翘嘴红鲌热休克蛋白60的基因克隆及其表达特征分析
2016-10-09杨雨虹
戴 东,杨雨虹
(东北农业大学动物科技学院,黑龙江哈尔滨 150030)
翘嘴红鲌热休克蛋白60的基因克隆及其表达特征分析
戴东,杨雨虹
(东北农业大学动物科技学院,黑龙江哈尔滨150030)
利用同源克隆和RACE技术克隆了翘嘴红鲌热休克蛋白HSP60 cDNA全长序列,并对HSP60基因的序列特征进行了分析。EiHSP60 cDNA序列长2 323 bp,开放阅读框为1 728 bp,编码575个氨基酸,其编码蛋白相对分子量为61.26 kD,等电点为5.27。BLAST分析结果表明EiHSP60与哺乳动物、鸟和鱼类等物种的CCT具有较高的同源性。生物信息学分析结果显示,EiHSP60蛋白序列中包含1个典型的分子伴侣蛋白60信号基序。实时荧光定量PCR检测结果显示,EiHSP60在翘嘴红鲌各个组织中均有表达,但在肝脏中的表达量为最高。使用脂多糖对翘嘴红鲌进行攻毒后,在不同时间段取其肝脏总RNA,利用实时荧光定量PCR技术进行EiHSP60 mRNA的表达水平分析。分析结果显示,经过脂多糖攻毒后翘嘴红鲌HSP60呈现时间依赖型的表达模式,且在肝脏中的表达水平呈现上调趋势。上述结果表明,EiHSP60为一种涉及免疫反应的诱导型蛋白,而高表达量的EiHSP60可能在提高翘嘴红鲌免疫反应过程中发挥重要作用。
翘嘴红鲌;热休克蛋白60;基因克隆;特征分析
Heat shock protein 60(HSP60)is a ubiquitous and highly conserved stress protein from bacteria to mam mals,including fish[1-3].It has been demonstrated to play crucial roles in protein folding,protein degradation,cellular response and the innate immune function[4-5].Recent reports suggest that HSP60 plays an important role in the development of the non-specific immune responses to bacterial and viral infections in aquatic animals[5-7]. However,previous study found that the expression of HSP60 remained unchanged throughout the disease process. And the expressional profiles of HSP60 are not only dependent on the induction,but also organ specific in Chinook salmon(Oncorhynchus tshawytscha)[6].Thus the research of HSP60 gene information,gene expression and regulation will give us a better understanding of HSP60s'complicated functions.
Erythroculter ilishaeformis is one of the important farmed fish species in China due to its high market value and high market demand[8].Despite its favorable growth traits,farmed topmouth culter are rather susceptible to various stresses,such as thermal stress.Outbreaks of disease associated with kinds of bacteria have caused high fish mortality,resulting in reduced production and considerable economic losses.Therefore,it would be useful to investigate relative immune responses induced by the components of bacteria,such as lipopolysaccharide(LPS),which could help us understand anti-infection regulatory effects.Considering that the anti-infection response at transcriptional level is an important part of innate immune functions,the aim of the present study was to clone and characterize the full-length cDNA sequence of HSP60 gene from topmouth culter(EiHSP60). In addition,evaluating the expression levels in the liver of topmouth culter is used to explore the immune responses to LPS challenge.
1 Materials and methods
1.1Animal and experiment designs
Two hundred topmouth culter(average weight:50±3.5 g)were obtained from a farm in October 2015 at Harbin Fisheries Co.,Heilongjiang,China.And topmouth culter were acclimated to laboratory conditions(pH 7.1±0.3 at 26±0.5°C)for 2 weeks.During the acclimation,topmouth culter were fed with an artificial feed made by our own lab and daily ration was equal to 3%w/w of the body weight.After feeding,feces and uneaten feed were removed to maintain water quality.After the acclimation,four topmouth culter were used for EiHSP60 gene isolation and tissue-specificity expression detection.After being anesthetized with ice,blood,gonad,liver,kidney,gill,muscle and intestine of topmouth culter were collected and immediately frozen in liquid nitrogen and stored at-80°C for RNA isolation and subsequent analyses.
