随机生存森林在小细胞肺癌预后分析中的应用
2016-08-25解瑞飞杭州市肿瘤医院信息科浙江杭州30002浙江省台州市中心医院放疗科浙江台州38000
解瑞飞 吴 波.杭州市肿瘤医院信息科,浙江杭州 30002;2.浙江省台州市中心医院放疗科,浙江台州 38000
随机生存森林在小细胞肺癌预后分析中的应用
解瑞飞1吴波2▲
1.杭州市肿瘤医院信息科,浙江杭州310002;2.浙江省台州市中心医院放疗科,浙江台州318000
目的 辨识与小细胞肺癌具有本质关联的基因变量,可以帮助临床医生制定个性化治疗方案,延长患者生存期,提高患者预后生活质量。方法 共入组 117例小细胞肺癌患者,含41000个基因变量,8个一般特征。利用随机生存森林方法结合基因表达谱及预后数据从一系列基因变量中探索与小细胞肺癌具有密切相关的基因变量。结果 一般特征及EGFR、K-ras、p53表达在预后上无明显差异;所挑选的前12个基因中,FTCD、BTC、PSMC4、SLC43A1与小细胞肺癌具有密切的关系,而UCHL5、PSMC4与PSMD7、PCSK4、VPS13D与VPS13A具有调控依赖关系。结论 随机生存森林可以高效的辨识与预后具有密切相关的本质基因。
小细胞肺癌;随机生存森林;基因表达谱;生存分析;基因调控
[Abstract]Objective To distinguish the genetic variables with essential relevance with small cell lung cancer,which is able to help clinical physicians to formulate customized therapeutic protocols,prolong patients'survival time,and improve patients'prognosis and life quality.Methods A total of 117 patients with small cell lung cancer were included,with 41000 genetic variables and 8 general characteristics.Random survival forests were applied,combined with gene expression profile and prognostic data,genetic variables closely related to small cell lung cancer were explored in a series of genetic variables.Results General characteristics and EGFR,K-ras and