濒危特有水生植物波叶海菜花的遗传多样性研究
2016-08-01李一贤余艳红张晓芸郭建玲孙文光张永洪
李一贤, 余艳红, 张晓芸, 郭建玲, 孙文光, 张永洪*
(1.云南师范大学生命科学学院,云南 昆明 650500;2:云南省环境科学研究院,云南 昆明 650034)
濒危特有水生植物波叶海菜花的遗传多样性研究
李一贤1,余艳红2,张晓芸1,郭建玲1,孙文光1,张永洪1*
(1.云南师范大学生命科学学院,云南 昆明 650500;2:云南省环境科学研究院,云南 昆明 650034)
摘要:波叶海菜花是一种濒危的水生植物,仅分布于云南省泸沽湖境内。采用AFLP分子标记技术对波叶海菜花11个居群进行遗传多样性以及遗传结构,空间分布及遗传变异的研究,并提出保护策略。结果表明:波叶海菜花具有较低的遗传多样性水平(多态百分比为44.2%)。平均Nei's基因多样性指数(HE)为0.138 0±0.189 2,平均Shannon信息指数(I)为0.207 9±0.278 3。分子方差分析(AMOVA)表明:有64%的遗传变异来自居群之间,而36%的遗传变异来自居群内部,基因流的限制(Nm = 0.314 8)可能是导致居群内遗传变异较低的原因。Mantel检验表明:波叶海菜花遗传距离与地理距离之间没有明显相关性。贝叶斯分配将全部居群分为2个遗传群组,PCoA主成分分析的结果与贝叶斯分组结果一致。因此,波叶海菜花居群之间基因流的限制可能是导致波叶海菜花遗传多样性较低以及濒危的主要原因。
关键词:波叶海菜花;遗传多样性;遗传结构;AFLP
Plant diversity of isolated and tiny distribution is often affected by human activities and environment changes to a great extent.Empirical studies complied that the rate of extinction of rare species constrained to a small scale which was more vulnerable to demographic distribution,anthropological devastating,environmental fluctuation and gene flow stochasticity,was approximately 100 to 1 000 times more than natural rates[1].The Lugu Lake located in a unique ecoregion of Yunnan plateau. During the past decades,the ecoregion has suffered destructions such as land reclamation,eutrophication, and over exploration[2].Losing suitable habitat may cause species distinction, and early reports shows many aquatic plants has shrinked drastically with population size declining and distribution scale decreasing[3].
The perennial diploid(2n=22)Otteliaacuminata(Gaghep.)Dandy var. crispa(Hand.-Mazz.) H.Li is a variety ofO.acuminata(Gaghep.)Dandy belongs to Hydrocharitaceae family.This species are grow along the lake offshore about 10 meters with root growth on the basin in depth of 0.2-4.5 m submerged the water accompanied withMyriophyllumspicatumL.andPotamogetonlucensL[4].O.acuminatavar.crispais a dioecism herb with unisexual flowers float on the surface of water with 0.2-4.5 meters long inflorescence axis.The pollination may rely on both hydrochory and pollination insects[5-6].
O.acuminatavar.crispais a representative submerged plants of Lugu Lake and often formed dominant community.This plant is sensitive to water pollution with important ecological values for environmental oscillation monitoring[7].In addition,local people usually collected the fresh flowers as vegetable.Recent years,natural populations ofO.acuminatavar.crispabecome rarer due to water pollution as well as the extensive collection.It confronted the risk of distinction due to the anthropological devastating and environmental fluctuation.It had been reported that individuals ofO.acuminatahad disappeared in several plateau lakes,such as Dianchi Lake in Kunming City,Erhai Lake in Dali City,and Qilu Lake in Tonghai county.In 2004,it was categorized as a critically endangered species by IUCN[8].ToO.acuminatavar.crispa,which restricted only in Lugu Lake,rapid development of local tourism in recent years may increase potential extinction risks of this variant due to water pollination caused by increasing tourist.