1.2Immune challenge
To determine the immune responses of EiHSP60,LPS(Sigma,USA)was used as immune-stimulant in time course experiments.For LPS challenge,one hundred Top mouth culter were intraperitoneally injected with LPS (10滋g/mL)suspended in phosphate buffered saline(PBS;100滋L/animal).Sixty healthy Black carp were injected with the same amount of PBS and then kept separately as a control group.The liver samples(n=4 for each time frame)were excised at 0,1.5,3,6,12,24 and 48 h from the challenge and the control group,respectively.And these samples were also immediately frozen in liquid nitrogen and stored at-80°C for RNAisolation and subsequent analyses.
1.3RNA extraction,cDNA synthesis and cloning of EiHSP60 gene
Total RNA was extracted from tissues using Trizol Reagent(Invitrogen,USA),spectrophotometrically quantified,and electrophoresed on a 1%denaturing agarose gel to test the integrity.For each reverse transcription (RT)reaction,3 μg of total RNA was firstly treated with RQI RNase-Free DNase(Takara,Japan)to remove DNA contaminant,and then subjected to cDNA synthesis by SuperScriptTMII RT reverse transcriptase(Takara,Japan)in 25 μL volume according to reagent′s instructions.Generally,an Oligo(dT)-adaptor primer(Tab.1)was used as RT primer to introduce an adaptor.The degenerate primer pairs of EiHSP60 01 and EiHSP60 02(Tab. 1)were designed based on multiple alignments by using the Clustal W 1.83 multiple sequence alignments program.The polymerase chain reaction(PCR)to get the fragment of EiHSP60 was conducted on an Eppendorf Mastercycler gradient(Eppendorf,German)to amplify EiHSP60 cDNA fragment.The PCR program was followed by 35 cycles of 94°C for 1 min,56°C for 1min,72°C for 1.5 min followed by a 10 min extension at 72°C. The PCR fragments were subjected to electrophoresis on a 1.5%agarose gel for length difference and cloned into the pMD-18T vector(Takara,Japan).After transforming into the competent cells of Escherichia coli DH5α,the recombinants were identified through blue-white color selection in ampicillin-containing LB plates and confirmed by PCR with M13-RVM and M13-47(Tab.1).Positive clones were sequenced in both directions and these resulting sequences were verified and subjected to cluster analysis in NCBI.
The 5′-end of EiHSP60 cDNA was obtained by RACE technique.One specific reverse primers,EiHSP60 03(Tab.1)were designed based on the partial sequence amplified by degenerated primers.The PCR amplification was performed using the same reaction system as described before with Abridged Anchor Primer(AAP)(Tab.1)and EiHSP60 03 by the 5′RACE system(Invitrogen,Carlsbad,CA,USA),and then a nested PCR was carried out using AUAP and EiHSP60 03.The 3′end of EiHSP60 was amplified by using sense primer EiHSP60 04 and Oligo(dT)-adaptor primer(Tab.2)with 1 μL of cDNA library mix as template,and then a nested PCR was carried out using NUP and EiHSP60 04.The full-length of the sequence was verified by sequencing the fragments.And all the PCR products were sequenced following the procedures described above.

Tab.1 Sequences of the primers used in this work
1.4Sequence analysis and phylogenetic analysis
The cDNA sequence of EiHSP60 was analyzed for similarity with other known sequences using theBLAST program at web servers of NCBI(http://www.ncbi.nlm.nih.gov/BLAST/).Alignment of multiple sequences was performed using the CLUSTALW program at the European Bioinformatics Institute(http://www.ebi.ac.uk/clustalw/)and Multiple Alignment show(http://www.bio-soft.net/sms/index.html).SMART program (http://smart.embl-heidelberg.de/)and PROSITE program(http://kr.expasy.org/prosite/)were used to predict the functional sites or domains in the amino acid sequence.Phylogenetic and molecular evolutionary analyses were conducted according to the amino acid sequences of the selected HSP60s by programs of CLUSTAL X1.83 and MEGA 4.0[9].An unrooted phylogenetic tree among these species was determined using the neighbour-joining distance method with the Kirumma two parameter.The relative importance of branching order was evaluated by the bootstrapping method(1 000 replications).
