MiR-125a-5p is Upregulated in Plasma of Residents from An Electronic Waste Recycling Site
2016-12-12LiRanQiuXinghuaYangQiaoyunLiKeqiuLiGuang
Li Ran, Qiu Xinghua,*, Yang Qiaoyun, Li Keqiu, Li Guang,#
1. State Key Joint Laboratory for Environmental Simulation and Pollution Control, College of Environmental Sciences and Engineering and Center for Environment and Health, Peking University, Beijing 100871, P. R. China 2. Basic Medical College, Tianjin Medical University, Tianjin 300070, P. R. China
MiR-125a-5p is Upregulated in Plasma of Residents from An Electronic Waste Recycling Site
Li Ran1, Qiu Xinghua1,*, Yang Qiaoyun1, Li Keqiu2, Li Guang2,#
1. State Key Joint Laboratory for Environmental Simulation and Pollution Control, College of Environmental Sciences and Engineering and Center for Environment and Health, Peking University, Beijing 100871, P. R. China 2. Basic Medical College, Tianjin Medical University, Tianjin 300070, P. R. China
Received 18 November 2015 accepted 30 December 2015
The mechanism of health effects caused by organohalogen pollutants, e.g., toxins from electronic waste(e-waste), is poorly understood. We supposed that microRNAs (miRNAs), an important post-transcriptional regulator, could play a role in this process. In this study, fasting peripheral blood samples were collected from residents living at an e-waste site in northern China and a nearby reference population. Concentrations of e-waste related organohalogen pollutants in plasma from the exposure group were higher than the corresponding measurement in the reference group. Correspondingly, sixty miRNAs in plasma showed > 2-fold change between the two groups in microarray analysis. Among them, miR-125a-5p was confirmed to be upregulated by qRT-PCR and its validated targets were enriched in responses to xenobiotics and cancer related pathways. Furthermore, significant positive correlations were found between levels of miR-125a-5p in plasma and reactive oxygen species (ROS) in polymorphonuclear neutrophil leukocytes (P < 0.05). These evidences suggested oxidative stress might be an intermediate between e-waste related POPs exposure and alteration of plasma miRNA.
e-waste originated pollutants; microRNA; miR-125a-5p; epigenetics; oxidative stress
Persistent organic pollutants (POPs) are a group of important environmental contaminants. They have attracted considerable attention during the past decades particularly because of their ubiquity in the environment and their potential toxicity to humans[1]. Some typical POPs, such as polybrominated diphenyl ethers (PBDEs), polychlorinated biphenyls (PCBs), polychlorinated dibenzo-p-dioxins and dibenzofurans (PCDD/Fs), and polychlorinated naphthalenes (PCNs) are characteristic pollutants derived from electronic waste (e-waste)[2-3]. High levels of these pollutants have been observed in the environment of e-waste dismantling and recycling sites[3], and elevated levels have been documented in the blood of dismantling workers and even local residents[4-5], which could cause adverse health effects such as DNA damage and thyroid dysfunction[6-7].
However, the mechanism of the health effects caused by e-waste originated pollutants is not yet fully understood. The epigenetic regulation of specific genes expression, e.g., by microRNA, is one potential mechanism. MicroRNAs (miRNAs) belong to a class of non-coding, single-stranded short RNAs that have 19-25 nucleotides[8]. Currently, more than 2,500 miRNAs have been found in humans according to the Sanger miRBase database[9]. They play pivotal roles in various biological processes such as cell differentiation, proliferation, and death through their negative regulation of target genes during the post-transcription stage[10]. By base pairing to the 3' untranslated region (UTR) of target messenger RNAs (mRNAs), miRNAs can cause mRNA degradation and/or translational repression[8,10].
In previous toxicological studies, pulmonary miRNA profiles were altered in mice when exposed to benzo[a]pyrene, a typical environmental mutagen[11], and miR-101a and miR-122 were downregulated in the liver after exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD)[12]. In addition, epidemiological studies have demonstrated changes in miRNAs in response to environmental toxins such as tobacco smoke[13]and metal-rich particulate matter[14]. However, few studies have reported such changes in people exposed to POPs, particularly e-waste originated pollutants.
The expression profiles of miRNAs vary among tissues[15], and these tissue-specific miRNAs could be transported to other parts of the organism by exosomes, high density lipoproteins (HDL), or other microvesicles[16]. Hence, numerous miRNAs are present in human body fluids, especially in the plasma or serum[17]. Plasma or serum miRNAs are relatively stable, and previous studies have suggested that they are novel biomarkers for tissue injury[18], cancers[19], and cardiovascular diseases[20].
