配合物;晶体结构;DNA相互作用
Recently,thiosemicarbazones(TSCs)and their transition metal complexes,have attracted intensity attention in the coordination chemistry because of their high biological and pharmaceutical activities, such as antibacterial,antiviral,antifungal,antitumor activity and so on[1-4].Recently,there is increasinginterest in the structure design of thiosemicarbazones derivatives with the purpose of improving the pharmaceutical properties and functions:a heterocyclic ring in the synthesized TSCs plays a major role in extending their pharmacological properties[2-3];the presence of a bulky group at the terminal nitrogen N(4)of TSCs could increase the biological activity[5-6,8].Furthermore,in most cases, coordination to metal ions results in improvement of the pharmacological activity of TSCs.Therefore,a large amount of TSCs metals containing six-member heterocycles have been found to possess considerable antitumor activity[6-8].However,the studieson antitumor activities of the complexes with TSCs bearing fivemember heterocycles,especially pyrrole,are relatively few[9-10].
In fact,our previous work shows that some pyrrole acylhydrazone Cucomplexes have certain antitumor activity[9-10].As a continuation of our research on TSC-metals,in this paper,two Cuand two Nicomplexeswith two pyrrole TSC ligands have been synthesized and structural determined by singlecrystal X-ray diffraction.In addition,the interactions between all compounds and ct-DNA have been studied by ethidium bromide(EB)fluorescence probe.
1 Experimental
1.1 M aterials and measurements
Solvents and startingmaterials for syntheseswere purchased commercially and used as received. Elemental analyses were carried out on an Elemental Vario EL analyzer.1H NMR spectra were recorded on a Bruker AV400 NMR spectrometer in DMSO-d6solution,with TMS as internal standard.The IR spectra(ν=4 000~400 cm-1)were determined by the KBr pressed disc method on a Bruker V70 FTIR spectrophotometer.The UV spectra were recorded on a Purkinje General TU-1800 spectrophotometer.The interactions between the compounds and ct-DNA are measured using literature method[11].Briefly,upon the addition of varying concentrations of the investigated complexes in 5mmol·L-1Tris-HCl+50mmol·L-1NaCl (pH=7.35)into the ethidium bromide(EB)-DNA solution(6.25μmol·L-1EB and 50μmol·L-1ct-DNA),emission spectra were recorded on a Varian CARY Eclipse spectrophotometer with the excitation wavelength set at 490 nm.
1.2 Preparations of HL1and HL2
As shown in Scheme 1,the ligand HL1was prepared by condensation of ethyl 5-formyl-3,4-dimethyl-1H-pyrrole-2-carboxylate(1.95 g,11 mmol) with thiosemicarbazide(0.91 g,10 mmol)in ethanol solution(30 mL)under reflux condition for 4 h.The yellow powder was filtered and washed three times with ethanol.Yield:2.48 g(84%).m.p.258~262℃. Anal.Calcd.For HL1(C11H16N4O2S)(%):C,49.24;H, 6.01;N,20.88;Found(%):C,49.19;H,6.08;N,20.91.1H NMR(400 MHz,DMSO-d6):δ11.306(1H,s,NH), 11.113(1H,s,NH),8.152(2H,s,NH),7.882(1H,s, CH=N),4.090~4.150(2H,q,CH2CH3),2.437(3H,s, CH3),2.109(3H,s,CH3),1.076~1.081(3H,t,CH3CH2). FTIR(cm-1):ν(O=C)1 656,ν(N=C)1 536,ν(C=S) 897.
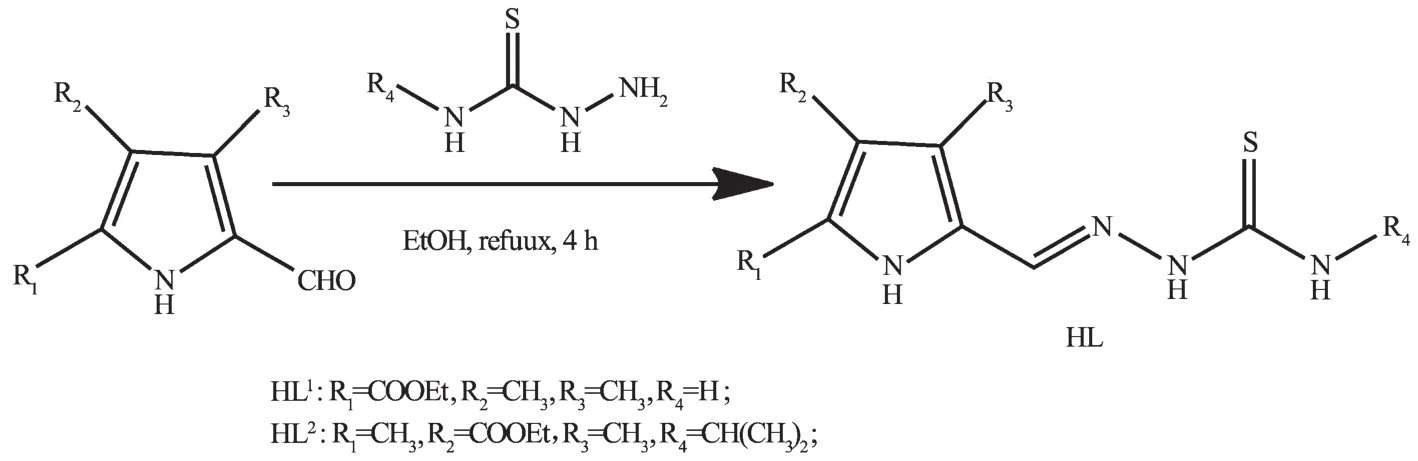
Scheme 1 Synthetic procedure for two TSC ligands HL1and HL2
HL2was synthesized by same method as that of HL1,while using ethyl 5-formyl-2,4-dimethyl-1H-pyrrole-3-carboxylate and N(4)-isopropyl thiosemicarbazide as startingmaterials.Yield:2.759 g(89%).m. p.:238~242℃.Anal.Calcd.for HL2(C14H22N4O2S)(%):C,54.17;H,7.14;N,18.05;Found(%):C,53.98; H,7.24;N,18.13.1H NMR(400 MHz,DMSO-d6):δ 11.29(1H,s,NH),11.26(1H,s,NH),8.17~8.19(1H, d,NH),7.97(1H,s,CH=N),4.41~4.49(1H,m,CH), 4.20~4.26(2H,q,CH2),2.14(3H,s,CH3),1.96(3H, s,CH3),1.24~1.28(3H,t,CH3CH2),1.18~1.19(6H,d, CH3CH2).FTIR(cm-1):ν(O=C)1675,ν(N=C)1 565,ν (C=S)930.