Thus,it is essential to protect this endemic and endangered species and its genetic diversity as well. Assessment of genetic diversity and population structure is not only necessary for species conservation but also useful for population dynamics and evolution process[9-10].However,empirical studies didn’t depict a refined delineation of plateau lake rare plants[11-12]and information about genetic analysis ofO.acuminatavar.crispawas limited. In this study,we focused on 1) Investigating the genetic diversity ofO.acuminatavar.crispacovering the Lugu Lake.2) Elucidating the genetic structure ofO.acuminatavar.crispa.3) Suggesting appropriate and feasible strategy for conservation.
Materials and methods
Population sampling
O.acuminatavar.crispahad been exhaustively searched on September 2013 based on the records specimens in Chinese herbarium and updated information.11 populations were collected during the flowering season (Figure 1). The sampled populations were separated apart approximately at a straight distance of 2 kilometers. In total,127 individuals were collected with sampled individuals being separated by at least 20 m.8-15 individuals were accessed in each population,young leaves were dried immediately in silica gel and stored at~20 centigrade for the experiment.

图1 波叶海菜花的居群分布地点
DNA isolation and AFLP procedure
Totalgenomic DNA was extracted from approximate 20 mg of dried leaf tissues using plant genomic DNA rapid extraction kit (TIANGEN, Beijing). Genomic DNA was quantified on 1% TAE-agarose gel using λDNA labeled, about 100 ng of DNA was used for AFLP procedure recommended by Vos[13]with a detailed describe by Lauterbach[14]. 0.15 μL EcoR I (20.000 U/μ) and 0.3 μL Mse I(20.000 U/μL)(New England Biolabs, Ipswich,MA,USA)endonuclease mixture were applied to digestion the 100ng DNA in a total volume of 40 μL for 1.5 h at 37 ℃,and digestion results was electrophorese on 1.5% agarose gel. 10 μL of mixture contained 3 μL EcoR I adaptor(5 μM), 3 μL Mse I adaptor (50 μM),1μL T4 ligase Buffer(10×),0.5 μL T4 DNA ligase(400.000 U/mL),2.5 μL ddH2O were added into digestion in total of 50 μL,ligation was incubated over night at 4 centigrade in ABI veriti 96 thermocycler(ABI Co.Ltd,).The product was then diluted 10 fold in ddH2O for PCR reaction.
For preselective DNA amplification,5 μL of diluted DNA restriction-ligation product was added to a 20 μL cocktail containing 2 μL 10×Ex - Taq buffer,1.2 μL Mg2+(25 m M),1.6 μL dNTPs (2.5 mM),1 μL primer(10 μM/L)E+A(Invitrogen),1μLprimer(10 μM/L)M+C(Invitrogen),0.1 μL Ex - Taq(5 U/μL), polymerase chain reaction was performed at 65 ℃ 5 min,followed by 30 cycles of 30 s of denaturation at 94 ℃,30 s of annealing at 56 ℃ and 1 min of elongation at 72 ℃,and a final 5 min at 72 ℃ step for complete extension,ending with a final cool down to 4 ℃.For selective PCR amplification,25 μL 10 fold diluted Pre-PCR product was used as template added to 15 μL cocktail contained 2 μL 10×Ex - Taq buffer,2 μL Mg2+(25 mM), 1.6 μL dNTPs(2.5 mM), 0.2 μL 5-FAM*EcoR I(10 μM/L),1.2 μL Mse I(10 μM/L),0.1 μL Ex-Taq(5 U/μL).The PCR parameters used were 2 min at 94 ℃,13 cycles of 30 s of denaturation at 94 ℃, 30 s of annealing at 65 ℃ and 1 min of elongation at 72 ℃,where the annealing temperature was reduced every subsequent step by 0.7 ℃, an additional 23 cycles of 30 s of denaturation at 94 ℃,30 s of annealing at 56 ℃ and 1min of elongation at 72 ℃, completed by a following 5-min step at 72 ℃ and a cool down to 4 ℃.All reagents for PCR amplification was applied by TAKARA. An initial selective polymerase chain reaction of eight individuals cross four populations was carried out with 12 primer combinations, only primers provides clear and reproducible bands with sufficient polymorphic variations between populations was used. We finally screened three most informative primer combinations (E-AGC/M-CAT,E-AGC/M-CTC,E-ACT/M-CAC)for this research.