In this study, we investigated the expression levels of miRNAs in plasma of residents from an e-waste recycling site and explored the putative functional networks of a certain miRNA that was significantly affected. Meanwhile, the relationship between miRNAs and oxidative stress was also discussed since high level of cellular reactive oxygen species (ROS) was observed in these residents in our previous study[21].
1 Materials and methods
1.1 Subjects and sample collection
A cross-sectional rural population was enrolled in October 2011, as described previously[21]. Participants in this study were randomly selected from this population. In brief, eighteen participants living at an e-waste recycling site (38.825° N, 116.777° E) that contained many dismantling workshops was defined as the exposure group, and another seventeen participants living in a region without any e-waste dismantling activities (38.636° N, 117.135° E) was considered as the reference group. The distance between the two sites, which are both located in the same county of Tianjin, is approximately 40 km. The e-waste recycling site has a history of e-waste dismantling and recycling of more than 20 years[5]. Participants in each group had been living at their respective areas for nearly 20 years on average. The two groups shared similar environmental conditions and personal lifestyles, except that none of the participants in the reference group had worked in or lived near a polluted area before.
Fasting peripheral blood was collected in EDTA-coated vacuum tubes. Plasma was separated by two subsequent steps: centrifugation at 670 × g for 10 min to remove cells, followed by additional centrifugation at 16 000 × g for 3 min to remove cell debris. All plasma samples were stored at -80 ℃ until analysis. Demographic information for all of the subjects, including age, sex, occupation, and personal lifestyle, was obtained through face-to-face interviews. The institutional review board of Tianjin Medical University approved this study, and informed consent was obtained from every participant.
1.2 Pollutants analysis and ROS measurement
Organohalogen pollutants including polychlorinated biphenyls (PCBs), polybrominated diphenyl ethers (PBDEs), and dechlorane plus (DP) in plasma were measured as described in our previous publications[5,21]. ROS levels in peripheral polymorphonuclear neutrophil leukocytes of each subject were detected by using the fluorescence probe dihydrorhodamine 123 (DHR123) as mentioned in our earlier paper[21].
1.3 Plasma RNA isolation
Total RNA was extracted using mirVana PARIS kits (Ambion, Life Technologies, Carlsbad, CA, USA) following manufacturer’s instructions for liquid samples. Briefly, each 400 μL plasma sample was lysed with 400 μL 2× denaturing solution. After spiking with synthetic C. elegans miRNAs cel-miR-39, cel-miR-54, and cel-miR-238 (RNA oligonucleotides synthesized by Invitrogen, Life Technologies, Shanghai, China) as normalization controls, the denatured samples were extracted with 800 μL acid-phenol: chloroform mixture and the aqueous phase containing total RNA was recovered. Finally, the total RNA extract was purified using a glass-fiber filter and eluted in 100 μL nuclease-free water (Sigma-Aldrich, St. Louis, MO, USA).
6)存储模块使用控制器中集成的数据存储单元,结合MySQL数据库完成巷道数据的存储、机身和截割头位置数据的存储。
1.4 Screening for differentially expressed miRNAs in plasma
To screen for differentially expressed miRNAs, six randomly selected plasma samples from the exposure group and six from the reference group were subjected to microarray analysis. For each sample, about 100 ng total RNA was taken and analyzed using Agilent human miRNA (8×60 K) V16.0 which contained probes for 1,205 human and 142 human viral miRNAs. After RNA labeling and hybridization, slides were scanned on an Agilent G2565BA microarray scanner and data were extracted using Agilent Feature Extraction Software (Agilent Technologies, Santa Clara, CA, USA). The microarray assay was conducted by National Engineering Center for Biochip at Shanghai (Shanghai Biochip Co., Shanghai, China).
1.5 Confirmation of differentially expressed miRNAs in plasma
To verify the results of the microarray analysis, four miRNAs (miR-191-3p, miR-933, miR-125a-5p, and miR-494) were selected for real-time reverse transcription polymerase chain reaction (qRT-PCR) validation following a previous method[22]with some modifications. Briefly, RNA was reverse transcribed using a TaqMan miRNA reverse transcription kit and miRNA-specific stem-loop primers (both Applied Biosystems, Life Technologies) in a 15 μL reverse transcription reaction. Each reaction contained 4.16 μL nuclease-free water (Sigma-Aldrich), 1.5 μL 10× reverse-transcription buffer, 0.15 μL dNTPs (100 mmol·L-1), 0.19 μL RNase inhibitor (20 U·μL-1), 1 μL MultiScribeTMreverse transcriptase (50 U·μL-1), 3 μL 5× RT primer, and 5 μL RNA sample. The reaction tubes were loaded into a thermal cycler (Eppendorf Mastercycler Gradient, Eppendorf, Hamburg, Germany) and the reverse transcription reaction was run with the following temperature program: 16 ℃ for 30 min, 42 ℃ for 30 min, 85 ℃ for 5 min, and hold at 4 ℃. After reverse transcription, 1.33 μL product was used for real-time PCR in a 20 μL reaction that contained 7.67 μL nuclease-free water, 10 μL TaqMan 2× Universal PCR Master Mix, and 1 μL miRNA-specific primer/probe mix (Applied Biosystems, Life Technologies). The real-time reactions were performed in triplicate on a Mastercyclerep realplex2 system (Eppendorf) with the following temperature cycles: 95 ℃ for 10 min, followed by 40 cycles of 95 ℃ for 15 s and 60 ℃ for 1 min, and finally holding at 4℃.
