Novel nuclear SSR markers in the large frond tree fern Alsophila gigantea and its congeneric species Alsophila spinulosa*
2017-05-18,,,,4
, , , ,4
(1. School of Life Sciences, Sun Yat-sen University, Guangzhou 510275, China; 2. College of Life Sciences, Nanjing Agricultural University, Nanjing 210095, China; 3. College of Life Science, South China Agricultural University, Guangzhou 510642, China; 4. Research Institute of Sun Yat-sen University in Shenzhen, Shenzhen 518057, China)
Novel nuclear SSR markers in the large frond tree fern Alsophila gigantea and its congeneric species Alsophila spinulosa*
RUANXiaoxian1,WANGZhen2,WANGTing3,SUYingjuan1,4
(1. School of Life Sciences, Sun Yat-sen University, Guangzhou 510275, China; 2. College of Life Sciences, Nanjing Agricultural University, Nanjing 210095, China; 3. College of Life Science, South China Agricultural University, Guangzhou 510642, China; 4. Research Institute of Sun Yat-sen University in Shenzhen, Shenzhen 518057, China)
Alsophilagiganteais a large frond tree fern in family Cyatheaceae. The plant is a vulnerable species that prefers specific subtropical montane climate. Little work has been done so far to access the population genetic variation inA.gigantea. In this study, we developed 15 SSR markers using FIASCO protocol and evaluated these markers in four natural populations from Hainan Island, China. Seven loci proved to be polymorphic. The actual number of alleles ranged from one to nine, and the observed and expected heterozygosity varied from 0 to 0.769, and from 0 to 0.805, respectively. LociAG-12 andAG-23 were found to significantly deviate from the Hardy-Weinberg equilibrium in populations HNjfl and HNbwl, respectively. Four null alleles were identified. Linkage disequilibrium was further detected at three pairs of loci. Moreover, four SSR markers were verified to be successfully transferred inAlsophilaspinulosa. The novel polymorphic SSR markers characterized here will be used to survey population genetic variation and local adaptation inA.gigantea, which helps design effective conservation strategies.
Alsophilagigantea; SSR markers; cross-species amplification; genetic variation; local adaptation
Alsophilagiganteais a large tree fern belonging to family Cyatheaceae with fronds 2 to 3 meter long[1]. Its fronds are bright green and thinly textured. Unbranched trunk reaches 2~4 m in height and 12~13 cm in diameter. Long stipes are densely covered with shiny, dark brown scales.Alsophilagiganteaforms a characteristic V-shaped soral arrangement in each pinnule lobe and margin of pinnae lobed 2~5 mm towards the costa[2]. The species is widely distributed in India, Sri Lanka, Burma, Thailand, Laos, Vietnam, and China. In China, it mainly occurs in Guangdong, Guangxi, Hainan, and Yunnan.Alsophilagiganteagrows near streams, river banks, and on mountain slopes in dense forests at an altitude of 200~1 200 m[1]. Traditionally, this species has been used as an important medicinal plant to treat white discharges[3-4]. Its trunk is also exploited for starch and used for growing epiphytic orchids[4]. Overexploitation and human activities have greatly affected the populations ofA.gigantea. Populations are drastically fragmented, and population sizes have diminished[5]. As a result,Alsophilagiganteahas been declared as a vulnerable species and listed under IUCN Red data book and the Appendix-II of CITES (Convention of International Trade in Endangered Species of Wild Fauna and Flora)[6-7].
Simple sequence repeats (SSRs) are short, tandemly repeated sequences of 1~6 nucleotides[8]. SSRs are powerful PCR-based markers extensively used in assessing genetic variation of plants due to codominant inheritance, polymorphism, and abundant coverage. Currently, there are a number of methods used to isolate SSR markers. However, Fast Isolation by AFLP of Sequences COntaining repeats (FIASCO) represents an efficient technique to develop SSR markers[9]. The method is simple, fast, and economic[10]. Moreover, no genomic information is needed.