Crystals of HL1·0.5H2O and HL2·EtOH suitable for X-ray diffraction analysis were obtained by recrystallization of both TSC ligands from the ethanol solution,respectively.
1.3 Preparations of the com plexes
The complexes 1~2 and 3~4 were generated by reaction of HL1(5 mmol)with equimolar of Ni(OAc)2· 6H2O in MeOH/DMF(20mL,3∶1,V/V)and Cu(OAc)2· 2H2O in MeOH/THF(20 mL,1∶1,V/V),HL2(5 mmol) with equimolar of Ni(OAc)2·6H2O in MeOH/THF (20 mL,2∶1,V/V)and Cu(OAc)2·2H2O in EtOH/THF (20mL,2∶1,V/V),respectively.
1:Brown blocks.Anal.Calcd.for(C28H44N10O6S2Ni) (%):C,45.47;H,6.00;N,18.94.Found(%):C,45.52; H,6.07;N,18.81.FTIR(cm-1):ν(O=C)1661,ν(N=C-S) 1586,ν(C=N)1 518,ν(C-S)863.
2:Brown blocks.Anal.Calcd.for (C26.25H43.5CuN8O7.5S2)(%):C,43.86;H,6.10;N,15.59. Found(%):C,44.02;H,6.19;N,15.55.FTIR(cm-1):ν (O=C)1 632,ν(N=C-S)1558,ν(C=N)1 508,ν(C-S)862.
3:Brown blocks.Anal.Calcd.for(C30H50N8O6S2Ni) (%):C,48.59;H,6.80;N,15.11.Found(%):C, 48.64;H,6.73;N,15.14.FTIR(cm-1):ν(O=C)1 663, ν(N=C-S)1 597,ν(C=N)1 548,ν(C-S)910.
4:Brown blocks.Anal.Calcd.for(C32H54N8O6S2Cu) (%):C,49.62;H,7.03;N,14.47.Found(%):C,49.53;H, 7.09;N,14.51.FTIR(cm-1):ν(O=C)1 662,ν(N=C-S) 1 621,ν(C=N)1 512,ν(C-S)908.
1.3 X-ray crystallography
The X-ray diffraction measurement for HL1· 0.5H2O,HL2·EtOH and complexes 1~4 were performed on a Bruker SMART APEXⅡCCD diffractometer equipped with a graphite monochromatized Mo Kαradiation(λ=0.071 073 nm) by usingφ-ωscan mode.Semi-empirical absorption correction was applied to the intensity data using the SADABS program[12].The structures were solved by directmethods and refined by fullmatrix least-square on F2using the SHELXTL-97 program[13].All nonhydrogen atoms were refined anisotropically.H atoms for O3(situated on special position),O4 and C12-C15 (occupancy value of each atom being 0.5),O5,O6 and C16(occupancy value of each atom being 0.125)of 2 were not added but directly included in themolecular formula.All the other H atoms were positioned geometrically and refined using a riding model. Details of the crystal parameters,data collection and refinements for HL1·0.5H2O,HL2·EtOH and complexes 1~4 were summarized in Table 1.
CCDC:1424498,HL1·0.5H2O;1424499,1; 1424500,2;1427468,HL2·EtOH;1427469,3;1427 470,4.