1.5Real-time PCR analysis of EiHSP60 mRNA expression
First strand cDNA synthesized in Section 2.2 was diluted by 10 times.For each PCR reaction,1μL of this dilution was added as template in a final volume of 20 μl.Reactions were carried out on a Rotor Gene 3 000 (Corbett Life Science Australia)using SYBR green I as fluorescent dye.Gene-specific primers EiHSP60 05 F/R (Tab.2)were applied to evaluate the mRNA level of EiHSP60 in topmouth culter.And a housekeeping gene,β-actin gene(Tab.2)was used as an endogenous reference to normalize the template amount.The real-time PCR temperature profile for EiHSP60 was 95°C for 2 min followed by 35 cycles of 5 s at 95°C,15 s at 59° C,20 s at 72°C.Fluorescent data were acquired during each annealing phase.During the detection,each sample was run in duplicate,and PCR-grade water which replaced the template was the negative control.The primer amplification efficiency was optimized for each pair of primers,resulting in:1.038 for EiHSP60 and 1.025 for β-actin.The absolute ΔCTvalue(EiHSP60 CT–β-actin CT)of the slope is 0.013 and close to zero,which indicated the ΔΔCTcalculation for the relative quantification of target genes might be used.Results were presented as folds of changes relative to the calibrator according to Livak and Schmittgen[10].
1.6Statistical analysis
All data were given in terms of relative mRNA expressed as mean±S.E.(N=4).And data of fold change of expression were logarithmic transformed before subjected to one-way analysis of variance(one-way ANOVA)using SPSS 17.0.When overall differences were significant,Tukey's test was conducted to compare the means between individual treatments.For statistically significant differences,P<0.05 was required.
2 Results
2.1cDNA cloning,sequencing and characterization of EiHSP60 gene
The PCR product amplified by the degenerated primers was 1 629 bp,and its nucleotide sequence was homogeneous to other known HSP60s.EiHSP60-specific primers were designed based on the above sequence,and used for the full-length cDNA cloning.By RACE and nested PCR approaches,two fragments corresponding to the 5′and 3′end of the EiHSP60 cDNA were amplified.The complete cDNA sequence of EiHSP60 was obtained by overlapping these fragments mentioned above.The complete sequence of EiHSP60 mRNA and the deduced amino acids were shown in Fig.1.The full-length cDNA of EiHSP60 was of 2 323 bp,including a 5′-terminal untranslated region(UTR)of 119 bp,a 3′-terminal UTR of 476 bp with a poly(A)tail,and an open reading frame(ORF)of 1 728 bp encoding a polypeptide of 575 amino acids with predicted molecular mass of 61.26 kDa and theoretical isoelectric point of 5.27.There was one Chaperonins cpn60 signature(430AAVEEGIVPGGG441)and a Cell attachment sequence(361RGD363)in the amino acid sequence of EiHSP60 according to PROSITE program,respectively.In addition,there are three putative N-glycosylation sites(103NNTN106,230NTTK233,426NATR429)in EiHSP60 according to the PROSITE program,which indicates that EiHSP60 is probably a glycoprotein.

Fig.1 Nucleotide and deduced amino acid sequences of EiHSP60.The nucleotides and amino acids are numbered along the left margin,respectively.The start(ATG)and stop(TAA)codons are marked in box.Shaded regions indicate the five HSP60 family signature sequences.The glycosylation sites are underlined.The Cell attachment sequence(RGD)is italic and double underlined. The Chaperonins cpn60 signature(AAVEEGIVPGGG)is in shade boxed.

Fig.2 Phylogenetic relationship between the amino acid sequences of EiHSP60 and 20 available HSP60s.The amino acid sequences were derived from the GenBank under the following accession numbers(in parentheses):Grass carp(Ctenopharyngodon idella,ADU34083);Wuchang bream(Megalobrama amblycephala,AGK38254);white cloud mountain minnow(Tanichthys albonubes,ADK27679);Common carp(Cyprinus carpio,KTG05155);Goldfish (Carassius auratus,DQ872653);Zebrafish(Danio rerio,AAH68415);Loach (Paramisgurnus dabryanus,AHN60091);Asian bonytongue(Scleropages formosus,KPP75440);Walking catfish(Clarias batrachus,AMA22413);Gilthead seabream(Sparus aurata,AGU38790);Orange spotted grouper(Epinephelus coioides,AIS72878);Northern snakehead (Channa argus,AMQ24770);Snakehead murrel(Channastriata,CCQ48609);HongKonggrouper (Epinephelusakaara,ADM73510);Salmon (Salmosalar,ACN11370);Japanese flounder(Paralichthys olivaceus,ABB76381);Large yellow croaker (Larimichthys crocea,KKF19941);Chicken(Gallus gallus,NP_001012934);Rat(Rattus norvegicus,CAA38564);Mouse(Mus musculus,NP_034607).Sequences were aligned using the CLUSTAL W algorithm and were analyzed by phylogenetic analysis using the neighbour-joining distance method.