1.6 miRNA targets networks analysis
We used miRTarBase[23], a database that curates experimentally validated miRNA-target interactions, to search the validated targets of differentially expressed miRNAs. And then we explored the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment and Gene Ontology (GO) enrichment of the validated target genes by applying the Database for Annotation, Visualization and Integrated Discovery (DAVID) tools[24].
1.7. Statistics
Raw data from the microarray were normalized by a quantile algorithm, Gene Spring Software 11.0 (Agilent technologies).Differentially expressed miRNAs between the two groups were determined by fold change (> 2) and Student’s t test (P < 0.05). For qRT-PCR data, the geometric mean cycle threshold (Ct) for the three C. elegans synthetic miRNAs in each sample was calculated as the normalization reference[25]. The relative expression levels of miRNAs in each group were calculated using the 2-ΔΔCTmethod[26]. Student’s t test was used to compare differences between the two groups. The association between either two variables was investigated by the Pearson correlation analysis. A P value < 0.05 was taken to indicate statistical significance (SPSS 16.0, SPSS Inc., Chicago, IL, USA).
2 Results
Demographic information and plasma levels of e-waste pollutants are shown in Table 1. Briefly, there were no statistically significant differences in age, body mass index (BMI), gender, or the number of smokers between the two groups, while plasma levels of some e-waste related pollutants such as PCBs and DPs were significantly higher in the exposure group than that in the reference group.

Table 1 Demographic characteristics and pollutants levels in plasma for the exposure and reference participants
Note:aStudent’s t test;bchi-square test;csum of PCB congeners -138, -153/132, -170, -171, -177, -180, -183, -187, -194, -195, -199, -202, -206, -208, and -209;dsum of PBDE congeners -28, -47, -153, -171, -183, -197, -201, -207, -208, and -209;esum of syn- and anti-isomers.

Fig. 1 Cluster analysis of 60 miRNAs that showed > 2-fold changes in plasma samples between exposure subjects (ES) and reference subjects (RS)
2.2 MiRNA profiles in plasma in the two groups
Plasma miRNA profiles for 12 randomly selected subjects were determined using microarrays. There was no difference (P = 0.8) in detection rate between the two groups (20.8% ± 4.2% for the exposure group and 19.5% ± 4.4% for the reference group). Among the detected miRNAs, sixty showed > 2-fold changes between the two groups (Fig.1) and two (miR-191-3p and miR-933) were downregulated in plasma samples of exposure group. Besides, miR-125a-5p and miR-494 were upregulated greatly (fold changes > 20) in exposure subjects, though 0.05 < P < 0.1.
2.3 Confirmation of miRNA expression
To verify the results of the microarray,qRT-PCR analysis was performed to quantify the levels of the two significantly downregulated miRNAs (miR-191-3p and miR-933) and two miRNAs with great fold changes (miR-125a-5p and miR-494) in all 35 subjects. As shown in Fig. 2, consistent with miRNA microarray analysis, miR-125a-5p levels in plasma samples were twice as high in the exposure group than in the reference group (P < 0.05). While for miR-191-3p or miR-494, although they were higher in the exposure group than those in the reference group, the differences were not statistically significant. As for miR-933, it was not detected in any samples in the qRT-PCR analysis. The results of miR-191-3p and miR-933 were different from microarray analysis, which indicated false-negative and false-positive results of microarray. Therefore, miR-125a-5p was selected for further study to investigate its possible biological functions.
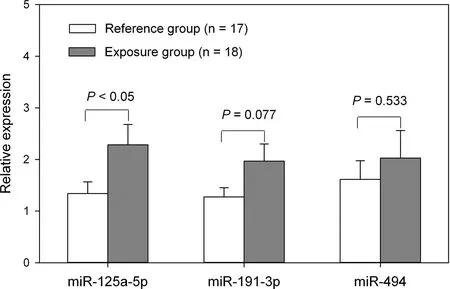
Fig. 2 qRT-PCR validation of miR-125a-5p, miR-191-3p, and miR-494 in plasma samples Note: Expression levels of miRNAs were normalized to spiked-in C. elegans synthetic miRNAs (cel-miR-39, cel-miR-54 and cel-miR-238).