Alsophilagiganteais perceived to be more adapted to subtropical montane climate in comparison to otherAlsophilaspecies[11]. However, little work has been done on its adaptive population genetic variation and structure. To aid in this field, we have developed the SSR markers forA.giganteausing the FIASCO approach.
1 Materials and methods
1.1 Plant materials and DNA extraction
A total of 49 individuals ofA.giganteawere collected from Diaoluoshan (N18°40′, E109°50′,N=13), Wuzhishan (N18°54′, E109°54′,N=11), Jianfengling (N18°44′, E108°52′,N=13), and Bawangling (N19°05′, E109°11′,N=12), Hainan Province, China. Two natural populations of the congeneric speciesA.spinulosawere sampled from Wuzhishan (Hainan, N18°54′, E109°40′,N=5) and Wulaishan (Taiwan, N24°52′, E121°33′,N=7) for cross-species amplication (Fig.1). Vouchers have been deposited at the Herbarium of Sun Yat-sen University (A.gigantea, YZ Ying 200606, DLS1, WZS2, JFL3, and BWL4;A.spinulosa, YZ Ying 200605, WZS6, Q Fan 200808, WLS3). Fresh and young leaves were collected and preserved in Ziplock plastic bags with silica gel. Total genomic DNA was extracted using a modified CTAB (cetyltrimethylammonium bromide) method with -20 ℃ propanone pretreatment to eliminate polysaccharides[12]. The quality of the DNA was checked by 0.8% agarose gel electrophoresis. DNA concentration was quantied by spectrophotometry. DNA was stored at -20 ℃ for further study.
1.2 Construction of SSR-enriched genomic library
The FIASCO approach was applied to construct SSR-enriched genomic library from one random individual ofA.gigantea. Approximately 250 ng DNA was digested withMseI restriction enzyme at 37 ℃ for 3 hours (h), following by incubation at 65 ℃ for 15 min to inactivate the enzyme. Digested DNA was ligated toMseI adaptors (F: 5′-TACTCAGGACTCAT-3′, R: 5′-GACGATGAGTCCTGAG-3′) with T4 DNA ligase. The adaptors-ligated DNA fragments were diluted ten folds and directly amplied in a total volume of 20 μL withMseI-N primer. After denaturation, PCR products were

Fig.1 The sampling location of Alsophila gigantea (solid dots) and Alsophila spinulosa (solid triangle)(a) The map shows a brief part of China;(b) Hainan;(c) Taiwan. WZS:Wuzhishan;DLS:Diaoluoshan;JFL:Jianfengling;BWL:Bawangling;WLS:Wulaishan
hybridized with 5′-biotinylated (AC)15probe at 95 ℃ for 3 minutes (min), then captured by Streptavidin-coated beads. To eliminate non-specific DNA, the captured products were treated by three non-stringent washes (1 mol/L Tris-HCl, 0.5 mol/L EDTA, 2 mol/L NaCl) and three stringent washes (20×SSC,w=10% SDS, ddH2O). The enriched products were eluted, precipitated, dissolved, and recovered by PCR withMseI-N primer. The PCR reaction program included 30 cycles of 30 seconds (s) at 94 ℃, 1 min at 53 ℃, and 1 min at 72 ℃. Amplified products purified by the E.Z.N.A. Cycle-Pure kit (Omega Bio-Tek, Guangzhou, China) were ligated into pMD-18T vector. The plasmids were transformed intoEscherichiacoliDH5α competent cells. The recombinant clones were screened on LB agar medium containing ampicillin and IPTG/X-galactosidase by blue-white selection.
1.3 Screening of positive clones and primer design
The recombinant clones were identified using PCR with M13 forward and reverse universal primers. Positive clones were sequenced using ABIPRISM 3730 Automated Sequencer. After the vector sequences were removed, sequences including SSRs and sufficient flanking region were screened for SSRs with SSRHunter. SSR primer pairs were designed using Primer Premier 5.0 software[13].