p53 expressions were not significantly different in prognosis;in the former 12 selected genes,FTCD,BTC,PSMC4,SLC43A1 were closely related to small cell lung cancer,but UCHL5,PSMC4 and PSMD7,PCSK4,VPS13D and VPS13A were in the dependent relation of regulation.Conclusion Random survival forests are able to effectively distinguish the essential genes closely related to the prognosis.
[Key words]Small cell lung cancer;Random survival forests;Gene expression profile;Survival analysis;Gene regulation
在全球范围,肺癌是最常见的恶性肿瘤之一,且死亡率较高,预后较差[1-3]。小细胞肺癌(small cell lung cancer,SCLC)较非小细胞肺癌 (non-small cell lung cancer,NSCLC)预后更差。在我国,超过80%的小细胞肺癌5年存活率不超过10%[4,5]。因此,寻找与SCLC发生发展相关的基因和分子,对于肿瘤的诊断和治疗尤为重要[2,6,7]。
近年来,转化医学的研究逐渐被重视,越来越多的研究者致力于基因组学的研究。高维基因组数据和生存信息的结合可以帮助研究者从全新的角度认识个体生物学过程以及疾病的发生、发展及预后过程。随机生存森林(random survival forest,RSF)[8-13]可以在高维基因组数据中有效地结合生存信息,提取与预后相关的基因变量,指导临床医生对患者进行个性化治疗[14]。
1 资料与方法
1.1临床资料
本文数据从117例小细胞肺癌患者中提取,共包含41000个基因,一般特征见表1。EGFR与性别、K-ras与性别及T分期具有较强的相关性。
1.2随机生存森林
随机生存森林是在随机森林(Random Forest)基础上,加入生存分析,采用bootstrap方法从原始数据中有放回的随机抽取N个样本,建立生存树模型,而袋外37%样本测试生存树模型。
