人环状RNA-0046366与肝细胞脂肪变性的相关性及其机制研究
2017-06-29郭行雅章瑞南何崇信范建高
郭行雅,章瑞南,何崇信,孙 芳,范建高,潘 勤
上海交通大学医学院附属新华医院消化内科,上海 200092
人环状RNA-0046366与肝细胞脂肪变性的相关性及其机制研究
郭行雅,章瑞南,何崇信,孙 芳,范建高,潘 勤
上海交通大学医学院附属新华医院消化内科,上海 200092
目的 探讨人环状RNA-0046366(circRNA-0046366)表达变化对肝细胞脂肪变性的影响及其机制。方法 将肝HepG2细胞株随机分为正常对照组(n=3)和脂肪变性模型组(n=3),分别给予高糖DMEM、高脂培养基(油酸∶棕榈酸=2∶1, 0.5 mmol/L)诱导24 h。采用油红O染色及甘油三酯(Triglyceride,TG)、丙二醛(Malonyldialdehyde,MDA)检测,观察肝细胞的脂肪变性及脂质过氧化。以实时荧光定量PCR(Real-Time PCR)法检测circRNA-0046366在脂肪变性前后的表达改变。在circBase数据库搜索、circRNA-miRNA互补序列分析的基础上,预测circRNA-0046366的靶miRNA及其下游基因。 通过Spearman相关性分析,揭示circRNA-0046366水平与靶miRNA表达、下游mRNA表达、肝细胞脂肪变性及脂质过氧化指标的相关性。结果 与正常对照组相比,脂肪变性模型组的肝细胞内脂滴、TG[(24.4±2.4) μmol/gvs(263.7±17.5) μmol/g,P<0.05]及MDA[(0.6±0.1) μmol/gvs(1.8±0.2) μmol/g,P<0.05]含量明显升高。circRNA-0046366表达水平在脂肪变性的肝细胞中则显著降低(P<0.05)。circRNA-0046366可与过氧化物酶体增殖物激活受体α(peroxisome proliferator-activated receptor α, PPARα)竞争性结合miR-34a,从而阻断miR-34a对PPARα翻译抑制作用。相关性分析显示,肝细胞的circRNA-0046366与PPARα表达水平呈显著正相关(r=0.78,P<0.05),与miR-34a表达水平(r=-0.83,P<0.05)、TG(r=-0.72,P<0.05)及MDA(r=-0.69,P<0.05)含量则呈显著负相关。结论 肝细胞的circRNA-0046366水平与脂肪变性及脂质过氧化呈负相关。其机制可能与靶向结合miR-34a,从而解除PPARα的表达抑制相关。
环状RNA;肝细胞;脂肪变性;微小RNA;过氧化物酶体增殖激活受体α
近年研究[1-2]显示,miRNA与肝细胞脂肪变性密切相关,miR-181d、miR-10b等11种miRNA均可靶向调节脂质代谢相关基因的表达水平,从而干预细胞内甘油三脂(Triglyceride,TG)的转运、合成、氧化,在此基础上影响肝细胞的脂肪变程度。 然而迄今为止,对肝细胞脂肪变相关miRNA的调控机制仍不明确。
20世纪70年代在RNA病毒中发现一类非编码RNA—环状RNA(circular RNA,circRNA)[3],近年来高通量测序结合转录组分析发现,circRNA大量存在于真核细胞中[4-6]。circRNA可通过碱基互补充当miRNA海绵(miRNA sponge)[7-8],与miRNA发生竞争性结合,进而解除其对下游靶基因的转录抑制作用。这提示circRNA可能成为miRNA调节因子,在多种病理生理过程发挥重要作用。
为探讨circRNA与肝细胞脂肪变性的关联,本研究采用油酸/棕榈酸诱导肝细胞脂肪变性模型。结合circBase[9]数据库搜索及circRNA-miRNA互补序列分析,预测对肝细胞脂肪变相关性miRNA具有调控作用的circRNA。 通过实时定量PCR检测TG及丙二醛(Malonyldia-ldehyde,MDA)水平定量,验证circRNA表达水平与肝细胞脂肪变性、脂质过氧化指标的相关性。 在miRNA “种子”序列与circRNA、mRNA互补配对的基础上,提出circRNA-miRNA-mRNA级联调控的可能作用机制。