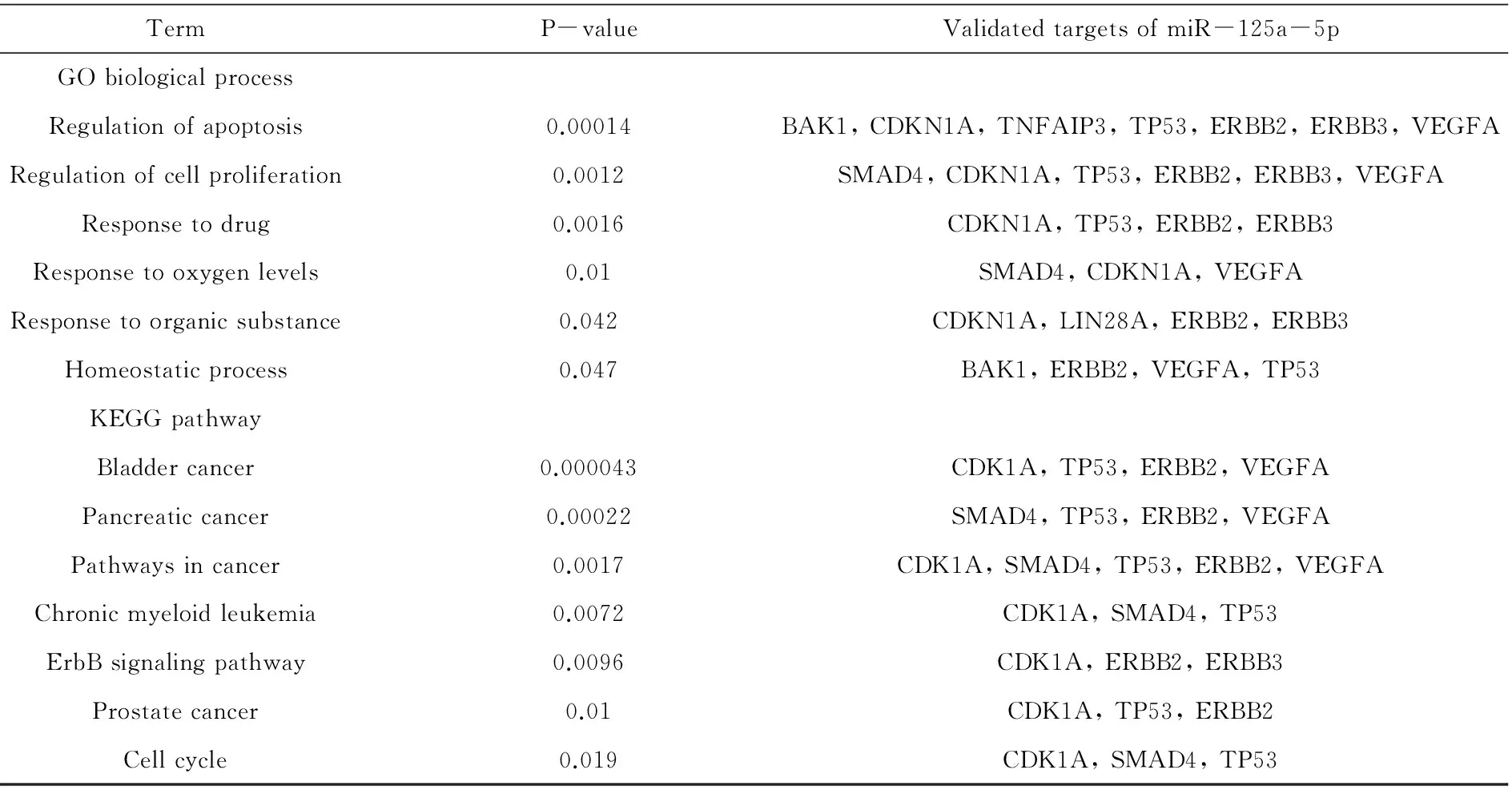
Table 2 Over-represented GO biological process and KEGG pathways annotations for validated targets of miR-125a-5p using DAVID[24]
Abbreviations:BAK1, BCL2-antagonist/killer 1; CDKN1A, cyclin-dependent kinase inhibitor 1A; TNFAIP3, tumor necrosis factor, alpha-induced protein 3; TP53, tumor protein p53; ERBB2, erythroblastic leukemia viral oncogene homolog 3; ERBB3, erythroblastic leukemia viral oncogene homolog 3; VEGFA, vascular endothelial growth factor A; SMAD4, SMAD family member 4; LIN28A, lin-28 homolog A.