假设在树节点h上有n(h)例样本,(T1,δ1),…,(Tn,δn)表示他们的生存时间和截尾信息,δ=0表示个体i在时间Ti时右截位,δ=1表示在时间Ti时死亡,则给定的一个变量Xj(j=1,2,…,m),在节点h处可以根据Xj≤c和Xj>c将生存数据分为两组数据。RSF在每棵树的节点处,随机选择M个变量作为分割节点的候选变量,选择使子节点生存差异最大的分支。树节点分裂准则采用Log-Rank分裂方法,计算生存函数采用Kaplan-Meier估计方法。为了选择极少最重要的基因变量,可以依据变量的重要性(VIMP)对变量进行筛选,VIMP值越大表明其预测能力越强。具体流程如下:第一步:清除缺失数据;第二步:对所有基因,使用Cox模型;第三步:选择P<0.005的基因变量;第四步:利用一般临床特征及最终选择的基因变量使用RSF,并根据其重要性对各变量进行排名。

表1 一般特征
2 结果
对于临床一般特征及EGFR、K-ras、p53突变,利用Kaplan-Meier和Log-rank进行数据分析,见封三图1。从封三图1可以看出,性别、年龄、EGFR、K-ras、p53在小细胞肺癌腺癌患者中,预后无明显差异。而在T、N和临床分期中,只有T1vs T4、N0vs N2、临床Ⅰ期 vs临床Ⅲ期所对应的P均<0.001,提示差异存在统计学意义。
从封三图2和表2可以看出,在建立模型过程中,随着生存树个数的增多,错误率趋于稳定。对于不同的根据对预后的影响进行排序,前12个变量分别为:FTCD、UCHL5、RANBP9、YWHAQ、LOC151878、PPP2R5C、C20orf96、NFKBIB、BTC、SUMO3、PSMC4、 C6orf64。通过 Genecard数据库分析,FTCD、BTC、PSMC4、SLC43A1与肿瘤具有密切的关系,而UCHL5、PSMC4与PSMD7,PCSK4、VPS13D与VPS13A具有调控依赖关系,如表3所示。与PCSK4、PSMC具有相关调控的基因关系如封三图3所示,各基因相互影响,相互控制,共同影响肿瘤的生成及演化过程。
为了进一步验证所获得的敏感基因和临床特征是否影响预后,对其采用Cox regression进行单因素和多因素分析,结果如表4所示。在单因素分析中,只有T、N、FTCD、UCHL5、BTC、PSMC4、PCSK4、SLC43A1具有统计学意义,对其进行多因素分析后,T、N、FTCD、UCHL5及PSMC4的P值小于0.05,具有统计学意义,即共同影响患者预后。
RSF方法利用基因表达谱,结合预后数据,可以有效地筛选出与肺癌具有密切关系的基因,指导临床医生制定个性化治疗方式,提高患者生活治疗,延长患者生存期。
3 讨论
随机生存森林有效地结合机器学习及临床生存数据,可以快速有效地识别与预后密切关系的本质基因。由于随机森林在挑选特征过程中考虑多个基因的联合作用,所挑选出的基因组具有较强的相关性或具有相互调控关系,为后期分析基因之间调控关系、建立基因调控网络奠定基础。