2.2Homology analysis of EiHSP60
The deduced amino acid sequence of EiHSP60wasclosematchedtoother HSP60s in vertebrates and mammals.It displayed high similarity to HSP60s of Ctenopharyngodon idella(99.65%),Megalobrama amblycephala(99.13%),Tanichthys albonubes(96.87%),Carassius auratus (94.27%),Danio rerio(96.52%),Cyprinus carpio(94.85%),Paramisgurnus dabryanus (94.97%),Rattus norvegicus(87.57%),Mus musculus(88.47%),Gallus gallus (87.46%)and so on.And multiple sequence alignment of EiHSP60 with other known HSP60 proteins revealed that they were highly conserved,especially in the regions of HSP60 family signatures.Based on the sequences of HSP60s,a phylogenetic tree was constructed using the programs of CLUSTAL X 1.83 and MEGA 4.0(Fig.2). HSP60s were separated and formed three distinct branches in the tree.HSP60s from all the fish were clustered together.Thebird and mammals were clustered together and formed a sister group to the branch of fish.The relationships displayed in the phylogenic tree were in a good agreement with traditional taxonomy.

Fig.3 Tissue specific expression levels of EiHSP60 mRNA in topmouth culter as detected by real time-PCR.EiHSP60 transcript levels in gill(G),kidney (kid),Adipose tissue(Ad),Brain(Br),Blood(Bl),liver (L),Muscle(M),Heart(H),Intestine(I),Skin(Sk)and Spleen(Sp).All values represent the mean±S.E.M. (n=4 replicates).
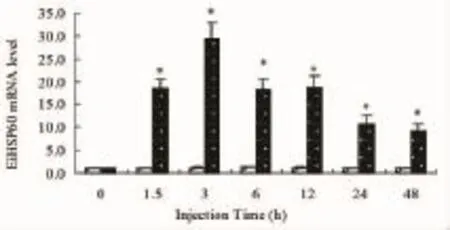
Fig.4 Relative EiHSP60 mRNA levels in liver of topmouth culter after LPS challenge.EiHSP60 mRNA levels were evaluated by real-time quantitative PCR. All values represent the mean±S.E.M.(n=4 replicates).Bars bearing different letters are significantly different(P<0.05;Tukey's test)
2.3Expression of EiHSP60 gene in different tissues
The tissue specific mRNA expression was determined by conducting real-time PCR assay with EiHSP60 gene specific primers(EiHSP60 05 F/R,Tab.2),and β-actin gene was used as an internal control.Constitutive EiHSP60 mRNA expression was detected in gill,kidney,adipose tissues,brain,blood,liver,muscle,heart,intestine,skin and spleen from healthy topmouth culter without any treatment.Relative EiHSP60 expression levels were calculated based on the lowest expression observed in gill,which is shown as 1 in Fig.3.The relative EiHSP60 mRNA expression levels were significantly higher in liver,muscle and heart than that in gill,kidney and spleen (P<0.01),which showed tissue specific variation of EiHSP60. And the highest relative EiHSP60 mRNA expression was detected in topmouth culter liver tissue by real-time PCR assays (P<0.01).
2.4Quantitative analysis of EiHSP60 gene expression in response to LPS challenge
The relative expression levels of the EiHSP60 in liver of topmouth culter challenged with LPS are presented in Fig.4.In liver,the expression levels of EiHSP60 transcript were upregulated and reached the maximum(28.48-fold higher than deficient groups)at 3h in topmouth culter challenged with LPS (P<0.05).And then EiHSP60 transcriptional levels all decreased at 6h,12h,24h and 48h in topmouth culter compared with these groups challenged with LPS at 3h (48.70 mg/kg)(P<0.01).