1.4 Polymorphism and cross-species amplification
1.5 Data analysis
GenAlEx software[15]was applied to assess genetic parameters including the effective number of alleles (Ne), actual number of alleles (Na), expected heterozygosity (He), and observed heterozygosity (Ho). The deviation from the Hardy-Weinberg equilibrium was evaluated using the same software. The null alleles and linkage disequilibrium were estimated using MICRO-CHECKER version 2.2.3[16]and GENEPOP version 4.0.10[17], respectively.
2 Results and discussion
2.1 Identification of the SSR markers
From 129 recombinant clones sequenced, 96 sequences were acquired, in which 89 sequences contained SSR motifs with enoughanking region for primer design. We designed 29 sets of SSR primers. Fifteen of these primer pairs amplied successfully in four populations ofA.gigantea. Of them, seven were polymorphic, whereas eight were monomorphic (Table 1).
2.2 Population genetic analysis
A total of 127 alleles were detected across the seven polymorphic SSR loci inA.gigantea(Table 2). The actual and effective number of alleles ranged from 1 to 9 and 1 to 5.121, respectively. The observed heterozygosity was from 0 to 0.769, with a mean of 0.489, and expected heterozygosity varied from 0 to 0.805, with the average of 0.594. Significant different levels of genetic diversity were observed in the four populations (Table 2). Population HNdls was found to possess the highest level of genetic variation, whereas Population HNjfl the lowest (Table 2). The high discriminatory power showed that these SSR loci are suitable for investigation of genetic variation inA.gigantea. All loci exceptAG-12 locus in HNjfl andAG-23 locus in HNbwl were in Hardy-Weinberg equilibrium (P<0.001) (Table 2). Four null alleles (AG-12,AG-40,AG-41, andAG-133) were detected. Three pairs of loci (AG-23 andAG-24;AG-23 andAG-105; andAG-24 andAG-105) were found to be in linkage disequilibrium (P<0.05).

Table 1 Characterization of 15 novel nuclear SSR markers developed in Alsophila gigantea1)
1)*: Polymorphic loci;Ta: annealing temperature
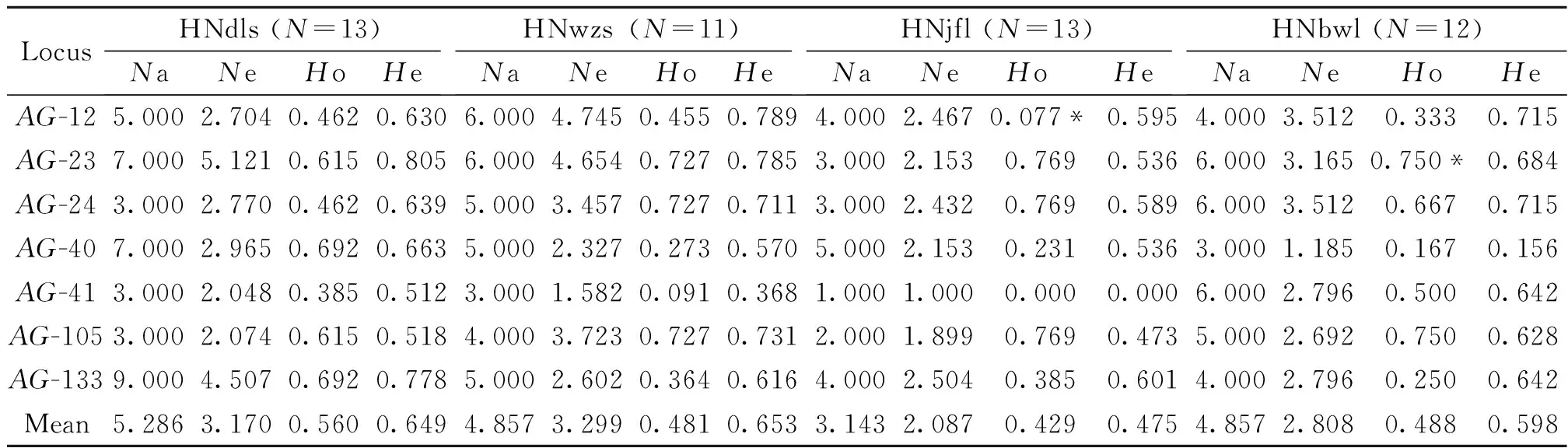
Table 2 Genetic parameters of seven polymorphic SSR markers in four Alsophila gigantea populations1)
1)N: The sample size for each population;Na: The actual number of alleles;Ne: The effective number of alleles;Ho: observed heterozygosity;He: expected heterozygosity;
*: Significant deviation from Hardy-Weinberg equilibrium atP<0.001 level
2.3 SSR markers cross-amplification
Cross-amplification inA.spinulosasuccessfully yielded PCR products, indicating transferability of the markers (Table 3). Two loci (AG-12 andAG-61) were polymorphic, which compensated for the pitfall that only three polymorphic loci were developed inA.spinulosa[14].