在众多的研究中,EGFR[27]、K-ras[28]、p53[29]位点是否发生突变影响着非小细胞肺癌的治疗手段和方法,如EGFR突变时,EGFR酪氨酸激酶抑制剂(EGFR Tyrosine Kinase Inhibitors,EGFR-TKIs)吉非替尼和厄罗替尼可以显著提高非小细胞肺癌患者的生存获益,已被FDA批准用于治疗晚期非小细胞肺癌(NSCLC)。然而,利用随机生存森林验证发现,EGFR、K-ras、p53并未进入影响小细胞肺癌预后的敏感基因中,而通过Kaplan-Meier及单因素也再次验证此3个突变位点并未影响患者的预后。因此,根据EGFR、K-ras、p53决定SCLC患者相应治疗方法的可能意义不大。
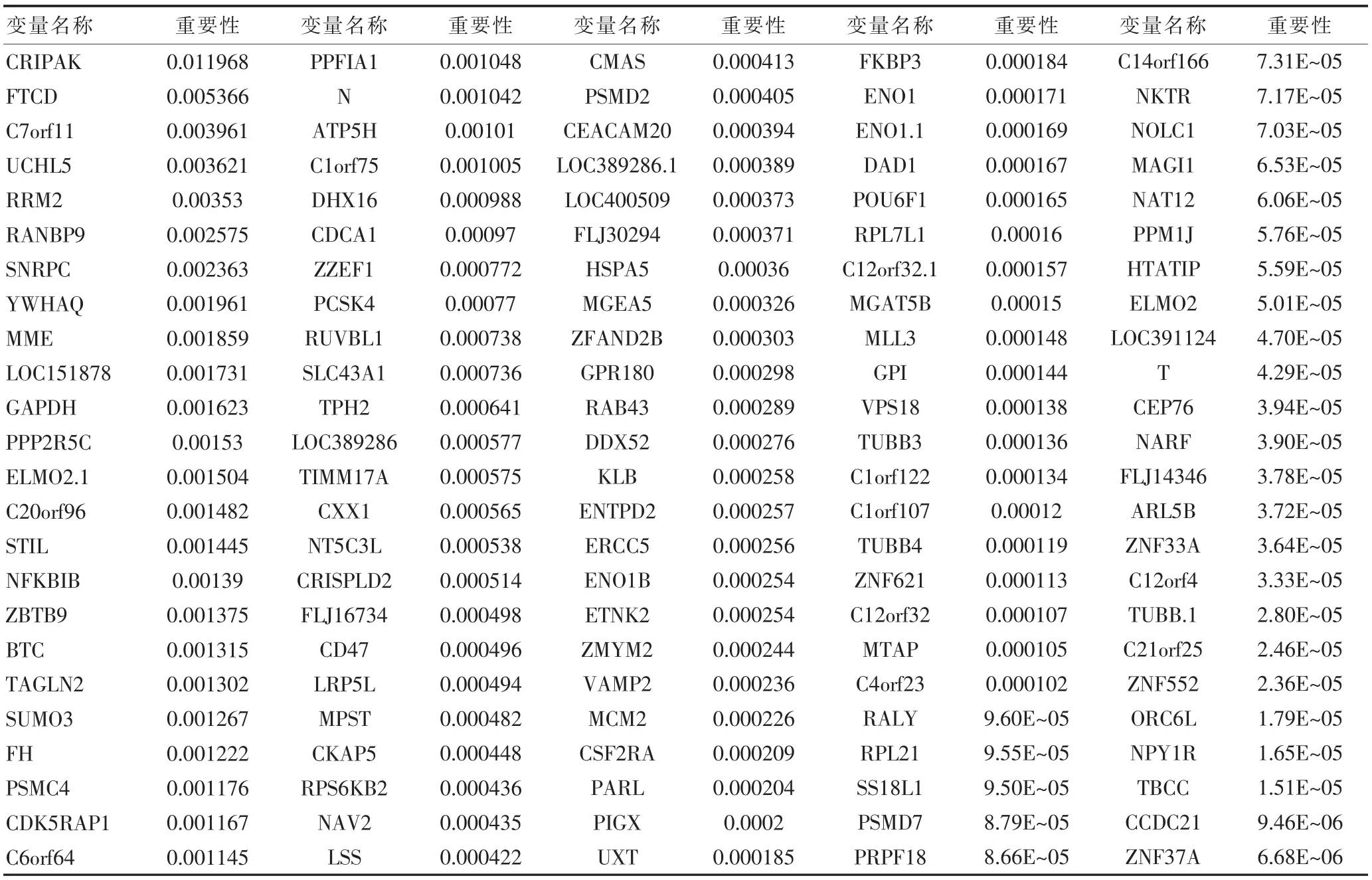
表2 变量重要性排名

表3 敏感基因的生物信息学
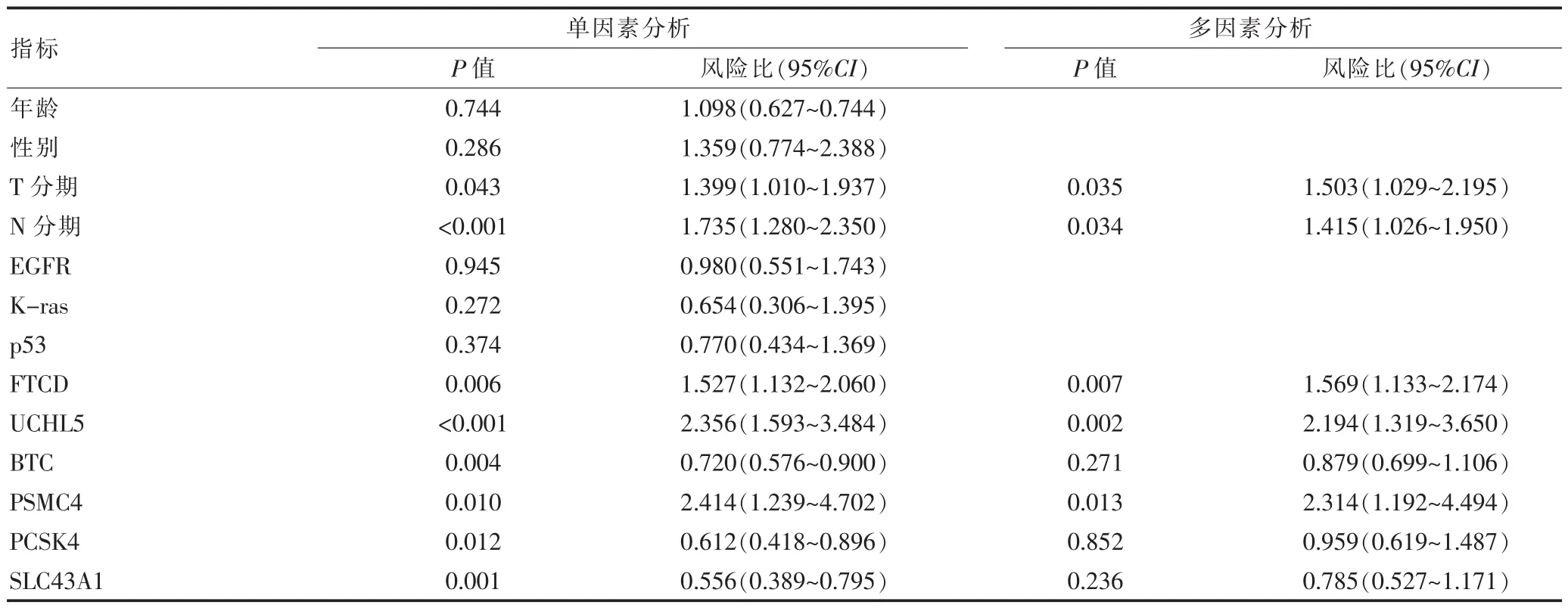
表4 临床特征及敏感基因单因素和多因素分析
对于进入随机生存森林的其他预后敏感基因,通过Cox回归模型进行单因素及多因素分析发现,T分期、N分期、FTCD、UCHL5、PSMC4具有统计相关性,并且与肿瘤预后具有较强的相关性。T分期为主要是通过肿瘤体积大小进行划分,N分期通过淋巴结转移位置及范围进行划分,肿瘤体积越小、淋巴结转移范围越小(即T、N分期越低),患者预后越好。而FTCD、UCHL5、PSMC4在发生突变的位点,预后风险比分别为1.569、2.194、2.314,与SCLC患者具有较显著的关联。有文献已经证实FTCD的敲除可以减少HIF-1α在低氧环境中的效果,加强HepG2在细胞中的化疗敏感性且FTCD和HIF之间的存在相互调控关系;同时,已经证实FTCD可作为一个靶基因用于治疗肝癌患者[30]。Randles等[31]已经证实UCHL5基因影响着细胞周期,UCHL5的蛋白缺失将导致细胞周期停止在G0/G1阶段,无法正常进行。PSMC4是ATP亚基酶的一种,亚基酶已经被证实与核激素受体的超高表达具有相互影响,在肝脏或肝脏蛋白中,并已经确定具有两个转录变异体[32]。对于临床医生,可以利用FTCD、UCHL5、PSMC4构建预后预测模型,可以针对发生FTCD、UCHL5、PSMC4发生基因突变的患者使用特定的靶向药物,抑制肿瘤的发展及恶化,提高患者预后。
随机生存森林可以快速、高效的辨识与预后具有较强相关性的基因,进一步促进SCLC患者的精准医疗,精确的寻找到SCLC的原因和治疗的靶点,并对不同状态和过程进行精确分类,最终实现对SCLC患者进行个性化精准治疗的目的,提高疾病诊治与预防的效益。
[1]Menachery A,Burt J,Chappell S,et al.Dielectrophoretic characterization and separation of metastatic variants of small cell lung cancer cells[J].Une,2016,(3):386-389.
