Development of polymorphic EST-SSR markers fromBradysiaodoriphaga(Diptera: Sciaridae), a serious agricultural pest in China
2016-12-07YunliTAOCunhuanZHANGShuleiTAILeiWANGYananGUOFengshanYANGChuanzhiZHAOFanghaoWANDongCHU
Yun-li TAO, Cun-huan ZHANG, Shu-lei TAI, Lei WANG, Ya-nan GUO, Feng-shan YANG, Chuan-zhi ZHAO, Fang-hao WAN,4, Dong CHU*
1KeyLaboratoryofIntegratedCropPestManagementofShandongProvince,CollegeofAgronomyandPlantProtection,QingdaoAgriculturalUniversity,Qingdao,Shandong266109,China;2CollegeofLifeSciences,HeilongjiangUniversity,Harbin,Heilongjiang150080,China;3BiotechnologyResearchCenter,ShandongAcademyofAgriculturalSciences,Jinan,Shandong250100,China;4StateKeyLaboratoryforBiologyofPlantDiseasesandInsectPests,InstituteofPlantProtection,ChineseAcademyofAgriculturarlSciences(CAAS),Beijing100081,China
Development of polymorphic EST-SSR markers fromBradysiaodoriphaga(Diptera: Sciaridae), a serious agricultural pest in China
Yun-li TAO1, Cun-huan ZHANG1, Shu-lei TAI1, Lei WANG1, Ya-nan GUO1, Feng-shan YANG2, Chuan-zhi ZHAO3, Fang-hao WAN1,4, Dong CHU1*
1KeyLaboratoryofIntegratedCropPestManagementofShandongProvince,CollegeofAgronomyandPlantProtection,QingdaoAgriculturalUniversity,Qingdao,Shandong266109,China;2CollegeofLifeSciences,HeilongjiangUniversity,Harbin,Heilongjiang150080,China;3BiotechnologyResearchCenter,ShandongAcademyofAgriculturalSciences,Jinan,Shandong250100,China;4StateKeyLaboratoryforBiologyofPlantDiseasesandInsectPests,InstituteofPlantProtection,ChineseAcademyofAgriculturarlSciences(CAAS),Beijing100081,China
【Background】 The chive gnat,BradysiaodoriphagaYang and Zhang (Diptera: Sciaridae), is a severe agricultural pest in China. Knowledge on the biology, dispersal, and other important aspects of this insect is limited. Filling this knowledge gap is hampered by the lack of suitable the genetic markers. The aim of the present study was to develop simple sequence repeat (SSR) markers from expressed sequence tags (ESTs) that can be used for genetic diversity and structure analysis ofB.odoriphaga. 【Method】 The SSRs primers were designed and tested based on the ESTs ofB.odoriphagaobtained in this study. 【Result】 A total of 3383 SSRs were identified from 42095 unigenes. Sixteen pairs of primers were designed and tested in 30B.odoriphagalarvae, of which nine primer pairs produced polymorphic amplicons. Thirty alleles were identified from 30 larvae using the nine markers, with an average of 3.33 alleles per locus (ranged from 3 to 4). The range of observed and expected heterozygosity was 0.0000~0.6875 and 0.0370~0.6877, respectively. Five of the nine loci exhibited significant departure from Hardy-Weinberg equilibrium. 【Conclusion and significance】 The nine polymorphic microsatellite loci developed in this study can be used to research the genetic diversity and structure ofB.odoriphagapopulations.
Bradysiaodoriphaga; expressed sequence tag; simple sequence repeat; genetic diversity; genetic structure
1 INTRODUCTION
The chive gnat,BradysiaodoriphagaYang and Zhang (Diptera: Sciaridae), is one of the most important pest of chives in China (Lietal.,2007; Taoetal.,2015). This pest is mainly destructive to Chinese chives (Alliumtuberosum) but can also attack other allium vegetables, as well as cabbage, radish, melon, celery, mushrooms, and various ornamentals (Meietal.,2003; Zhangetal.,2015). The larvae ofB.odoriphagacan feed on roots or stems underground, causing plants to be stunted or to even die and resulting in serious economic losses (Yangetal.,2015). This pest can cause severe yield losses of Chinese chives every year (Dangetal.,2001). Analysis of the genetic structure helps to elucidate the gene flow and genetic differentiation of many species (De Lucaetal.,2002). So far, the gene flow and genetic differentiation ofB.odoriphagaremain poorly understood.