3 Discussion
In the present study,the complete cDNA sequence and characterization of HSP60 gene from topmouth culter were reported.Conserved sequences and characteristic motifs were found in the deduced EiHSP60 amino acid sequence,such as ATP/ADP and Mg2+binding domains(76DGVTVAK82and109AGDGTTTATVL119)(Fig.1),which is the major structural and functional domains typically in HSP60[7,11,12].The cpn60 signature(430AAVEEGIVPGGG441)sequence in EiHSP60 was strictly conserved and shared with the other members of HSP60 family.
Searching for sequence similarities revealed that the deduced amino acid sequence of EiHSP60 shared high similarity with other known HSP60s(more than 80%similarity in all the matches),especially with those from fish species including Grass carp,Wuchang bream,white cloud mountain minnow,Goldfish,Zebrafish,Common carp and Loach(Fig.2).Based on the sequence alignment,structure comparison and phylogenetic analysis,EiHSP60 was concluded to be a member of HSP60 family[7].And the result of phylogenetic tree indicates that EiHSP60 and other known HSP60s are evolved from a same ancestor.The real-time RT-PCR analysis showed that EiHSP60 was ubiquitously expressed in the examined tissues of topmouth culter.The relativeEiHSP60 mRNA expression levels were significantly higher in liver,muscle and heart as compared with other tested tissues(gill,kidney and spleen)(Fig.3).This constitutive expression of HSP60 has also been reported in Chinook salmon[6],grass carp[7],broilers[13]and shrimp[14].The liver is an important tissue for immune defenses and is a key component of the innate immune system[15],which suggested that high expression of EiHSP60 could play an important role in the normal grass carp liver cell.
After challenged with LPS,the temporal expression profiles of EiHSP60 mRNA in liver were significantly upregulated at 1.5 h post injection(P<0.05),reached maximum level at 3h post injection,and then decreased at 6 h post injection(P<0.05)(Fig.4).Besides serving as a chaperone,HSP60 could activate macrophages as a signal and inhibit the pro-inflammatory activities,which could prompt the induction and release of more HSPs[16]. And LPS could serve as pro-inflammatory activator in animal[17].Thus higher expression levels of EiHSP60 might be used to inhibit the pro-inflammatory activities after LPS injection in the liver of topmouth culter.In conclusion,a HSP60 gene was cloned from the topmouth culter and constitutively expressed in various tissues,suggesting it might be involved in a multifunctional role in healthy fish.Moreover,the higher expression of EiHSP60 induced by LPS infection suggested its importance innate immune function.
References:
[1]CHENG M Y,HARTL F U,MARTIN J,et al.Mitochondrial heat-shock protein hsp60 is essential for assembly of proteins imported into yeast mitochondria[J].Nature,1989,337:585-674.
[2]MARTIN J,HORWICH A,HARTL F.Prevention of protein denaturation under heat stress by the chaperonin Hsp60[J]. Science,1992,258:995-998.
[3]LIANG P,MACRAE T H.Molecular chaperones and the cytoskeleton[J].J Cell Sci,1997,110:1431-40.
[4]BORGES J C,RAMOS C H I.Protein folding assisted by chaperones[J].Protein Pept Lett,2005,12:257-261.
[5]HUANG W J,LEU J H,TSAU M T,et al.Differential expression of LvHSP60 in shrimp in response to environmental stress[J]. Fish Shellfish Immunol,2011,30:576-82.
[6]EDER K J,LEUTENEGGER C M,KOHLER H R,et al.Effects of neurotoxic insecticides on heat-shock proteins and cytokine transcription in Chinook salmon(Oncorhynchus tshawytscha)[J].Ecotoxicol Environ Saf,2009,72:182-90.
[7]XU X Y,SHEN Y B,FU J J,et al.Molecular cloning,characterization and expression patterns of HSP60 in the grass carp (Ctenopharyngodon idella)[J].Fish Shellfish Immunol,2011,31(6):864-70.