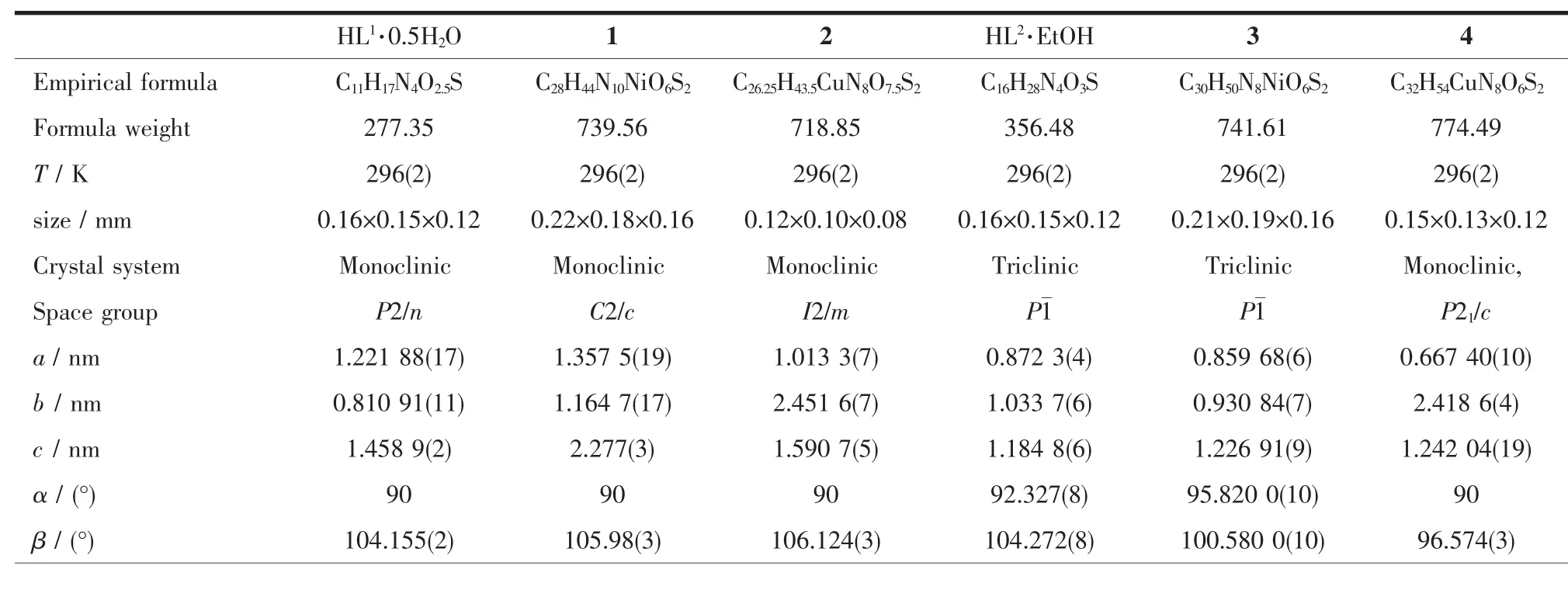
Table 1 Selected crystallographic data for HL1,HL2and com plexes 1~4
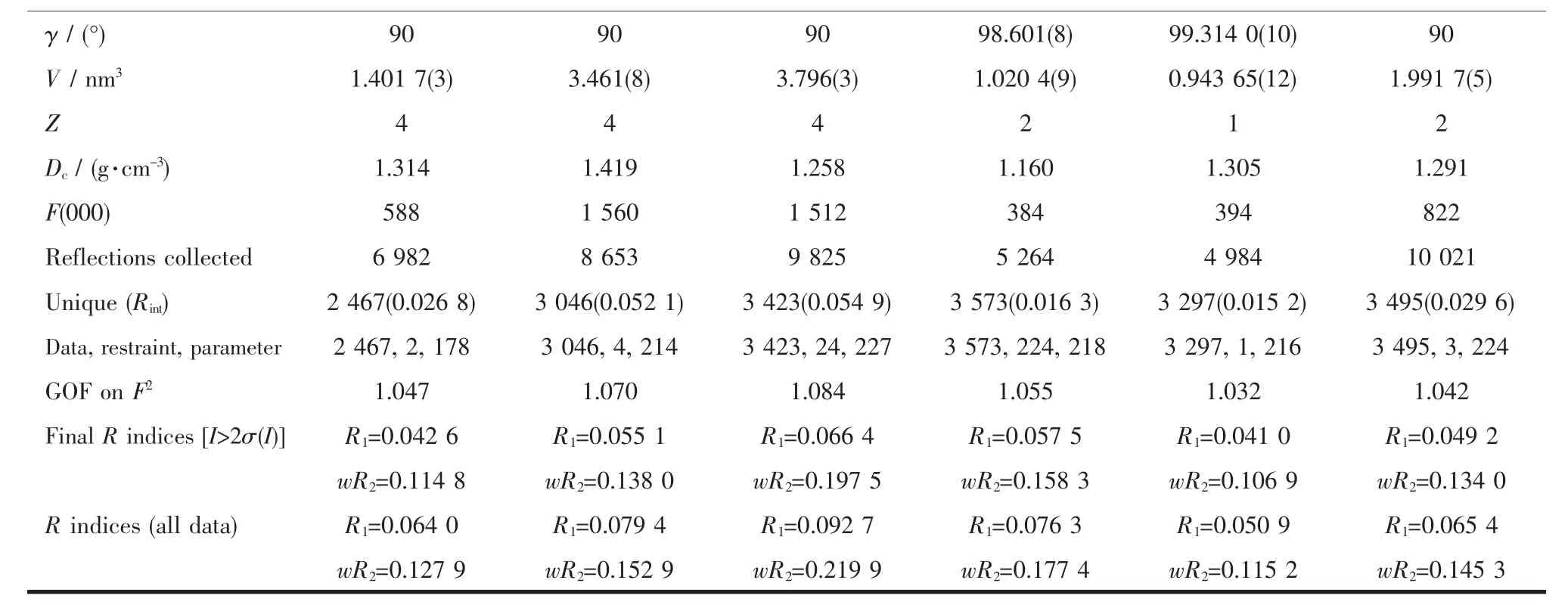
Continued Table 1
2 Result and discussion
2.1 Crystal structures description
A diamond drawing of HL1·0.5H2O,HL2·EtOH and complexes 1~4 is shown in Fig.1.Selected bond distances and angles are listed in Table 2.Hydrogen bonds information is in Table 3.As shown in Fig.1a and 1d,HL1and HL2in the structures of HL1·0.5H2O and HL2·EtOH,are in a thione form,respectively[11-12]. The lengths of S1-C11 in HL1·0.5H2O,HL2·EtOH are 0.168 7(2)and 0.169 5(3)nm,respectively.However, those change to 0.172 5(3),0.172 7(5),0.172 8(3)) and 0.174 5(3)nm in complexes 1~4,respectively, showing that the TSC ligands HL1and HL2have thiolated and deprotonated in the complexes[14]. Furthermore,the imine C=N bond(N2-C10)has E configuration in HL1and HL2,while has Z configuration in the complexes.In the crystal of HL1·0.5H2O,one lattice water molecule links two HL1molecules to form a centrosymmetric dimer via intermolecular N-H…O and O-H…O hydrogen bonds, which are further connected with each other through two pairs of intermolecular N-H…S hydrogen bonds (Fig.2a),giving an extended 3D structure.In the structure of HL2·EtOH,a similar dimer is formed by intermolecular O-H…O and N-H…O hydrogen bonds between HL2and crystal methanol molecule.Pairs of intermolecular N-H…S hydrogen are helpful to construct an extended one dimentional chain(Fig.2d).