Data analysis
The AFLP genotype was obtained from ABI sequence prism 377 using GENSCAN-ver. 3.7(ABI Co.Ltd,),obtained profiles we reanalyzed using Gene Marker ver-2.2.2[15](Available at http:www.softgenetics.com/genemarker),which was applied to identified fragments between 100-500 bp,only intense and unambiguous bands were scored as present (1) or absent(0),a 0/1 binary matrix was constructed for further analysis.
The number of polymorphic bands(NP),Nei's gene diversity(HE),Shannon's information index(I) were conducted by POPGENE ver-1.3.1[16],genetic differentiation among populations(Gst) was calculated based on Nei's and Li diversity statistics[17],the average level of gene flow(Nm) among populations was estimated by traditional method based on GST[Nm=(1-GST)/4GST][18].
Principal Coordinate Analysis(PCoA) and Mantel test were conducted using the program GENALEX ver-6.502(Available at http://biology-assets.anu.edu.au/GenAlEx) to examine the similarity of the populations included and identified if there was a correlation between geographic distance and genetic distance[19-20].Using the same software, genetic variation within and among populatons was partioned by the analysis of molecular variance(AMOVA)[21].Based on the Nei’s and Li genetic distance, an UPGMA dendrogram with 1 000 bootstraps was constructed in NTSYSpc-2.10[22].
Bayesian clustering was conducted with STRUCTURE ver-2.3.4[23-24](Available at analysis was performed using admixture model with unrelated allele frequencies, 106iterations with a 5×105burn-in iterations, Ten runs for each K were performed from 1-11, the K value of dataset was assessed to identify the appropriate number of clusters of individual, lnP(D)values was used to selected the correct number of groups by calculating ΔK[25-26].
Results
Population Genetic diversity
Weanalyzed 127 individuals from 11 populations ofO.acuminatavar.crispaand obtained a total of 138 AFLP bands based on three primer combinations.Of the 138 loci surveyed,61(44.2%) were polymorphic.The number of bands of different primer combinations varied fiercely (41-54),and the percentage of polymorphic fragments varied from 25.6% to 63.4 % (Table 1).
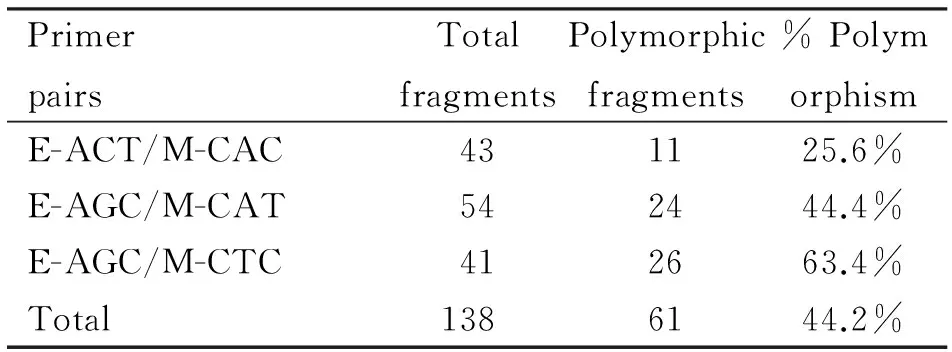
表1 3对引物得到的片段以及多态百分比
At species level, the percentage of polymorphic bands(PPB), Nei's gene diversity(HE) and Shannon’s information index(I) was 44.20%, 0.137 1 and 0.296 4 respectively.At the population level, the PPB of each population varied from 8.70%( XLS) to 19.57%(NSW),with an average of 14.42%. Mean values of HEand I were 0.136 7(SD 0.035 2) and 0.052 8(SD 0.009 9).Population NSW exhibited the highest level of genetic diversity and population XLS showed the lowest level of genetic diversity (Table 2).

表2 波叶海菜花居群采集信息及遗传多样性
注:NP, 多态位点数,PPB,多态百分比;HE,Nei’s遗传多样性指数,I Shannon 信息指数;S.D. ,标准差。
Note: NP, number of polymorphic; PPB, percentage of polymorphic bands;HE,Nei’s gene diversity;I,Shannon’s information index;S.D.,standard deviation.
Genetic structure
The coefficient of genetic differentiation among populations(Gst) was 0.613 6.The estimated gene flow(Nm)between populations obtained from Gstrevealed that the number of migrants per generation was 0.314 8.The AMOVA analysis further revealed a significant genetic differentiation across the sample distribution,64% of the total molecular variation was attributed to inter-population differentiation and 36% to individual differentiation within population.