[8]LIU B,XIE J,GE X P,et al.Effect of high dietary carbohydrate on growth,serum physiological response,and hepatic heat shock cognate protein 70 expression of the top-mouth culter Erythroculter ilishaeformis Bleeker[J].Fish Sci,2012,78(3):613-623.
[9]TAMURA K,DUDLEY J,NEI M,et al.MEGA4:Molecular Evolutionary Genetics Analysis(MEGA)software version 4.0[J].Mol Biol Evol,2007,24(8):1596-1599.
[10]LIVAK K J,SCHMITTGEN T D.Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)method[J].Methods,2001,25:402-408.
[11]FENTON W A,KASHI Y,FURTAK K,et al.Residues in chaperonin GroEL required for polypeptide binding and release[J]. Nature,1994,371:614-619.
[12]BROCCHIER L,KARLIN S.Conservation among HSP60 sequences in relation to structure,function,and evolution[J].Protein Sci,2000,9:476-486.
[13]YAN J,BAO E,YU J.Heat shock protein 60 expression in heart,liver and kidney of broilers exposed to high temperature[J]. Res Vet Sci,2009,86:533-538.
[14]ZHOU J,WANG W N,HE W Y,et al.Expression of HSP60 and HSP70 in white shrimp,Litopenaeus vannamei in response to bacterial challenge[J].J Invertebr Pathol,2010,103:170-178.
[15]LI Z,DIEHL A M.Innate immunity in the liver[J].Curr Opin Gastroenterol,2003,19:565-71.
[16]QUINTANA F J,COHEN I R.Heat shock proteins as endogenous adjuvants in sterile and septic inflammation[J].J Immunol,2005,175:2 777-2 782.
[17]LEE J K.Anti-inflammatory effects of eriodictyol in lipopolysaccharide-stimulated raw 264.7 murine macrophages[J].Arch Pharm Res,2011,34(4):671-679.
Molecular Cloning,Characterization and Expression Analysis of Heat Shock Protein 60 in Erythroculter ilishaeformis
DAI Dong,YANG Yu-hong
(College of Animal Science and Technology,Northeast Agricultural University,Harbin150030,China)
In the present study,the cDNA of heat shock protein 60 from Erythroculter ilishaeformis(EiHSP60)was cloned by the combination of homology cloning and rapid amplification of cDNA ends(RACE)approaches.The full-length cDNA of EiHSP60 was of 2 323 bp,including an open reading frame(ORF)of 1 728 bp encoding a polypeptide of 575 amino acids with predicted molecular weight of 61.26 kDa and theoretical isoelectric point of 5.27.BLAST analysis revealed that EiHSP60 shared high similarity with other known HSP60s,and one conserved Chaperonin 60 signature was also identified in EiHSP60,which indicated that EiHSP60 should be a member of the HSP60 family.The mRNA of EiHSP60 was constitutively expressed in all tested tissues of untreated topmouth culter,including brain,muscle,kidney,liver,skin,spleen,heart,gill and intestine with the highest expression level in the liver.Fluorescent real-time quantitative RT-PCR was used to examine the expression of the EiHSP60 gene in topmouth culter after the challenge with LPS.A clear time-dependent expression pattern of EiHSP60 was found after the LPS challenge,and the up-regulated mRNA expression of EiHSP60 in topmouth culter after LPS challenge indicates that the HSP60 gene is inducible and involved in immune responses.These results suggest that EiHSP60 plays an important role in immune responses in topmouth culter.
Erythroculter ilishaeformis;heat shock protein 60;gene cloning;characterization analysis
S917.4Document code:A
date:2016-01-10
1008-830X(2016)02-0105-08
2016-01-10
Fund project:The Program for Heilongjiang science and technology(GA06B203-4-3)
黑龙江省科技计划项目(GA06B203-4-3)
Biography:DAI Dong(1992-),Male,Shandong Zaozhuang,Research Direction:Fish Immunology.E-mail:2444362737@qq.com作者简介:戴东(1992-),男,山东枣庄人,研究方向:鱼类免疫学.E-mail:2444362737@qq.com
Author for correspondence:YANG Yu-hong,Professor.E-mail:yangyuhong_68@yahoo.com.cn
杨雨虹,教授.E-mail:yangyuhong_68@yahoo.com.cn