[2]Mitsudomi T.Molecular epidemiology of lung cancer and geographic variations with special reference to EGFR mutations[J].Transl Lung Cancer Res,2014,3(4):205-211.
[3]Jung KW,Won YJ,Kong HJ,et al.Cancer statistics in Korea:Incidence,mortality,survival,and prevalence in 2012[J]. Cancer Res Treat,2015,47(2):127-141.
[4]Chen W,Zheng R,Zeng H,et al.Epidemiology of lung cancer in China[J].Thorac Cancer,2015,6(2):209-215.
[5]Zhou C.Lung cancer molecular epidemiology in China:Recent trends[J].Transl Lung Cancer Res,2014,3(5):270-279.
[6]Chen W,Zheng R,Zeng H,et al.Geographic distribution and epidemiology of lung cancer during 2011 in Zhejiang province of China[J].Asian Pac J Cancer Prev,2014,15(13):5299-5303.
[7]Blakely CM,Pazarentzos E,Olivas V,et al.NF-kappaB-activating complex engaged in response to EGFR oncogene inhibition drives tumor cell survival and residual disease in lung cancer[J].Cell Rep,2015,11(1):98-110.
[8]Miao F,Cai YP,Zhang YX,et al.Risk prediction of oneyear mortality in patients with cardiac arrhythmias using random survival forest[J].Comput Math Methods Med,2015,2015:303250.
[9]Marino SR,Lin S,Maiers,et al.Identification by random forest method of HLA class I amino acid substitutions associated with lower survival at day 100 in unrelated donor hematopoietic cell transplantation[J].Bone Marrow Transplant,2012,47(2):217-226.
[10]Buhnemann C,Li S,Yu H,et al.Quantification of the heterogeneity of prognostic cellular biomarkers in ewing sarcoma using automated image and random survival forest analysis[J].PLoS One,2014,9(9):e107105.
[11]Choi JY,Kim SK,Lee WH,et al.A survival prediction model of rats in hemorrhagic shock using the random forest classifier[J].Conf Proc IEEE Eng Med Biol Soc,2012,2012:5570-5573.
[12]Shim JH,Jun MJ,Han S,et al.Prognostic nomograms for prediction of recurrence and survival after curative liver resection for hepatocellular carcinoma[J].Ann Surg,2015,261(5):939-946.
[13]Biesbroek S,vander ADl,Brosens MC,et al.Identifying cardiovascular risk factor-related dietary patterns with reduced rank regression and random forest in the EPICNL cohort[J].Am J Clin Nutr,2015,102(1):146-154.
[14]Kasinski AL,Kelnar K,Stahlhut C,et al.A combinatorial microRNA therapeutics approach to suppressing nonsmall cell lung cancer[J].Oncogene,2015,34(27):3547-3555.
[15]Seimiya Masanori,Tomonaga Takeshi,Matsushita Kazuyuki,et al.Identification of novel immunohistochemical tumor markers for primary hepatocellular carcinoma;clathrinheavychainandformiminotransferasecyclodeaminase[J].Hepatology,2008,48(2):519-530.
[16]Kawaguchi M,Hosotani R,Kogire,et al.Auto-induction and growth stimulatory effect of betacellulin in human pancreatic cancer cells[J].Int J Oncol,2000,16(1):37-41.