1 材料与方法
1.1 材料 HepG2细胞购自上海细胞库;棕榈酸(PA,P9767)、油酸(OA,07501)均购自SIGMA公司;TG检测试剂盒(北京普利莱基因技术有限公司);BCA蛋白浓度测定试剂盒(碧云天生物公司);MDA检测试剂盒(碧云天生物公司);Cell Counting Kit-8(CCK-8)试剂盒(碧云天生物技术公司);FastQuant RT Kit逆转录试剂盒(大连宝生物工程有限公司);HieffTMqPCR SYBR@Green Master Mix(上海翊圣生物科技有限公司));PCR仪(ABI,美国);酶标仪(Thermo,芬兰);紫外分光光度计(Eppendorf,德国)。
1.2 方法
1.2.1 建立肝细胞脂肪变性模型:将HepG2细胞随机分为正常对照组(n=3)、脂肪变性模型组(n=3)。各组细胞采用含100 g/L的高糖DMEM培养72 h后,脂肪变性模型组改为含有0.5 mmol/L游离脂肪酸(free fatty acid, FFA)混合物(OA∶PA=2∶1)的高脂培养基,经高脂诱导24 h,收集细胞待用。
1.2.2 油红O染色:采用70%的油红O溶液,浸染10~15 min,60%异丙醇/三蒸水冲洗,苏木素复染,镜检。
1.2.3 细胞活力检测:按上述分组方法,于96孔板中接种HepG2细胞(1×104个/孔)。分别采用正常或高脂培养基诱导24 h后,加入CCK-8溶液(10 μl/孔),37 ℃孵育4 h,采用酶标仪测定450 nm处吸光度值。
1.2.4 TG、MDA含量检测:收集正常对照组、脂肪变性模型组细胞,分别采用BCA法作蛋白定量,TG酶法作TG定量,TBA比色法作MDA定量。
1.2.5 circRNA-0046366下游靶miRNA、靶mRNA预测:通过circRNA与miRNA序列互补分析,并结合circBase数据库,以预测与circRNA-0046366靶向结合的miRNA。利用miRNA靶点预测工具TargetScan,发现miR-34a的下游靶基因。
1.2.6 circRNA-0046366、miR-34a及PPARα mRNA含量检测:分别提取各组细胞Total RNA,合成 cDNA第一链。设计并合成引物,引物序列如下(见表1),miR-34a qPCR引物由大连宝生物工程有限公司提供。 配制反应液,设置反应程序,应用ABI7500实时PCR仪获得目的基因扩增曲线及CT值,采用 2-ΔΔCT法对各组CT值进行结果分析。
1.2.7 相关性分析:采用Spearman法分析miR-34a、过氧化物酶体增殖激活受体α(peroxisome proliferation activated receptor α,PPARα)与circRNA-0046366的相关性,及TG、MDA与circRNA-0046366的相关性。
表1 目的基因引物序列
Tab 1 Target gene primer sequence
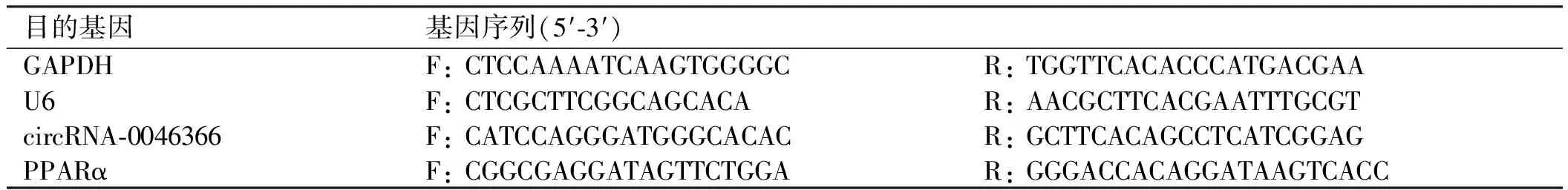
目的基因基因序列(5′⁃3′)GAPDHF:CTCCAAAATCAAGTGGGGCR:TGGTTCACACCCATGACGAAU6F:CTCGCTTCGGCAGCACAR:AACGCTTCACGAATTTGCGTcircRNA⁃0046366F:CATCCAGGGATGGGCACACR:GCTTCACAGCCTCATCGGAGPPARαF:CGGCGAGGATAGTTCTGGAR:GGGACCACAGGATAAGTCACC

2 结果
2.1 油红O染色结果 正常对照组HepG2细胞为不规则形梭形,胞质无明显红染或淡染。脂肪变性模型组细胞广泛染色,胞质含大量红色脂滴(见图1)。CCK-8结果显示两组细胞生长均良好, 细胞活力无明显差异。

图1 肝细胞油红O染色(200×) A:正常对照组;B:脂肪变性模型组