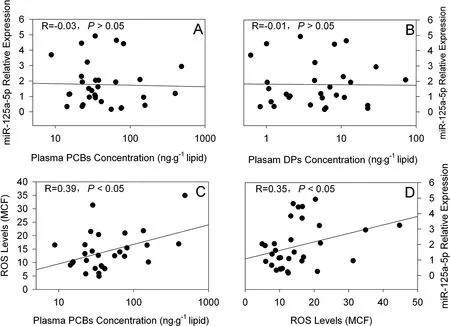
Fig. 3 Relationships among PCBs and DPs concentrations in plasma, plasma miRNA levels and oxidative stress Note: MCF represented mean channel fluorescence.
2.4 Enriched biological functions for validated targets of miR-125a-5p
By searching the miRTarBase[23], a database that covers experimentally validated miRNA-target interactions, we got targets for miR-125a-5p. Among the 138 validated target genes of miR-125a-5p, 16 targets were validated by more reliable methods such as reporter assay and western blot. As shown in Table 2, these 16 targets are enriched in many biological processes and pathways by applying the DAVID tools[24]. Most of the targets participate in regulation of apoptosis and cell proliferation, while some targets are enriched in responses to xenobiotics including drugs and organic substances. In addition, these targets are involved in cancer related pathways, such as bladder cancer, pancreatic cancer, chronic myeloid leukemia and prostate cancer.
2.5 Association between e-waste related POPs exposure and plasma miRNAs
Since high concentration of e-waste related POPs (PCBs and DPs) and miR-125a-5p were detected in plasma among residents at the e-waste site, correlation analysis was conducted to explore the association between POPs and miR-125a-5p. In Pearson correlation analysis, no significant correlations were observed between plasma concentrations of PCBs, DPs and miR-125a-5p levles in plasma (Fig.3A and 3B). However, positive correlations were found between plasma PCBs concentration and levels of ROS in peripheral polymorphonuclear neutrophil leukocytes (P < 0.05, Fig. 3C), and such correlation was also found between levels of plasma miR-125a-5p and those of ROS in peripheral polymorphonuclear neutrophil leukocytes (P < 0.05, Fig. 3D).
3 Discussion
POPs are ubiquitous in the environment and are thought to be associated with a series of adverse health effects[1]. However, the trace concentrations present in the environment make it difficult to investigate their effects in general populations. Pollutants such as PCBs, PBDEs, PCDD/Fs, and PCNs, which are released directly from mechanical disposal processes or are formed during the burning of e-waste, are key pollutants at e-waste recycling sites[2-3]. Therefore, we conducted this study in the highly polluted area of an e-waste recycling site in northern China.
Exposure to e-waste originated pollutants can bring about a series of adverse health effects; we have detected increased oxidative stress and weakened innate immunity in a previous study[21]. In addition, many studies have demonstrated that exposure to these pollutants was also associated with altered neuropsychological[27]and endocrine functions[7]. However, the underlying mechanisms are not yet fully understood, and possible epigenetic effects have attracted considerable attention in recent years. For instance, Li et al. reported that miRNA expression profiles in the spermatozoa of men living in e-waste areas were different from controls[28]. In addition,plasma/serum miRNAs have been regarded as new biomarkers for tissue injury[29], cancers[19], and cardiovascular diseases in recent years. Combined with Ago2 protein complexes and packaged by exosomes, HDL or other microvesicles[16], these circulation miRNAs could avoid RNase digestion and maintain stability after freezing-thawing processes[22]. Investigating the changes of plasma miRNAs and the potential function among people living at the high polluted region will contribute to our understanding of the health effects caused by these pollutants. In the present study, we found different plasma miRNA profiles between subjects living at the e-waste recycling region and the reference region. Especially, miR-125a-5p levels was significantly higher in plasma samples from exposed people than in reference individuals. This is the first study to show that exposure to e-waste related pollutants can alter plasma miRNAs, and this might indicate the mechanistic basis of certain health effects.