The optimal division withinO.acuminatavar.crispawas found at K=2 in the STRUCTURE ver-2.3.4 analysis(as indicated by ΔK
values)(Figure 2).Accordingly,the genetic groups were assigned to 2 groups(Figure 3).
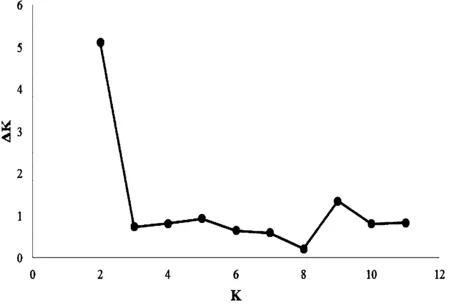
图2 基于ΔK的K值估算
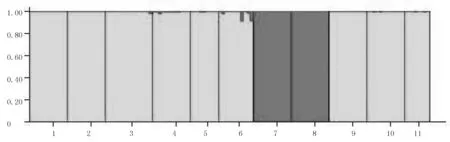
图3 K=2时的波叶海菜花居群分组图
PCoA analysis subdivided all sampled individuals into two major groups(Figure 4) with the first three axis accounted for 20.69%,12.82% and 9.87% of the total variation.Genetic variances of which UPGMA(Figure 5) analysis also supported the result of division.Mantel test shows that there was no significant correlation between genetic distance and geographic distance among 11 populations(Figure 6).
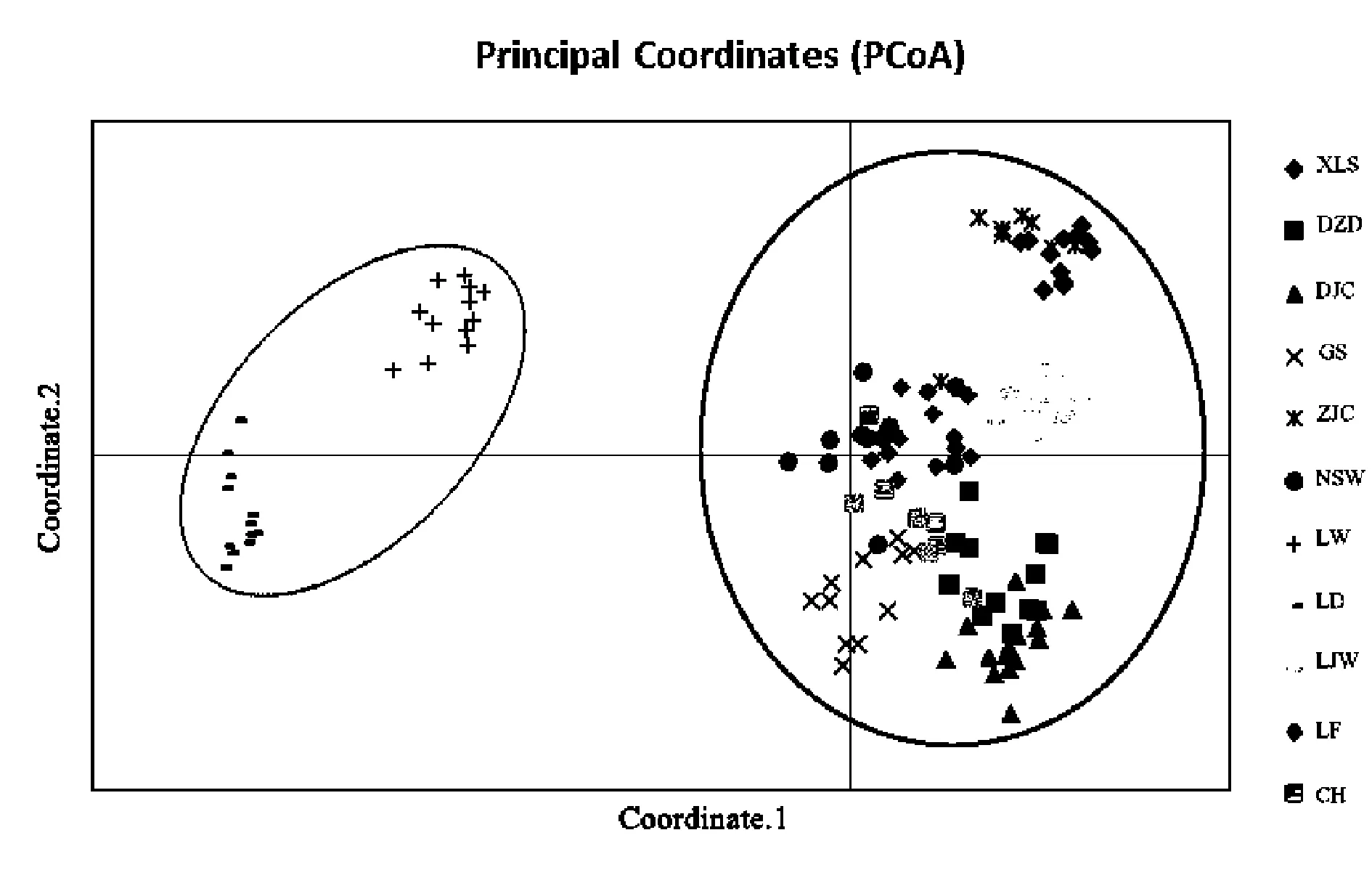
图4 波叶海菜花11个居群主成分分析结果
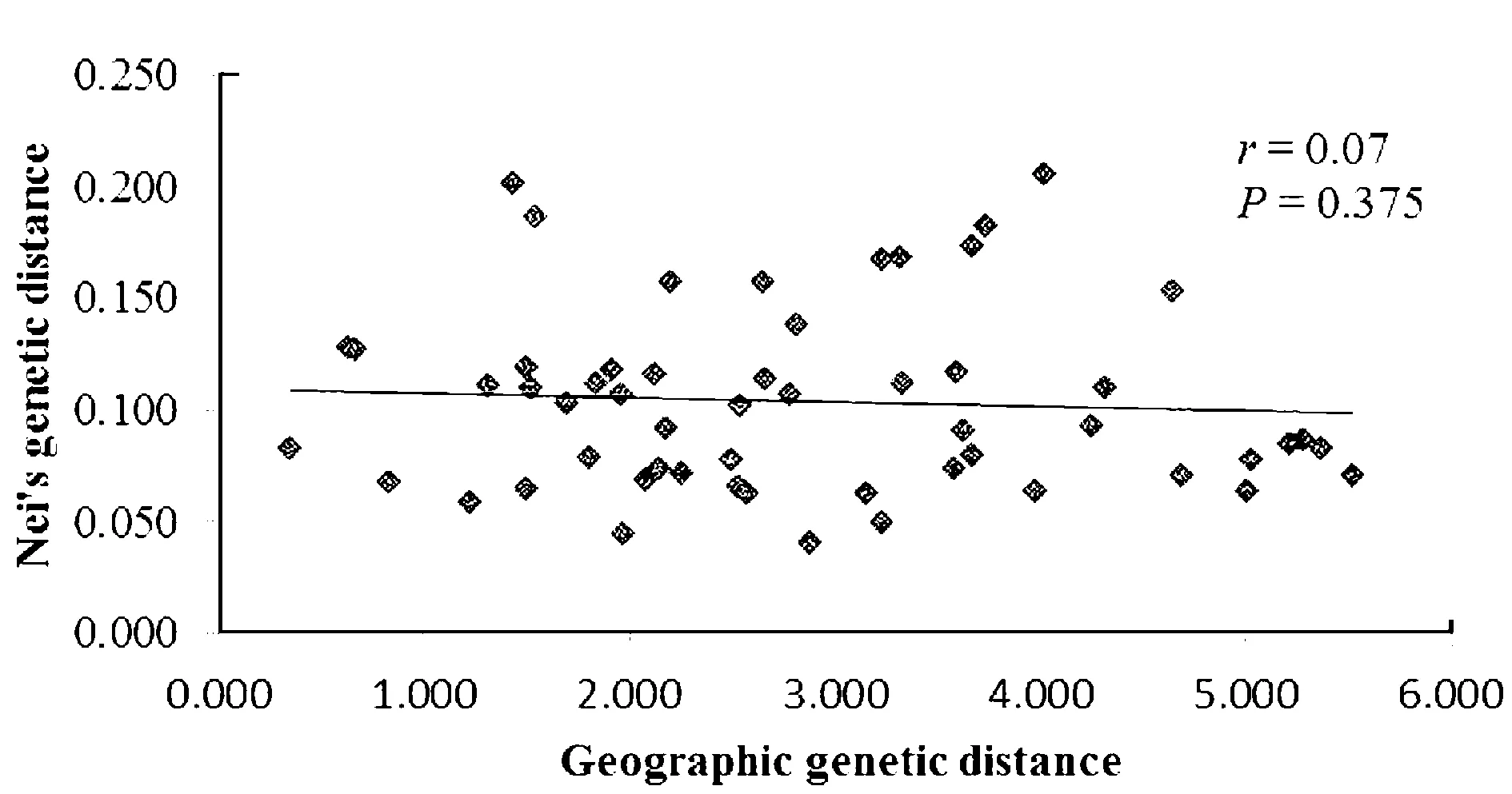
图5 波叶海菜花遗传距离与地理距离的相关性分析

图6 基于遗传距离的UPGMA聚类树
Discussion
Genetic diversity within O. acuminata var. crispa
O.acuminatavar.crispaappeared to retain a low level of genetic diversity (PPB=44.2 %,HE=0.138,I=0.207 9)as compared to the same taxa in Hydrocharitaceae[28-29].Similar results have been documented in other aquatic species like fern Ceratopteris pteridoides[30](PPB=44.5%,HE=0.141 and I=0.21)as compared to the similar distribution pattern and habitats.However,high genetic diversity also documented by Kang[31]with 61.2% polymorphic bands inferred from 8 pairs of AFLP primers in the study of endangered Isoetes sinensis (Isoetaceae).Therefore, based on pervious study and the present results,the estimated level of genetic diversity in angiosperm was consistent with the gymnosperm.