Fig 1 Oil Red O staining of hepatocytes (200×) A: normal control group; B: steatosis model group
2.2 TG含量、MDA水平比较 正常对照组和脂肪变性模型组的TG含量分别为(24.4±2.4) μmol/g 和(263.7±17.5) μmol/g。与正常对照组相比,脂肪变性模型组的TG含量明显升高(t=13.6,P<0.05)。正常对照组和脂肪变性模型组的MDA含量分别为(0.6±0.1) μmol/g 和(1.8±0.2) μmol/g,两者比较,差异有统计学意义(t=5.2,P<0.05,见表2)。
2.3 circRNA-0046366的靶miRNA、靶mRNA 生物信息学分析结果显示,miR-34a为circRNA-0046366下游靶miRNA(见图2),PPARα为miR-34a下游靶基因(见图3)。

图2 circRNA-0046366下游靶miRNAFig 2 Downstream target miRNA of circRNA-0046366

图3 miR-34a下游靶基因预测Fig 3 Predicting on downstream target gene of miR-34a
2.4 circRNA-0046366、miR-34a及PPARα mRNA的表达水平 正常对照组circRNA-0046366水平显著高于脂肪变性模型组[(3.43±0.55)倍,P<0.05];脂肪变性模型组miR-34a水平显著高于正常对照组[(1.92±0.30)倍,P<0.05];正常对照组PPARα mRNA水平显著高于脂肪变性模型组(1.75±0.19)倍(P<0.05,见图4)。
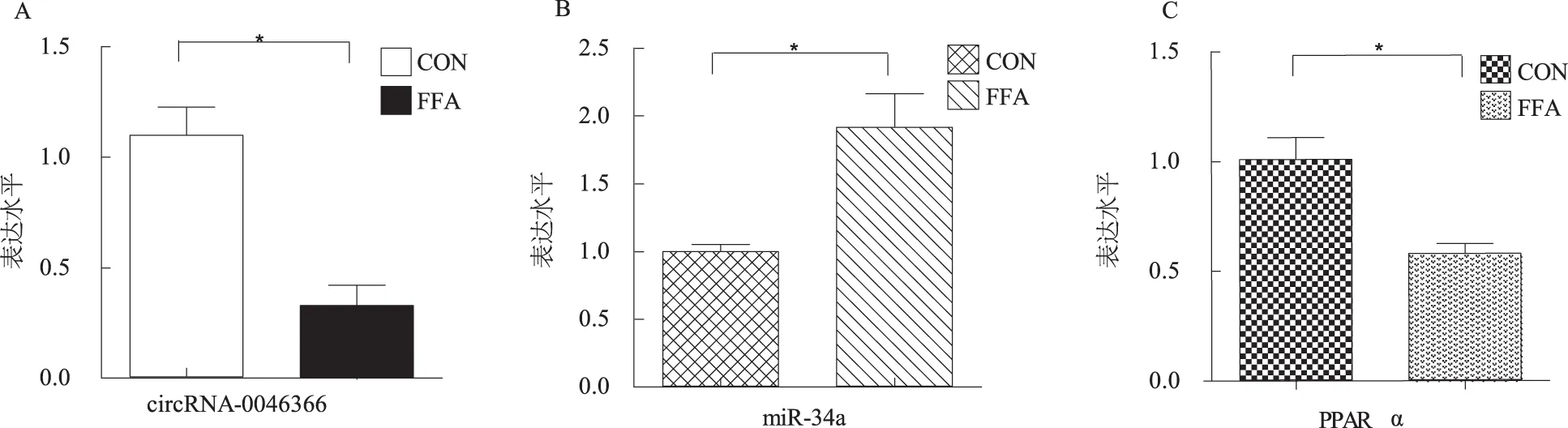
注:与正常对照组比较,*P<0.05。
图4 两组细胞circRNA-0046366、miR-34a及PPARα mRNA水平比较 A: circRNA-0046366; B: miR-34a; C: PPARα
Fig 4 The expression levels of circRNA-0046366, miR-34a and PPARα mRNA between two groups A: circRNA-0046366; B: miR-34a; C: PPARα mRNA
2.5 circRNA-0046366与miR-34a、PPARα相关性分析 脂肪变性模型组circRNA-0046366与miR-34a呈显著负相关(r=-0.83,P<0.05);脂肪变性模型组 circRNA-0046366与PPARα呈显著正相关(r=0.78,P<0.05,见图5)。
2.6 circRNA-0046366与TG及MDA相关性分析 脂肪变性模型组circRNA-0046366与TG呈显著负相关(r=-0.72,P<0.05);脂肪变性模型组circRNA-0046366与MDA呈显著负相关(r=-0.69,P<0.05,见图6)。
3 讨论
目前研究已表明,circRNA具有较强的转录后调控作用[7-8]。多种circRNA均携带miRNA应答元件(miRNA response element,MRE),并能够与miRNA竞争性结合,从而成为竞争性内源RNA(competing endogenous RNA,ceRNA)。这一作用可解除miRNA对其靶基因的表达抑制,从而调控心力衰竭、骨关节炎及肿瘤等多种病理生理过程[10-14]。本研究在circBase数据库的基础上开展circRNA与miRNA“种子”序列的互补配对分析,发现脂肪酸合成酶基因的差异剪切产物可环化形成circRNA-0046366,且其177~183 bp序列携带miR-34a特异性 MRE。 Ding等[15]报道,miR-34a通过抑制PPARα的翻译水平,可下调CPT1等脂质氧化相关的下游基因表达,导致肝细胞脂肪变性。由此推测,circRNA-0046366可能通过与miR-34a竞争性结合,消除其对下游靶基因的抑制作用,进而发挥拮抗肝细胞脂肪变性的效应。