According to the microRNA org resource[30], human miR-125a-5p is mainly expressed in normal tissues such as spleen, thyroid and pancreatic islet, and tumor cells. Located at human chromosome19q13, miR-125a-5p belongs to the miR-125 family. Previous studies suggested that miR-125 family had important roles in cell proliferation, differentiation, and apoptosis[31]. By targeting to MMP11 (matrix metalloproteinase 11) and VEGFA (vascular endothelial growth factor A), miR-125a was proved to inhibit the proliferation and metastasis of hepatocellular carcinoma[32]. The expression of miR-125a-5p was downregulated in non-small cell lung cancer, and contributed to the invasion and migration of lung cancer cells[33]. In contrast, miR-125a-5p was upregulated in basal cell carcinoma[34], and was identified as potential invasiveness enhancer in urothelial carcinomas[35]. The inconsonant functions indicated miR-125a-5p plays distinctive roles under different conditions. Actually, the validated targets of miR-125a-5p were linked to multiple cancer related pathways and regulation of apoptosis. Besides, it was noteworthy that the validated targets were enriched in responses to xenobiotics including drugs and organic substances (Table 2). This suggested that the upregulated expression of miR-125a-5p was probably due to exposure to e-waste related POPs.
Although high concentrations of PCBs and DPs were detected in plasma among residents at the e-waste site, no significant correlation was found between concentrations of these pollutants and those of miR-125a-5p in plasma (P > 0.05, Fig.3A and 3B), indicating a complicated process probably mediated by other mechanism(s) between pollutants and miRNA alternation. In a large number of previous studies, oxidative stress had been proposed as the cause of altered miRNA expression[36-38]. While in our previous work[21], levels of ROS in peripheral polymorphonuclear neutrophil leukocytes were significantly higher among residents at the e-waste site, which were significantly correlated with plasma levels of pollutants (e.g., PCBs, Fig.3C). In the present study, a significant positive correlation was also found between levels of plasma miR-125a-5p and those of ROS in peripheral polymorphonuclear neutrophil leukocytes (P < 0.05; Fig.3D). This evidence indicated that elevated miR-125-5p might be a result of oxidative stress after exposing to e-waste originated POPs. This hypothesis agreed with previous studies. For instance, Chen et al. reported that the expression of miR-125a-5p in human primary monocytes was upregulated after exposing to oxidized low density lipoprotein (oxLDL)[39], and levels of miR-125a-5p in vascular endothelial cells was also regulated by oxLDL[40]. In addition, cellular miR-125a-5p levels were upregulated in human alveolar epithelial cells (A549) treated with aldehydes, a group of volatile organic compounds (VOCs) that are able to cause oxidative stress[41]. More importantly, the expression of miR-125a-5p in LPS treated macrophages[42]and neutrophils of patients with major traumatic injury[43], in which both of the leukocytes had high levels of ROS, were increased. In combination with the evidence that levels of ROS in peripheral polymorphonuclear neutrophil leukocytes were significantly correlated with e-waste originated pollutants such as PCBs, these results suggested oxidative stress was an intermediate between e-waste related POPs exposure and alteration of plasma miRNAs.
In summary, this preliminary study found altered plasma miRNA profiles in local residents from a highly polluted e-waste recycling region by applying miRNAs microarray assay. Through qRT-PCR, plasma miR-125a-5p was demonstrated to be upregulated in local residents. The validated targets of miR-125a-5p were involved in responses to xenobiotics and cancer related pathways. The plasma miR-125a-5p and miR-191-3p levels were positively correlated with ROS levels in peripheral polymorphonuclear neutrophil leukocytes, which suggested that oxidative stress might be an intermediate between e-waste related POPs exposure and alteration of plasma miRNA. However, restricted to the small number of study subjects, we didn’t find the direct correlation between e-waste related POPs and plasma miRNA levels. Hence, further studies were warranted to reveal the mechanism of miRNAs expression alteration caused by exposure to e-waste originated pollutants, and the role of oxidative stress.
Acknowledgements and grant information: This work was supported by National Natural Science Foundation of China (21322705, 41121004, 21190051, and 21177091), and the Collaborative Innovation Center for Regional Environmental Quality. The authors declare no competing financial interests.