Simple sequence repeats (SSR), also known as microsatellites, have become one of the most important genetic markers in the analysis of population genetic structure (De Lucaetal.,2002). SSRs contain tandem repeat genetic loci of 1~8 bp (Richardetal.,2008). Because of their abundance in genomes, co-dominant inheritance, and high level of polymorphism (Melnikovaetal.,2012; Mengetal.,2012; Yakovinetal.,2011), SSRs are useful for gene mapping and population genetic structure analysis (Jungetal.,2006). Expressed sequence tags (ESTs) have been applied to discover SSR markers rather than using genomic sequences (Wangetal.,2014). Compared with conventional genomic SSR markers, EST-based SSRs have several intrinsic advantages such as reduced costs, faster development of suitable markers, and broad transferability between species (Yu & Li,2007).
The aim of the present study was to develop EST-SSRs that can be used for the analysis of genetic diversity and structure ofB.odoriphaga. A total of 42095B.odoriphagaunigene sequences were used firstly mined for analyzing SSR. Then the primer pairs were designed to select the polymorphic loci. Due to lack of suitable the genetic markers, knowledge on the biology, dispersal, and other important aspects of this insect is limited, and the polymorphic EST-SSRs obtained forB.odoriphagacould be helpful for genetic studies of this organism.
2 MATERIALS AND METHODS
2.1 Samples and EST mining
Samples collected from Tianjin, China in 2013 were used to construct the EST library ofB.odoriphaga. A total of 42095B.odoriphagaunigene sequences (data not presented) were mined for analyzing SSR. The DNAstar software (DNASTAR, Madison, WI, USA) was used to pretreat theB.odoriphagaEST sequences and the vector sequences and sequences shorter than 150 bp were removed. The software MISA (http:∥pgrc.ipk-gatersleben.de/misa/) was used to search SSRs with the criteria as follows: mononucleotide repeats at least 12 times, dinucleotide repeats at least six times, trinucleotide repeats at least five times, tetranucleotide repeats at least five times, and pentanucleotide and hexanucleotide repeats at least four times.
2.2 DNA extraction, primer design and testing
Genomic DNA was extracted fromB.odoriphagalarvae collected using the lysis method described by Frohlichetal.(1999). The DNA-containing supernatant was stored at -20 ℃. Sixteen pairs of primers were designed using PRIMER PREMIER 5.0 software (Lalitha,2000), and 30B.odoriphagalarvae collected from Shouguang, Shandong Province, China in 2013 were used to test the validity. The samples were freshly collected and stored at -20 ℃. The DNA-containing supernatant of each individual larva was used for subsequent PCR amplification. PCR amplification was performed in 13 μL volumes containing 0.13 μL of Taq DNA polymerase (5 U·μL-1), 1.3 μL of 10×Easy Taq PCR Buffer, 0.26 μL of each dNTP (10 μmol·L-1), 0.26 μL of each primer (10 μmol·L-1), and 2 μL of DNA. The PCR conditions were as follows: initial denaturation at 94 ℃ for 5 min; 35 cycles of 30 s at 94 ℃, 45 s at the primer-specific annealing temperature (50~56 ℃: the different primer sets had different PCR reactions) , and 30 s at 72 ℃; and a final elongation step at 72 ℃ for 7 min. The allele size was determined according to Gaoetal.(2014). Briefly, the products were run on an ABI 3730 xl DNA analyzer (Sangon, Shanghai, China) and the allele size was determined by comparing the mobility of the PCR products to the GeneScanTM400HD size standard using GeneMapper software version 3.2 (Applied Biosystems, Shanghai, China).
2.3 Data analysis
The average number of alleles per locus (Na), the observed heterozygosity (Ho), and the expected heterozygosity (He) were calculated using POPGENE v.1.31 (Wangetal.,2014; Yehetal.,1999). GENEPOP v.3.4 was used to test for deviation from Hardy-Weinberg equilibrium (Mortegaetal.,2015; Raymond & Rousset,1995) and to calculate FIS values (Weir & Cockerham,1984).