图5 circRNA-0046366与miR-34a、PPARα相关性分析 A: circRNA-0046366与miR-34a; B: circRNA-0046366与PPARα; 图6 circRNA-0046366与TG、MDA含量相关性分析 A: circRNA-0046366与TG; B: circRNA-0046366与MDA
Fig 5 The correlation analysis of circRNA-0046366 level and miR-34a, PPARα expression A: circRNA-0046366 level and miR-34a expression; B: circRNA-0046366 level and PPARα expression; Fig 6 The correlation analysis of circRNA-0046366 level and TG, MDA content A: circRNA-0046366 level and TG content; B: circRNA-0046366 level and MDA cotent
为阐明circRNA-0046366与肝细胞脂肪变性的相关性,本研究采用高脂培养基(OA∶PA=2∶1,0.5 mmol/L)诱导肝细胞脂肪变性模型,并定量检测circRNA-0046366在脂肪变性前后的表达变化。结果显示,脂肪变性模型组肝细胞含有大量脂滴,且TG含量显著高于正常对照组,脂肪变细胞内的MDA水平也明显升高。因而,高脂培养可诱导肝细胞发生脂肪变性及脂质过氧化。Real-Time PCR检测表明,肝细胞的circRNA-0046366表达水平伴随脂肪变性而明显下调。Spearman相关性分析提示,circRNA-0046366水平与肝细胞的TG、MDA含量呈显著负相关。上述结果证实circRNA-0046366与肝细胞脂肪变性密切相关,可能在脂肪变性和脂质过氧化过程中发挥负调控作用。
近年多项研究提出,circRNA、miRNA及其下游mRNA可构成“circRNA -miRNA-mRNA”调控网络,对肿瘤和阿尔兹海默症等[16]的发生、发展具有重要意义。在circRNA-0046366特征性低表达的基础上,本研究针对其靶miRNA进行定量检测,可见miR-34a表达水平在脂肪变性的肝细胞内大幅度上升。利用miRNA靶点预测工具TargetScan来分析miR-34a与mRNA的序列互补性,发现PPARα mRNA 3′ 非编码区1668~1674 bp、5080~5086 bp、8004~8010 bp均存在miR-34a 2-7位碱基的互补序列[17]。由于PPARα具有促进脂肪酸及脂蛋白代谢,改善胰岛素抵抗和非酒精性脂肪性肝炎(non-alcohol steatohepatitis,NASH)的作用[18],因此,高丰度miR-34a与PPARα靶向性结合能够阻碍脂质代谢,诱导肝细胞脂肪变性,这与Ding等[15]的报道相吻合。结合circRNA及miRNA的表达改变,提示circRNA-0046366在正常肝细胞中可能竞争性结合miR-34a,从而避免PPARα发生表达抑制,由此发挥拮抗脂肪变性的效应。而circRNA-0046366在脂肪变性的肝细胞中出现表达缺失,阻断了这一负向调节机制,结果导致miR-34a介导的PPARα抑制,随之引起TG蓄积和脂质过氧化。
综上所述,circRNA-0046366在肝细胞脂肪变性过程中呈明显低表达,并与TG含量、脂质过氧化水平呈显著负相关,其作用机制可能与竞争性结合miR-34a,继而解除肝细胞内PPARα的表达抑制相关。
[1]Zheng L, Lv GC, Sheng J, et al. Effect of miRNA-10b in regulating cellular steatosis level by targeting PPARα expression, a novel mechanism for the pathogenesis of NAFLD [J]. J Gastroenterol Hepatol, 2010, 25 (1): 156-163.