[1] Li Q Q, Loganath A, Chong Y S, et al. Persistent organic pollutants and adverse health effects in humans [J]. Journal of Toxicology and Environmental Health-Part A-Current Issues, 2006, 69(21): 1987-2005
[2] Lin Y, Zhao Y F, Qiu X H, et al. Spatial distribution of polychlorinated naphthalenes in the atmosphere across North China based on gridded field observations [J]. Environmental Pollution, 2013, 180: 27-33
[3] Liu H X, Zhou Q F, Wang Y W, et al. E-waste recycling induced polybrominated diphenyl ethers, polychlorinated biphenyls, polychlorinated dibenzo-p-dioxins and dibenzo-furans pollution in the ambient environment [J]. Environment International, 2008, 34(1): 67-72
[4] Bi X H, Thomas G O, Jones K C, et al. Exposure of electronics dismantling workers to polybrominated diphenyl ethers, polychlorinated biphenyls, and organochlorine pesticides in South China [J]. Environmental Science & Technology, 2007, 41(16): 5647-5653
[5] Yang Q Y, Qiu X H, Li R, et al. Exposure to typical persistent organic pollutants from an electronic waste recycling site in Northern China [J]. Chemosphere, 2013, 91(2): 205-211
[6] Yuan J, Chen L, Chen D H, et al. Elevated serum polybrominated diphenyl ethers and thyroid-stimulating hormone associated with lymphocytic micronuclei in Chinese workers from an e-waste dismantling site [J]. Environmental Science & Technology, 2008, 42(6): 2195-2200
[7] Zhang J Q, Jiang Y S, Zhou J, et al. Elevated body burdens of PBDEs, dioxins, and PCBs on thyroid hormone homeostasis at an electronic waste recycling site in China [J]. Environmental Science & Technology, 2010, 44(10): 3956-3962
[8] Ha M, Kim V N. Regulation of microRNA biogenesis [J]. Nature Reviews Molecular Cell Biology, 2014, 15(8): 509-524
[9] Kozomara A, Griffiths-Jones S. miRBase: Annotating high confidence microRNAs using deep sequencing data [J]. Nucleic Acids Research, 2014, 42(D1): D68-D73
[10] Orellana E A, Kasinski A L. MicroRNAs in cancer: A historical perspective on the path from discovery to therapy [J]. Cancers, 2015, 7(3): 1388-1405
[11] Halappanavar S, Wu D, Williams A, et al. Pulmonary gene and microRNA expression changes in mice exposed to benzo(a)pyrene by oral gavage [J]. Toxicology, 2011, 285(3): 133-141
[12] Yoshioka W, Higashiyama W, Tohyama C. Involvement of microRNAs in dioxin-induced liver damage in the mouse [J]. Toxicological Sciences, 2011, 122(2): 457-465
[13] Schembri F, Sridhar S, Perdomo C, et al. MicroRNAs as modulators of smoking-induced gene expression changes in human airway epithelium [J]. Proceedings of the National Academy of Sciences of the United States of America, 2009, 106(7): 2319-2324
[14] Bollati V, Marinelli B, Apostoli P, et al. Exposure to metal-rich particulate matter modifies the expression of candidate microRNAs in peripheral blood leukocytes [J]. Environmental Health Perspectives, 2010, 118(6): 763-768
[15] Yin J Q, Zhao R C, Morris K V. Profiling microRNA expression with microarrays [J]. Trends in Biotechnology, 2008, 26(2): 70-76
[16] Chen X, Liang H W, Zhang J F, et al. Secreted microRNAs: A new form of intercellular communication [J]. Trends in Cell Biology, 2012, 22(3): 125-132
[17] Weber J A, Baxter D H, Zhang S, et al. The microRNA spectrum in 12 body fluids [J]. Clinical Chemistry, 2010, 56(11): 1733-1741
[18] Wang K, Zhang S, Marzolf B, et al. Circulating microRNAs, potential biomarkers for drug-induced liver injury [J]. Proceedings of the National Academy of Sciences of the United States of America, 2009, 106(11): 4402-4407
[19] Mitchell P S, Parkin R K, Kroh E M, et al. Circulating microRNAs as stable blood-based markers for cancer detection [J]. Proceedings of the National Academy of Sciences of the United States of America, 2008, 105(30): 10513-10518
[20] Gupta S K, Bang C, Thum T. Circulating micrornas as biomarkers and potential paracrine mediators of cardiovascular disease [J]. Circulation-Cardiovascular Genetics, 2010, 3(5): 484-488
[21] Li R, Yang Q Y, Qiu X H, et al. Reactive oxygen species alteration of immune cells in local residents at an electronic waste recycling site in Northern China [J]. Environmental Science & Technology, 2013, 47(7): 3344-3352
[22] Kroh E M, Parkin R K, Mitchell P S, et al. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR) [J]. Methods, 2010, 50(4): 298-301
[23] Hsu S D, Lin F M, Wu W Y, et al. miRTarBase: A database curates experimentally validated microRNA-target interactions [J]. Nucleic Acids Research, 2011, 391: D163-D169