3 RESULTS
3.1 Microsatellite loci identified
A total of 3383 SSR loci from 42095 EST sequences were identified. The 3383 SSRs mainly comprised mononucleotide (1408), dinucleotide (693), and trinucleotide (1210) repeats, with tetranucleotide (41), pentanucleotide (28) and hexanucleotide (3) repeats being uncommon (Table 1).
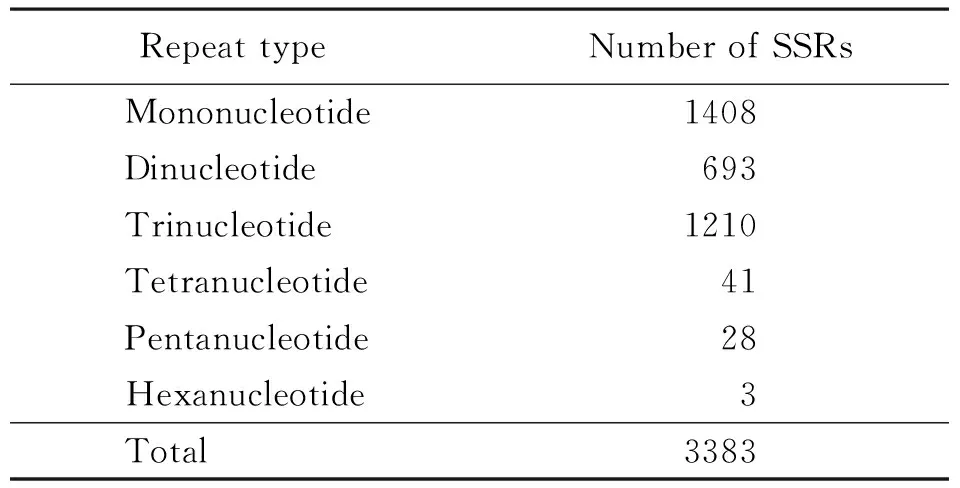
Table 1 Occurrence frequency of repeat motifs of EST-SSRs inBradysiaodoriphaga
Among the mononucleotide repeats, A/T motifs were most common (Table 2). Among the dinucleotide repeats, AG/CT motifs dominated, followed by AC/GT and AT/AT (Table 2). Among the trinucleotide repeats, AAC/GTT motifs were most common , followed by ATC/ATG and AAG/CTT (Table 2). Tetranucleotide, pentanucleotide, and hexanucleotide repeats had relatively low occurrence frequencies (<1.00%).

Table 2 Occurrence frequency of main repeat motifs of EST-SSRs inBradysiaodoriphaga
3.2 Microsatellite primer pairs designed and testing
Sixteen primer pairs were designed using PRIMER PREMIER 5.0 software. We tested 16 primer pairs in the 30B.odoriphagalarvae however, only nine primer pairs produced amplicons and revealed polymorphisms. We used the nine markers to identify 30 alleles, with 3~4 alleles detected per locus. The observed and expected heterozygosity was 0.0000~0.6875 and 0.0370~0.6877, respectively. Five loci (code: 3477, 3373, 2479, 2745, and 2312) exhibited significant departure from Hardy-Weinberg equilibrium (Table 3).

Table 3 Expressed sequence tag-simple sequence repeats used to identify polymorphisms in oneBradysiaodoriphagapopulation
SSR: Simple sequence repeat; F: Forward; R: Reverse;Ta: Annealing temperature;Na: Number of detected alleles;Ho: Observed heterozygosity;He: Expected heterozygosity;FIS: Estimator of the fixation index;P: Probability value of Hardy-Weinberg equilibrium (*Significance atP<0.05).
4 DISCUSSION
In China,B.odoriphagais an important pest in agricultural ecosystems. However, due to lack of suitable the genetic markers, knowledge on the biology, dispersal, and other important aspects of this insect is limited. The population spread and dynamics in the field, which is important for the sustainable regional management of this pest. The potential spread model of a pest insect can be revealed via genetic markers including microsatellite loci. The design and selection of polymorphic microsatellite loci are important to reveal the gene flow, genetic differentiation, and the population dynamics of insect pests. Using microsatellite markers, Gaoetal.(2014) demonstrated that the spread of the greenhouse whitefly,Trialeurodesvaporariorum(Westwood), in some regions of China was through secondary introduction from regions where it was initially introduced.