[2]Whittaker R, Loy PA, Sisman E, et al. Identification of microRNAs that control lipid droplet formation and growth in hepatocytes via high-content screening [J]. J Biomol Screen, 2010, 15(7): 798-805.
[3]Wilusz JE, Sharp PA. A circuitous route to noncoding RNA [J]. Science, 2013, 340(6131): 440-441.
[4]Wu Q, Wang Y, Cao M, et al. Homology-independent discovery of replicating pathogenic circular RNAs by deep sequencing and a new computational algorithm [J]. Proc Natl Acad Sci U S A, 2012, 109(10): 3938-3943.
[5]Stoffelen R, Jimenez MI, Dierckxsens C. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types [J]. PLoS One, 2012, 7: e30733.
[6]Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats [J]. RNA, 2013, 19(2): 141-157.
[7]Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency [J]. Nature, 2013, 495(7441): 333-338.
[8]Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges [J]. Nature, 2013, 495(7441): 384-388.
[9]Glazar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs [J]. RNA, 2014, 20(11): 1666-1670.
[10]Wang K, Long B, Liu F, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223 [J]. Eur Heart J, 2016, 37(33): 2602-2611.
[11]Liu Q, Zhang X, Hu X, et al. Circular RNA related to the chondrocyte ECM regulates MMP13 expression by functioning as a MiR-136′Sponge′ in human cartilage degradation [J]. Sci Rep, 2016, 6: 22572.
[12]Xie H, Ren X, Xin S, et al. Emerging roles of circRNA-001569 targeting miR-145 in the proliferation and invasion of colorectal cancer [J]. Oncotarget, 2016, 7(18): 26680-26691.
[13]Zheng Q, Bao C, Guo W, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs [J]. Nat Commun, 2016, 7: 11215.