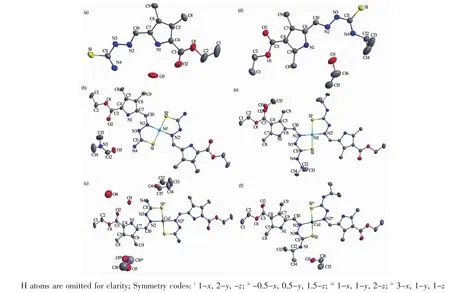
Fig.1 Diamond drawing of HL1·0.5H2O(a),HL2·EtOH(d)and complexes 1~4(b,c,e and f,respectively)with 30% thermal ellipsoids
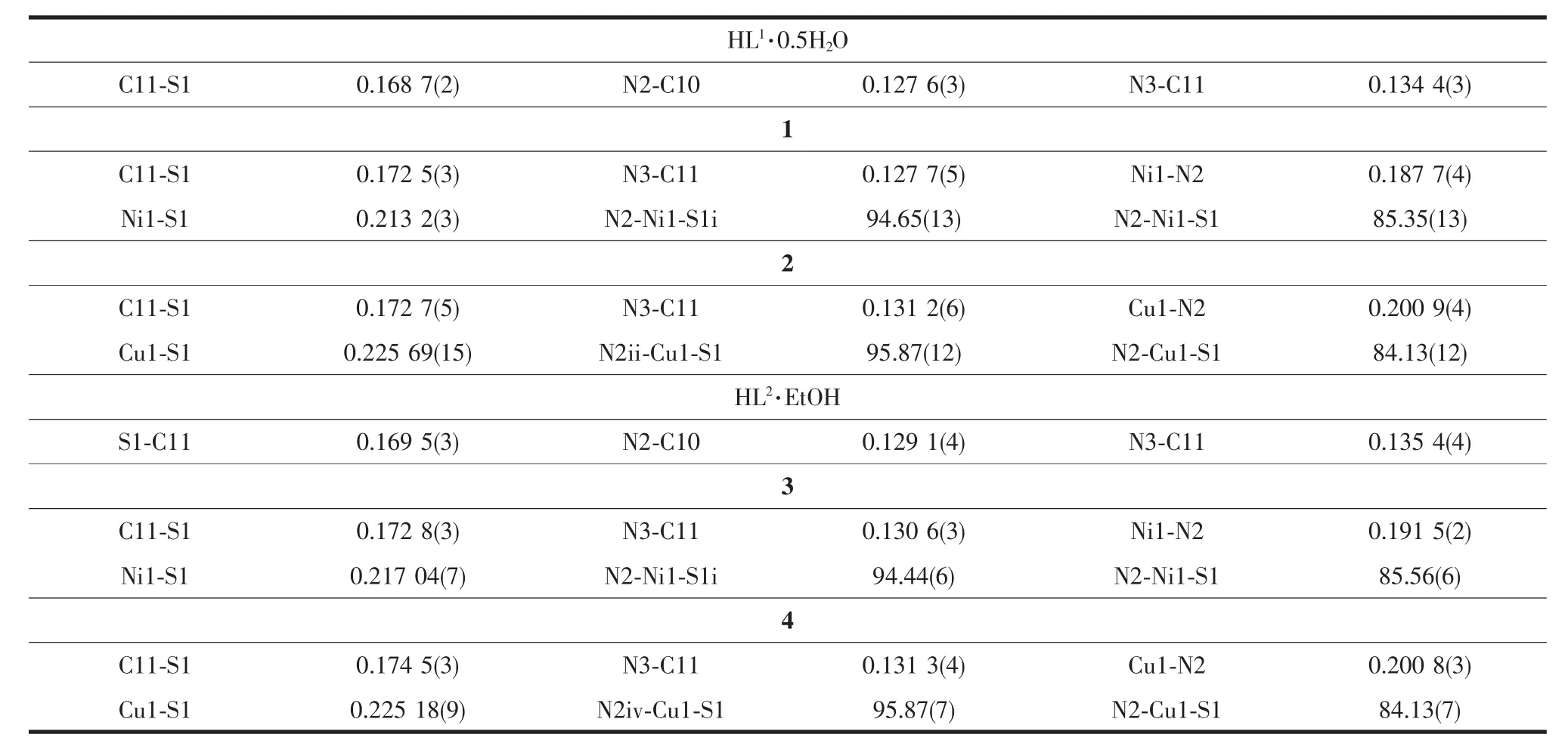
Table 2 Selected bond lengths(nm)and angles(°)in HL1·0.5H2O,HL2·EtOH and comp lexes 1~4
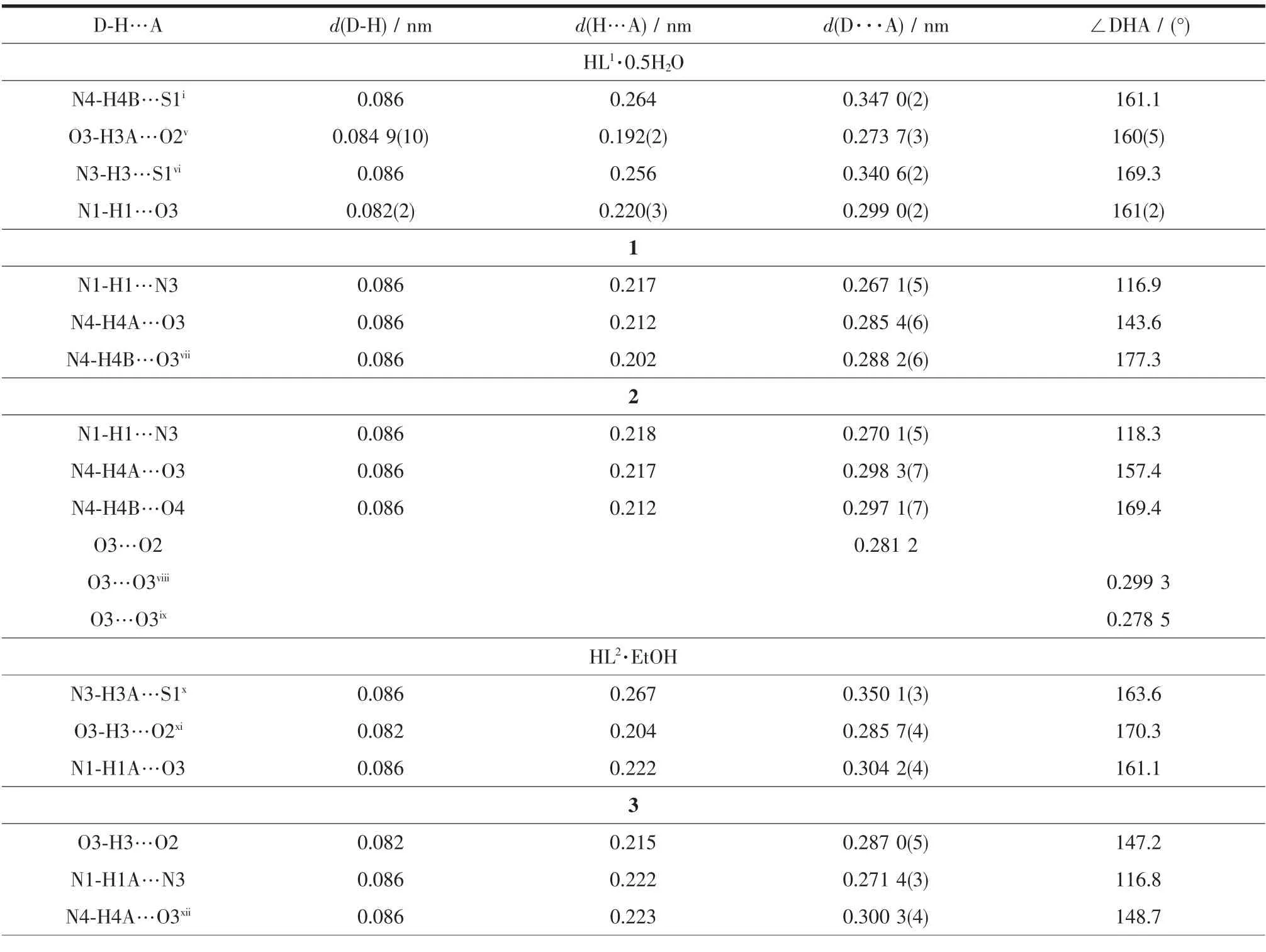
Table 3 Hydrogen bonds information in HL1·0.5H2O,HL2·EtOH and com plexes 1~4
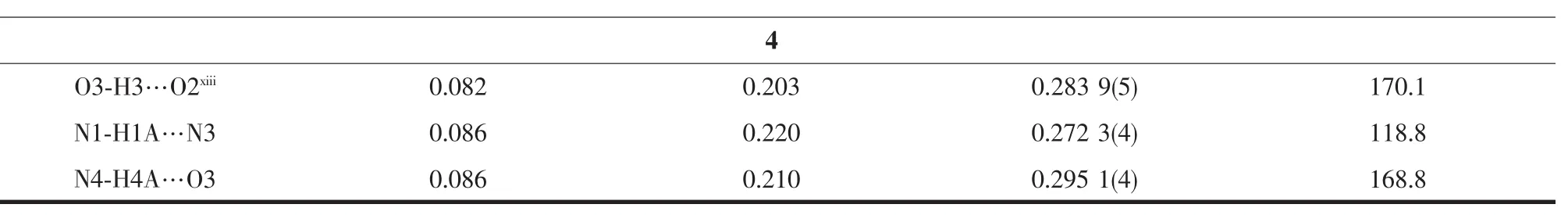
Continued Table 3
The structures of complexes 1~4 are similar while with different solvent molecules.As shown in Fig.1,the central metal ion in each complex is coordinated with two independent thiolated TSC ligands with N2S2donor set,thus giving a distorted square planar geometry.The coordination bond lengths of M-N/S of complexes 1~4 are comparable with those of some reported complexes with similar donor set[8,15].In complex 1(Fig.2b)and 3(Fig.2d), crystal DMF and methanol molecules link the complexes into chains via the intermolecular N-H…O (additional O-H…O for 3)hydrogen bonds, respectively.In complex 2(Fig.2c),an extended 3Dstructure is formed by intermolecular N-H…O and NH…O hydrogen bonds involving the complex,lattice water and THF molecules.However,in complex 4, crystal ethanol molecules link the complexes into an extended 2D supramolecular structure through intermolecular O-H…O and N-H…O hydrogen bonds (Fig.2f).Intramolecular N-H…N hydrogen bonds between the nitrogen atom ofpyrrole ring and the imine nitrogen atom are also present in complexes1~4.