In this study, the constructed ESTs ofB.odoriphagashowed that the proportion of different motifs of SSRs in this species varied greatly. The most common motifs were mononucleotide repeats, similar to the western flower thrips,Frankliniellaoccidentalis(Duanetal.,2012). However, the proportion of the repeat motif types differs according to the species (see Duanetal.,2012), which indicates that the proportion of motif types is species-specific.
Among the 42095B.odoriphagaunigene sequences, a total of 3383 SSR loci were revealed, which suggests that SSR loci are distributed widely in the ESTs of this species. Compared with conventional genomic SSR markers, EST-based SSRs have several intrinsic advantages (Yu & Li,2007). Previous study showed that the ESTs can be useful for the development of SSR markers (Wangetal.,2014). The new SSR markers developed could be helpful to reveal the gene flow and genetic differentiation ofB.odoriphagain China.
Acknowledgments: We appreciate Wei-hong XU (Tianjin Academy of Agricultural Sciences) for providingBradysiaodoriphagasamples, and anonymous reviewers for their helpful comments on the manuscript.
Dang Z H, Dong J Z, Gao Z L, Jia H M, Zhang K J and Pan W L, 2001. Biology and injury ofBradysiaodoriphagaon leek in different types of cultivation.JournalofAgriculturalUniversityofHebei, 24(4): 65-68 (in Chinese).
De Luca F, Reyes A, Veronico P, Di Vito M, Lamberti F and De Giorgi C, 2002. Characterization of the (GAAA) microsatellite region in the plant parasitic nematodeMeloidogyneartiellia.Gene, 293(1/2): 191-198.
Duan H S, Zhang A S, Zhao C Z, Yu Y and Chu D, 2012. Characterization and molecular marker screening of EST-SSRs and their polymorphism compared with Genomic-SSRs inFrankliniellaoccidentalis(Thysanoptera: Thripidae).ActaEntomologicaSinica, 55(6): 634-640 (in Chinese).
Frohlich D R, Torres-Jerez I, Bedford I D, Markham P G and Brown J K, 1999. A phylogeographical analysis of theBemisiatabacispecies complex based on mitochondrial DNA markers.MolecularEcology, 8(10): 1683-1691.
Gao R R, Zhang W P, Wu H T, Zhang R M, Zhou H X, Pan H P, Zhang Y J, Brown J K and Chu D, 2014. Population structure of the greenhouse whitefly,Trialeurodesvaporariorum(Westwood), an invasive species from the Americas, 60 years after invading China.InternationalJournalofMolecularScience, 15(8): 13514-13528.
Jung J, Lee E and Kim W, 2006. Isolation and characterization of polymorphic microsatellite markers ofAnophelessinensis, a malaria vector mosquito in the East Asia region.MolecularEcologyNotes, 6(4): 1272-1274.
Lalitha S, 2000. Primer premier 5.BioTechSoftware&InternetReport, 1(6): 270-272.
Li H J, He X K, Zeng A J, Liu Y J and Jiang S R, 2007.Bradysiaodoriphagacopulatory behavior and evidence of a female sex pheromone.JournalofAgriculturalandUrbanEntomology, 24(1): 27-34.
Mei Z X, Wu Q J, Zhang Y J and Hua L, 2003. The biology, ecology and management ofBradysiaodoriphaga.EntomologicalKnowledge, 40(5): 396-398 (in Chinese). Melnikova M N, Petrov N B, Lomov A A, La Porta N and Politov D V, 2012. Testing of microsatellite primers with different populations of Eurasian sprucesPiceaabies(L.) Karst. andPiceaobovataLedeb.RussianJournalofGenetics, 48(5): 562-566.Meng Y, Zhang Y, Liang H W, Xiao H B and Xie C X, 2012. Genetic diversity of Chinese giant salamander (Andriasdavidianus) based on the novel microsatellite markers.RussianJournalofGenetics, 48(12): 1227-1231.
Mortega K G, Horsburgh G J, Illera J C and Dawson D A, 2015. Characterization of microsatellite markers forSaxicolaspecies.ConservationGeneticsResources, 7(1): 273-278.