[14]Geng H, Li R, Su Y, et al. The circular RNA Cdr1as promotes myocardial infarction by mediating the regulation of miR-7a on its target genes expression [J]. PLoS One, 2016, 11(3): e151753.
[15]Ding J, Li M, Wan X, et al. Effect of miR-34a in regulating steatosis by targeting PPARα expression in nonalcoholic fatty liver disease [J]. Sci Rep, 2015, 5: 13729.
[16]Chen Y, Li C, Tan C, et al. Circular RNAs: a new frontier in the study of human diseases [J]. J Med Genet, 2016, 53(6): 359-365.
[17]Peterson SM, Thompson JA, Ufkin ML, et al. Common features of microRNA target prediction tools [J]. Front Genet, 2014, 5: 23.
[18]朱梦飞, 施军平, 范建高. 非酒精性脂肪性肝炎治疗新靶点和分子靶向药物[J] . 实用肝脏病杂志, 2012, 15(4): 282-283.
(责任编辑:王全楚)
The association and related mechanisms between human circRNA-0046366 and hepatocyte steatosis
GUO Xingya, ZHANG Ruinan, HE Chongxin, SUN Fang, FAN Jiangao, PAN Qin
Department of Gastroenterology, Xinhua Hospital Affiliated to Shanghai Jiao Tong University School of Medicine, Shanghai 200092, China
Objective To investigate the association and related mechanisms between human circular RNA-0046366 (circRNA-0046366) and hepatocyte steatosis.Methods HepG2 cells were randomly divided into groups of normal control (n=3) and steatosismodel group (n=3), respectively. The normal control group was treated with DMEM, while the steatosis-model group was administrated with high-fat medium (OA∶PA=2∶1, 0.5 mmol/L) for 24 hours. Both oil red O staining and triglyceride (TG) concentration were employed to evaluate the hepatocyte steatosis. Hepatic level of malonaldehyde (MDA) reflected the lipid peroxidation, which occurred on the basis of hepatocyte steatosis. Real-time PCR exhibited the alternation of circRNA-0046366 expression during hepatic steatosis. Bioinformatic analysis demonstrated the target miRNA, and downstream mRNAs, of circRNA-0046366 by means of circBase searching and circRNA-miRNA complementation. Finally, the association among circRNA-0046366 level, miRNA, mRNAs, hepatocyte steatosis, and lipid peroxidation was assessed by Spearman correlation analysis.Results Compared with normal control group, steatosismodel group exhibited significant increase in lipid droplets, and hepatic contents TG [(24.4±2.4) μmol/gvs(263.7±17.5) μmol/g,P<0.05] and MDA [(0.6±0.1) μmol/gvs(1.8±0.2) μmol/g,P<0.05]. The expression level of circRNA-0046366 underwent statistically decrease (P<0.05) during the hepatocyte steatosis. Both circRNA-0046366 and peroxisome proliferator-activated receptor α (PPARα) shared the sequence complementary to miR-34a, which may abolish the translational inhibition of miR-34a on PPARα by binding competition. In result, the circRNA-0046366 level correlated with PPARα expression (r=0.78,P<0.05), but negatively correlated with miR-34a level (r=-0.83,P<0.05), and concentrations of TG (r=-0.72,P<0.05) and MDA (r=-0.69,P<0.05) in the steatotic HepG2 cells. Conclusion circRNA-0046366 level negatively correlates with hepatic steatosis and lipid peroxidation. Abolishment of the PPARα inhibition by miR-34a may underlie its effect.
Circular RNA; Hepatocyte; Steatosis; miRNA; PPARα

科技部“973”项目(2012CB517501);国家自然科学基金项目(81470859、81270492、81070346)
郭行雅,在读硕士,研究方向:消化系统疾病的研究。E-mail: 13856041526@163.com
潘勤,副研究员,研究方向:消化系统疾病的研究。E-mail: pan_qin@yeah.net
10.3969/j.issn.1006-5709.2017.06.022
R575
A
1006-5709(2017)06-0713-05
2016-10-10