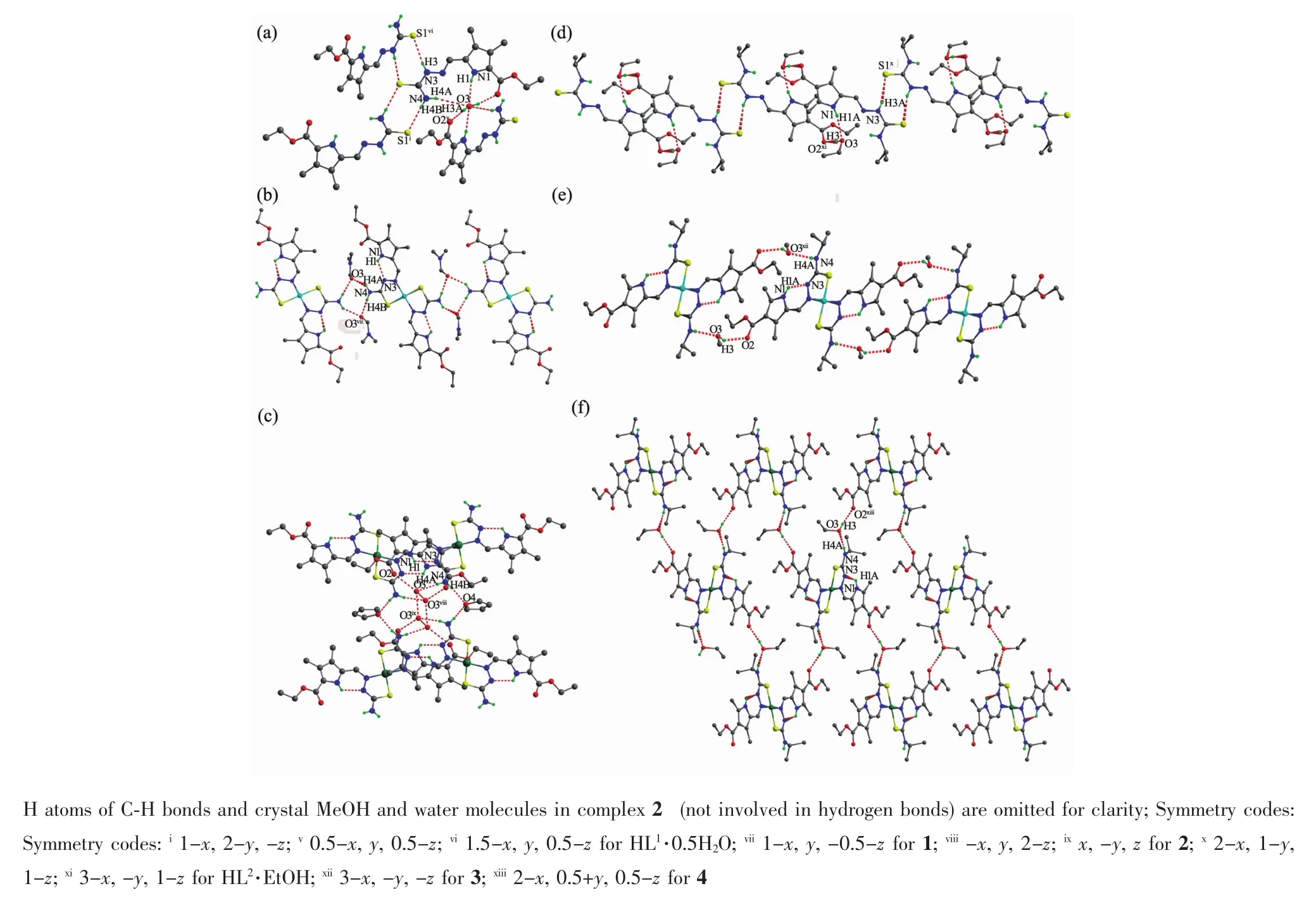
Fig.2 Extend supramolecular structures in HL1·0.5H2O(a),HL2·EtOH(d)and complexes 1~4(b,c,e and f,respectively)
2.2 IR spectra
The IR spectra for complexes 1~4 are more or less similar due to the similarity in coordination modes of the ligands with themetal centre.Theν(C= S)vibration of the free ligand HL1and HL2are at 896 and 930 cm-1,respectively,while it is absent in the complexes.Meanwhile,new(N=C-S)and(C-S) stretching vibration absorption are observed at 1 586 and 863,1 558 and 862,1 597 and 910,1 621 and 908 cm-1in complexes 1~4,respectively,inferring that the C=S bond has thiolated to N=C-S moiety and the sulphur atom coordinates to the centralmetal ion in the complexes[16-17].Theν(N=C)vibration of the free ligand HL1and HL2is at 1 536 and 1 565 cm-1,respectively, while at 1 518,1 508,1 548 and 1 512 cm-1in complexes 1~4,respectively,showing the N=C bond participates in the coordination in each complex[18].It is in accordance with the crystal structure study.
2.3 UV spectra