Table 3 Genetic estimation of cross-species amplification for two polymorphic SSR loci in the congeneric species Alsophila spinulosa1)
1)N: Population sample size;Na: The actual number of alleles;Ne: The effective number of alleles;Ho: observed heterozygosity;He: expected heterozygosity
3 Conclusions
It is the first to develop seven polymorphic SSR loci inA.giganteaand to examine transferability in its congeneric speciesA.spinulosa. The markers will be used to explore the population genetic adaptation. In addition, the high discriminatory power of SSR loci also allows for an accurate estimation of genetic variation. All of these are important factors for developing effective conservation strategies.
Acknowledgments:We gratefully thank Zhanming Ying, Qiang Fan, Lu Huang, Qi Deng, Ning Li, and Limin Cui of School of Life Science, Sun Yat-sen University for assistance and guidance in material collection, experiments, and data analysis. This work was supported by the National Natural Science Foundation of China (31370364, 31570652, and 31670200), the National Natural Science Foundation of Guangdong Province, China (2016A030313320), and Project of Department of Science and Technology of Shenzhen City, China (JCYJ20160425165447211).
[1] ZHANG X C, NISHIDA H. Cyatheaceae[M] //WU Z Y, RAVEN P H, HONG D Y. Flora of China. Beijing: Science Press, 2013: 134-138.
[2] DUDANI S N, MAHESH M K, CHANDRAN M D S, et al.CyatheanilgirensisHolttum (Cyatheaceae: Pteridophyta): a threatened tree fern from central Western Ghats, India[J]. Journal of Threatened Taxa, 2014, 6 (1): 5413-5416.
[3] TALUKDAR A D, CHOUDHURY M D, CHAKRAB ORTY M, et al. Phytochemical screening and TLC profiling of plant extracts ofCyatheagigantea(Wall. ex. Hook.) Haltt. andCyatheabrunoniana. Wall. ex. Hook. (Cl. & Bak.)[J]. Assam University Journal of Science & Technology, 2010, 5 (1): 70-74.
[4] KIRAN P M, RAJU A V, RAO B G. Investigation of hepatoprotective activity ofCyatheagigantea(Wall. ex. Hook.) leaves against paracetamol-induced hepatotoxicity in rats[J]. Asian Pacific Journal of Tropical Biomedicine, 2012, 2 (5): 352-356.
[5] AO G H. Research advance in Cyatheaceae[J]. Journal of Neijiang Teachers College, 2004, 19 (6): 79-82.
[6] FU L G R. List of China-rare and endangered plants[M]. Beijing: Science Press, 1992.
[7] KHOLIA B S, JOSHI R, PUNETHA R. Extended distribution ofCyatheaspinulosaWall ex Hook in Uttarakhand Himalaya with a note on distribution and diversification of Himalayan ferns in relation to recent climate change[J]. NeBio, 2013, 4 (2): 40-45.