Raymond M and Rousset F, 1995. GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism.TheJournalofHeredity, 86: 248-249.Richard G F, Kerrest A and Dujon B, 2008. Comparative genomics and molecular dynamics of DNA repeats in eukaryotes.MicrobiologyandMolecularBiologyReviews, 72(4): 686-727.Tao Y L, Guo Y N, Wang J, Li L L, Yu Y and Chu D, 2015. Detection and identification ofWolbachiainBradysiaodoriphaga(Diptera: Sciaridae) populations from Shandong Province, China.ActaEntomologicaSinica, 58(4): 454-459 (in Chinese). Wang H M, Zhao H H, Zhao C Z and Chu D, 2014. EST-SSR markers fromHeteroderaglycinesIchinohe.RussianJournalofGenetics, 50(10): 1117-1119.Weir B S and Cockerham C C, 1984. Estimating F-statistics for the analysis of population structure.Evolution, 38(6): 1358-1370.Yakovin N A, Fesenko I A, Isachkin A V and Karlov G I, 2011. Polymorphism of microsatellite loci in cultivars and species of pear (PyrusL.).RussianJournalofGenetics, 47(5): 564-570.Yang Y T, Li W X, Xie W, Wu Q J, Xu B Y, Wang S L, Li C R and Zhang Y J, 2015. Development ofBradysiaodoriphaga(Diptera: Sciaridae) as affected by humidity: an age-stage, two-sex, life-table study.AppliedEntomologyandZoology, 50: 1-8.Yeh F C, Yang R C and Boyle T, 1999.POPGENEVersion1.31,MicrosoftWindows-BasedFreeWareforPopulationGeneticAnalysis. Edmonton, AB, Canada: University of Alberta.Yu H and Li Q I, 2007. EST-SSR markers from the Pacific oyster,Crassostreagigas.MolecularEcologyNotes, 7(5): 860-862.Zhang P, Liu F, Mu W, Wang Q H and Li H, 2015. Comparison ofBradysiaodoriphagaYang and Zhang reared on artificial diet and different host plants based on an age-stage, two-sex life table.Phytoparasitica, 43(1): 107-120.
2016-04-07 接受日期(Accepted): 2016-09-20
农业行业公益性专项(201303027); 山东省泰山学者建设工程专项
陶云荔, 女, 硕士研究生。 研究方向: 农业昆虫与害虫防治。 E-mail: tucson2011@sina.com
中国重要农业害虫韭菜迟眼蕈蚊多态性EST-SSR标记的开发
陶云荔1, 张存环1, 台术蕾1, 王 磊1, 郭雅男1, 杨峰山2, 赵传志3, 万方浩1,4, 褚 栋1*
1青岛农业大学农学与植物保护学院,山东省植物病虫害综合防控重点实验室,山东 青岛 266109;2黑龙江大学生命科学学院,黑龙江 哈尔滨 150080;3山东省农业科学院生物技术研究中心,山东济南 250100;4中国农业科学院植物保护研究所,植物病虫害生物学国家重点实验室,北京 100081
【背景】韭菜迟眼蕈蚊是我国重要的农业害虫,然而它的遗传资源有限。本研究旨在开发韭菜迟眼蕈蚊EST-SSR标记,为研究不同地区的韭菜迟眼蕈蚊种群结构和遗传多样性奠定基础。【方法】从韭菜迟眼蕈蚊的表达序列标签(EST序列)中设计16对简单重复序列(SSR)引物,进一步筛选出9对具有多态性的SSR引物。【结果】从42095条unigene中确定了3383个SSR位点。利用查找到的SSR位点共设计出16对引物,进一步检测筛选发现9对引物具有多态性,引物的每个位点平均有3.33个等位基因。利用9对引物对30头韭菜迟眼蕈蚊进行检测,共获得30个等位基因,观测杂合度和期望杂合度的范围分别为0.0000~0.6875和0.0370~0.6877;其中,9个位点中有5个位点显著偏离Hardy-Weinberg平衡。【结论与意义】本研究成功从迟眼蕈蚊EST序列中筛选出9个具有多态性的微卫星位点,这为进一步分析该害虫种群的遗传结构和遗传多样性奠定了基础。
韭菜迟眼蕈蚊; 表达序列标签; 简单重复序列; 遗传多样性; 遗传结构
*通讯作者(Author for correspondence), E-mail: chinachudong@qau.edu.cn
10. 3969/j.issn.2095-1787.2016.04.004