[17]Yamamoto T,Akisue T,Marui T,et al.Expression of betacellulin,heparin-binding epidermal growth factor and epiregulin in human malignant fibrous histiocytoma[J]. Anticancer Res,2004,24(3b):2007-2010.
[18]Moon WS,Park HS,Yu KH,et al.Expression of betacel-lulin and epidermal growth factor receptor in hepatocel lular carcinoma:Implications for angiogenesis[J].Hum Pathol,2006,37(10):1324-1332.
[19]Watanabe T,Shintani A,Nakata M,et al.Recombinant human betacellulin:Molecular structure,biological activities,and receptor interaction[J].J Biol Chem,1994,269 (13):9966-9973.
[20]Ocharoenrat P,Modjtahedi H,Rhys-Evans P,et al.Epidermal growth factor-like ligands differentially up-regulate matrix metalloproteinase 9 in head and neck squamous carcinoma cells[J].Cancer Res,2000,60(4):1121-1128.
[21]Sakon M,Kishimoto S,Aoki T,et al.A patient with HCC successfully treated by ethanol injection therapy with etoposide[J].Gan To Kagaku Ryoho,1996,23(11):1585-1587.
[22]Lu Z,Hu X,Li Y,et al.Human papillomavirus 16 E6 oncoprotein interferences with insulin signaling pathway by binding to tuberin[J].J Biol Chem,2004,279(34):35664-35670.
[23]Szabo A,Perou CM,Karaca M,et al.Statistical modeling for selecting housekeeper genes[J].Genome Biol,2004,5 (8):R59.
[24]Kokkinakis DM,Liu X,Chada S,et al.Modulation of gene expression in human central nervous system tumors under methionine deprivation-induced stress[J].Cancer Res,2004,64(20):7513-7525.
[25]Bassi DE,Mahloogi H,Klein-Szanto AJ.The proprotein convertases furin and PACE4 play a significant role in tumor progression[J].Mol Carcinog,2000,28(2):63-69.
[26]Cole KA,Chuaqui RF,Katz K,et al.cDNA sequencing and analysis of POV1(PB39):A novel gene up-regulated in prostate cancer[J].Genomics,1998,51(2):282-287.
[27]Paez J Guillermo,Jänne Pasi A,Lee Jeffrey C,et al.EGFR mutations in lung cancer:Correlation with clinical response to gefitinib therapy[J].Science,2014,304(5676):1497-1500.
[28]Johnson Leisa,Mercer Kim,Greenbaum Doron,et al.Somatic activation of the K-ras oncogene causes early onset lung cancer in mice[J].Nature,2001,410(6832):1111-1116.
[29]Denissenko Mikhail F,Pao Annie,Tang Moon-shong,et al. Preferential formation of benzo[a]pyrene adducts at lung cancer mutational hotspots in P53[J].Science,1996,274 (5286):430-432.
[30]Yu Zhenhai,Ge Yingying,Xie Lei,et al.Using a yeast two-hybrid system to identify FTCD as a new regulator for HIF-1α in HepG2 cells[J].Cellular signalling,2014,7(26):1560-1566.
[31]Randles L,Anchoori RK,Roden RB,et al.Proteasome Ubiquitin Receptor hRpn13 and its Interacting Deubiquitinating Enzyme Uch37 are Required for Proper Cell Cycle Progression[J].J Biol Chem,2016,M115:694588.
[32]Choi HS,Seol W,Moore DD.A component of the 26S proteasome binds on orphan member of the nuclear hormone receptor superfamily[J].J Steroid Biochem Mol Biol,1996,56(6):23-30.
Application of random survival forests in the analysis of small cell lung cancer prognosis
XIE Ruifei1WU Bo2
1.Department of Information,Hangzhou Tumor Hospital,Hangzhou310002,China;2.Department of Radiology,Taizhou Central Hospital in Zhejiang Province,Taizhou318000,China
R734
A
1673-9701(2016)17-0004-05
浙江省科技厅公益技术研究社会发展项目(2015C33268);浙江省医药卫生科技项目(2014KYA181);浙江省杭州市卫生科技计划(一般)项目(2014A33)
▲
(2016-04-29)