[8] HOU X G, GUO D L, CHENG S P, et al. Development of thirty new polymorphic microsatellite primers forPaeoniasuffruticosa[J]. Biologia Plantarum, 2011, 55 (4): 708-710.
[9] ZHANG Y, HE J, ZHAO P X, et al. Genome-wide identification of microsatellites in white clover (TrifoliumrepensL.) using FIASCO and phpSSRMiner[J]. Plant Methods, 2008, 4 (1): 331-339.
[10] ZANE L, BARGELLONI L, PATARNELLO T. Strategies for microsatellite isolation: a review[J]. Molecular Ecology, 2002, 11 (1): 1-16.
[11] SINGH S, SAHU T R. Tree ferns of Pachmarhi Biosphere Reserve, Madhya Pradesh, India: taxonomy, ethnobotany and conservation[J]. International Journal of Advanced Research, 2015, 3 (8): 566-577.
[12] SU Y J, WANG T, ZHENG B, et al. Genetic differentiation of relictual populations ofAlsophilaspinulosain southern China inferred from cpDNAtrnL-F noncoding sequences[J]. Molecular Phylogenetics & Evolution, 2005, 34 (2): 323-333.
[13] CLARKE K R, GORLEY R N. PRIMER Version 5: User Manual/Tutorial [J]. Plymouth, United Kingdom:Primer-E Ltd, 2001.
[14] ZHOU Y, CHEN G P, WANG T. Isolation and characterization of microsatellite loci in the tree fernAlsophilaspinulosa[J]. American Fern Journal, 2008, 98 (1): 42-45.
[15] PEAKALL R O D, SMOUSE P E. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research[J]. Molecular Ecology Notes, 2006, 6 (1): 288-295.
[16] OOSTERHOUT C V, HUTCHINSON W F, WILLS D P M, et al. MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data[J]. Molecular Ecology Notes, 2004, 4 (3): 535-538.
[17] RAYMOND M, ROUSSET F. GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism[J]. Journal of Heredity, 1995, 86 (3): 248-249.
2016-06-29 基金项目:国家自然科学基金(31370364, 31570652, 31670200);广东省自然科学基金(2016A030313320);深圳市科技创新 (JCYJ20160425165447211)
阮效先、王桢并列第一作者;阮效先 (1990年生),女;研究方向:植物遗传适应性; E-mail:11011445012@qq.con
苏应娟(1965年生),女;研究方向:植物遗传适应性; E-mail: suyj@mail.sysu.edu.cn
Q347
A
0529-6579(2017)01-0115-06
大叶黑桫椤及桫椤SSR标记的开发
阮效先1,王桢2,王艇3,苏应娟1,4
(1. 中山大学生命科学学院,广东 广州510275; 2. 南京农业大学生命科学学院,江苏 南京210095; 3. 华南农业大学生命科学学院,广东 广州510642; 4. 深圳市中山大学研究所,广东 深圳518057)
大叶黑桫椤是桫椤科的大型树蕨,多分布在亚热带山地,为渐危种。迄今为止,鲜有大叶黑桫椤种群遗传变异的报道。采用FIASCO法开发了大叶黑桫椤15个SSR标记,用来自海南岛的4个种群检测标记的多态性。其中,7个标记具多态性,每个标记的实际等位基因数为1~9,观察杂合度和期望杂合度范围分别为0~0.769和0~0.805。位点AG-12和AG-23分别在HNjfl和HNbwl种群显著地偏离哈代温伯格平衡。鉴定出4个哑等位基因。发现3对标记存在连锁不平衡现象。此外,4个SSR标记能成功地用于桫椤种群的遗传分析。这些新的多态性SSR标记将用于大叶黑桫椤的群体遗传变异、局部适应性的研究以及建立有效的保护策略。
大叶黑桫椤;SSR标记;跨种扩增;遗传变异;局部适应性
10.13471/j.cnki.acta.snus.2017.01.018