[24] Huang D W, Sherman B T, Lempicki R A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists [J]. Nucleic Acids Research, 2009, 37(1): 1-13
[25] Vandesompele J, De Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes [J]. Genome Biology, 2002, 3(0034.17)
[26] Livak K J, Schmittgen T D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method [J]. Methods, 2001, 25(4): 402-408
[27] Haase R F, Mccaffrey R J, Santiago-Rivera A L, et al. Evidence of an age-related threshold effect of polychlorinated biphenyls (PCBs) on neuropsychological functioning in a native American population [J]. Environmental Research, 2009, 109(1): 73-85
[28] Li Y, Li M C, Liu Y X, et al. A microarray for microRNA profiling in spermatozoa from adult men living in an environmentally polluted site [J]. Bulletin of Environmental Contamination and Toxicology, 2012, 89(6): 1111-1114
[29] Wang Z, Liu Y M, Han N, et al. Profiles of oxidative stress-related microRNA and mRNA expression in auditory cells [J]. Brain Research, 2010, 1346: 14-25
[30] Betel D, Wilson M, Gabow A, et al. The microRNA.org resource: Targets and expression [J]. Nucleic Acids Research, 2008, 36(SI): D149-D153
[31] Sun Y M, Lin K Y, Chen Y Q. Diverse functions of miR-125 family in different cell contexts [J]. Journal of Hematology & Oncology, 2013, 6(6). DOI: 10.1186/1756-8722-6-6
[32] Bi Q, Tang S H, Xia L, et al. Ectopic expression of miR-125a inhibits the proliferation and metastasis of hepatocellular carcinoma by targeting MMP11 and VEGF [J]. PLOS ONE, 2012, 7(e401696)
[33] Jiang L, Huang Q, Zhang S, et al. Hsa-miR-125a-3p and hsa-miR-125a-5p are downregulated in non-small cell lung cancer and have inverse effects on invasion and migration of lung cancer cells [J]. BMC Cancer, 2010, 10(318).
[34] Sand M, Skrygan M, Sand D, et al. Expression of microRNAs in basal cell carcinoma [J]. British Journal of Dermatology, 2012, 167(4): 847-855
[35] Ratert N, Meyer H A, Jung M, et al. Reference miRNAs for miRNAome analysis of urothelial carcinomas[J]. PLOS ONE, 2012, 7(e393096)
[36] Cheng Y H, Liu X J, Zhang S O, et al. MicroRNA-21 protects against the H2O2-induced injury on cardiac myocytes via its target gene PDCD4 [J]. Journal of Molecular and Cellular Cardiology, 2009, 47(1): 5-14
[37] Simone N L, Soule B P, Ly D, et al. Ionizing radiation-induced oxidative stress alters miRNA expression [J]. PLOS ONE, 2009, 4(e63777)
[38] Magenta A, Cencioni C, Fasanaro P, et al. miR-200c is upregulated by oxidative stress and induces endothelial cell apoptosis and senescence via ZEB1 inhibition [J]. Cell Death and Differentiation, 2011, 18(10): 1628-1639
[39] Chen T, Huang Z, Wang L, et al. MicroRNA-125a-5p partly regulates the inflammatory response, lipid uptake, and ORP9 expression in oxLDL-stimulated monocyte/macrophages [J]. Cardiovascular Research, 2009, 83(1): 131-139
[40] Li D, Yang P, Xiong Q, et al. MicroRNA-125a/b-5p inhibits endothelin-1 expression in vascular endothelial cells [J]. Journal of Hypertension, 2010, 28(8): 1646-1654
[41] Song M, Lee H, Ryu J. Integrated analysis of microRNA and mRNA expression profiles highlights aldehyde-induced inflammatory responses in cells relevant for lung toxicity [J]. Toxicology, 2015, 334: 111-121
[42] Banerjee S, Cui H, Xie N, et al. miR-125a-5p regulates differential activation of macrophages and inflammation [J]. Journal of Biological Chemistry, 2013, 288(49): 35428-35436
[43] Yang J, Liang Y, Han H, et al. Identification of a miRNA signature in neutrophils after traumatic injury [J]. Acta Biochimica et Biophysica Sinica, 2013, 45(11): 938-945
◆
10.7524/AJE.1673-5897.20151118001
biography:Li Ran, PhD, E-mail: ranli28@126.com;
* Corresponding authors:Qiu Xinghua, E-mail: xhqiu@pku.edu.cn;
s’ biography: Qiu Xinghua: PhD., assistant professor in College of Environmental Sciences and Engineering, Peking University, Beijing 100871, P. R. China. E-mail: xhqiu@pku.edu.cn.
And Li Guang: Professor in Basic Medical College, Tianjin Medical University, Tianjin 300070, P. R. China. E-mail: lig@tijmu.edu.cn.
Li R, Qiu X H, Yang Q Y, et al. MiR-125a-5p is upregulated in plasma of residents from an electronic waste recycling site [J]. Asian Journal of Ecotoxicology, 2016, 11(2): 134-143 (in Chinese)
Grant information:National Natural Science Foundation of China (21322705, 41121004, 21190051, and 21177091)
# Co-corresponding author:Li Guang, E-mail: lig@tijmu.edu.cn