The UV spectra of the ligands HL1,HL2and complexes 1~4 in DMF solution(c=10μmol·L-1)were measured at room temperature(Fig.3).The spectra of HL1and HL2features two main bands located around 352(ε=29 923 L·mol-1·cm-1)and 368(ε=27 954 L· mol-1·cm-1),340(ε=39 130 L·mol-1·cm-1)and 354 nm (ε=33 697 L·mol-1·cm-1),respectively,which could be assigned to characteristicπ-π*transitions centered on pyrrole rings and imine unit[14,19].However,in complexes 1~4,the two main bands were combined along with red shift to 378(ε=28 759 L·mol-1·cm-1), 372(ε=25 139 L·mol-1·cm-1),375(ε=23 566 L· mol-1·cm-1)and 381 nm(ε=25 389 L·mol-1·cm-1), respectively,showing that the N=C bond participates in the coordination in each complex.Furthermore, there is a new absorbance band of 2(452 nm,ε= 6 401 L·mol-1·cm-1)and 4(453 nm,ε=10 982 L· mol-1·cm-1),which is probably due to d-d transition of the centre Cuions[20].
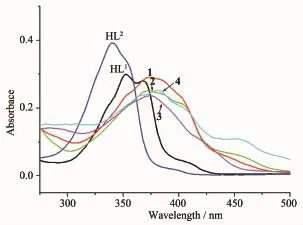
Fig.3 UV spectra of the ligands HL1,HL2and complexes 1~4 in the DMF solution at room temperature
2.4 EB-DNAbindingstudybyfluorescencespectrum It is well known that EB can intercalate nonspecifically into DNA,which causes it to fluoresce strongly.Competitive binding of other drugs to DNA and EB will result in displacement of bound EB and a decrease in the fluorescence intensity[15].The effects of the ligand and complexes on the fluorescence spectra of EB-DNA system are presented in Fig.3,the fluorescence intensities of EB bound to ct-DNA at about 600 nm show remarkable decreasing trendswith the increasing concentration of the tested compounds, indicating that some EB molecules are released into solution after the exchange with the compounds.The quenching of EB bound to DNA by the compounds is in agreement with the linear Stern-Volmer equation:
I0/I=1+Ksqr[11],where I0and I represent the fluorescence intensities in the absence and presence of quencher,respectively,Ksqis the linear Stern-
Volmer quenching constant,r is the ratio of the concentration of quencher and DNA.In the quenching plots of I0/I versus r,Ksqvalues are given by the slopes.The Ksqvalues are 0.586,1.487 and 1.601 for HL1,complexes 1 and 2,respectively,while those for HL2,complexes 3 and 4 are tested to be 0.753,2.971 and 1.385,respectively.The results indicate that interactions of the complexes with DNA are stronger than those of the corresponding TSC ligands,probablydue to the higher rigidity of the complexes,which is in accordance with the literature[11].In addition, complex 3 has the highest quenching ability among the tested complexes,this could be explained from three aspects:metal-ligand synergism effectmay be more effective in 3 than in other complexes[18];the isopropyl at the terminal nitrogen N(4)of HL2is helpful to enhance the quenching ability,which is in agreement with the fact that interaction of HL2with DNA is stronger than that of HL1;the positions of the substitutes in the pyrrole ring may be also responsible for the DNA interaction ability in some content.
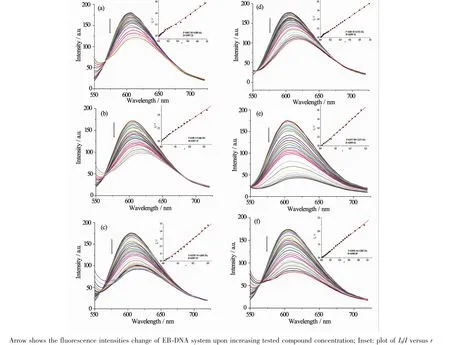
Fig.3 Emission spectra of EB-DNA system in the presence of HL1(a),HL2(d)and complexes 1~4(b,c,e and f,respectively)
[1]Chari M A,Shoba D,Prakash K M M S,et al.Asian J. Chem.,2012,24:11-14
[2]Demoro B,RossiM,Caruso F,et al.Biol.Trace Elem.Res., 2013,153:371-381
[3]Singh S,Athar F,Maurya M R,et al.Eur.J.Med.Chem., 2006,41:592-598
[4]BaldiniM,Belicchi-FerrariM,Bisceglie F,etal.Inorg.Chem., 2004,43:7170-7179
[5]Stanojkovic T P,Kovala-Demertzi D,Primikyri A,et al.J. Inorg.Biochem.,2010,104:467-476
[6]Sreekanth A,Fun H K,Kurup M R P.Inorg.Chem. Commun.,2004,7:1250-1253
[7]Paul P,Butcher R J,Bhattacharya S.Inorg.Chim.Acta, 2015,425:67-75
[8]Joseph M,Kuriakose M,Prathapachandra-Kurup M R,et al. Polyhedron,2006,25:61-70
[9]YE Xing-Pei(叶行培),WANG Guan-Jie(王冠杰),PAN Peng (潘鹏),etal.Chinese J.Inorg.Chem.(无机化学学报),2014, 30(12):2789-2795
[10]Ye X P,Zhu T F,Wu W N,et al.Inorg.Chem.Commun.,2014,47:60-62
[11]SHEN Wei(沈伟),HU Wei-Ji(胡未极),WU Xiao-Yong (吴小勇),et al.Chinese J.Inorg.Chem.(无机化学学报), 2016,32(6):1101-1110
[12]Sheldrick GM.SADABS,University of Göttingen,Germany, 1996.
[13]Sheldrick G M.SHELX-97,Program for the Solution and the RefinementofCrystal Structures,University ofGöttingen, Germany,1997.
[14]MuralisankarM,Sujith S,Bhuvanesh NSP,etal.Polyhedron, 2016,118:103-117
[15]Joseph M,Suni V,Prathapachandra-Kurup M R,et al. Polyhedron,2004,23:3069-3080
[16]Xu Z H,Zhang X W,Zhang W Q,et al.Inorg.Chem. Commun.,2011,14:1569-1573
[17]DENG Han-Qin(邓汉芹),WANG Ming-Xiong(王明雄), J.Hubei Univ.:Nat.Sci.Ed.(湖北大学学报:自然科学版), 1992,14(2):155-160
[18]Wang Y,Yang Z Y,Chen Z N.Bioorg.Med.Chem.Lett., 2008,18:298-303
[19]Mala N,Sharma N,Sharma C L.Synth.React.Inorg. Met.-Org.Chem.,1989,19:339-356
[20]Forster D,Goodgame D M L.Inorg.Chem.,1965,4:823-829
LIXiao-Jing MAO Pan-Dong WUWei-Na*KOU Kai LIU Shu-Yang WANG Yuan*
(College of Chemistry and Chemical Engineering,Henan Polytechnic University,Jiaozuo,Henan 454000,China)
Two Niand two Cucomplexes,namely,[Ni(L1)2]·2DMF(1),[Cu(L1)2]·THF·0.25MeOH·2.25H2O (2),[Ni(L2)2]·2MeOH(3)and[Cu(L2)2]·2EtOH(4)(where HL1=5-formyl-3,4-dimethyl-1H-pyrrole-2-carboxylate thiosemicarbazone,HL2=5-formyl-2,4-dimethyl-1H-pyrrole-3-carboxylate N(4)-isopropyl thiosemicarbazone)have been synthesized and characterized by single crystal X-ray diffraction,elemental analysis and IR spectroscopy.X-ray diffraction analysis results show that the structures of all complexes are similar while with different solvent molecules.Themetal ion in each complex with a distorted square planar geometry is surrounded by two anionic thiosemicarbazone ligands with N2S2donor set.In addition,the fluorescence spectra indicate that the interactions of the complexes with DNA are stronger than those of the corresponding ligands.CCDC:1424498,HL1·0.5H2O; 1424499,1;1424500,2;1427468,HL2·EtOH;1427469,3;1427470,4.
pyrrole;thiosemicarbazone;complex;crystal structure;DNA interaction
O614.81+3;O614.121
A
1001-4861(2017)07-1257-09
10.11862/CJIC.2017.151
2016-11-30。收修改稿日期:2017-05-18。
国家自然科学基金(No.21001040)、河南省科技厅基础与前沿项目(No.162300410011)、河南省教育厅自然科学基金(No.12B15001, 14B150029)和河南省青年骨干教师项目(No.2014GGJS-045)资助。
*通信联系人。E-mail:wuwn08@hpu.edu.cn,wangyuan08@hpu.edu.cn;会员登记号:S06N6704M1112。