Generally,species with small geographic distribution range tended to maintain a low level of genetic diversity than geographically widespread species[32-34].And for aquatic species,genetic diversity was usually infinite by ecological fluctuation and mating system[35-38].O.acuminatavar.crispawas a dioecious perennial aquatic herb endemic to Lugu Lake.Pollination limitation and allele effects related to population size and sex ratio has been the major factors of species. Fruits ofO.acuminatavar.crispawere only produced by allogamy, and insects visits was needed for cross-pollination[39].Early reports showed that breeding restriction resistance the species variance[40-41].The present results combined with empirical studies revealed that restricted pollination might be an important factor to influence the low genetic diversity ofO.acuminatavar.crispa.
In the case of circle lake arrangedpopulations,hypothesis of migration patterns could be fairly constructed based on its micro ecological characters,In the patches of specific subpopulation such as lay on the outlet,a relatively high level of genetic diversity according to its stenotic ecology might be maintained.Simolutinously,populations lied in inlet might retain a lower genetic diversity related to the lacking if source-sink,this equilibrium of natural set a pattern which cannot be ignorantly considering and delimited[42-44].The results here seemed to meet the case:the population XLS being located in the inlet of the lake maintained the lowest level of genetic diversity;the population CH being located in the outlet maintained the high genetic diversity.
Genetic structure ofO.acuminatavar.crispa
Although the limited variabilityof the markers may affected the analysis,the primer pairs used in this study were sufficient to reveal some genetic structure ofO.acuminatavar.crispa. Significant genetic variance was found inO.acuminatavar.crispaby the value of Nei’s Gst(0.613 6).Genetic variation among populations which was almost identical to the result of AMOVA analysis.Previous study indicated that outcrossing species tended to weaken the genetic structure and inbreeding enhance the genetic structure[45-48].High level of genetic variance amongO.acuminatavar.crispapopulations might be attributed to the inbreeding system.
The sampled populations ofO.acuminatavar.crispawere portioned into 2 genetic groups monitored by STRUCTURE and PCoA,additional evidence from UPGMA clustered results also sustained the division of 2 groups,in the case of the geographic,the two groups were not significantly divided but overlapped each other. As an aquatic herb,gene flow was restrict limited between population even at a micro geographical[49-50].According to the theory of isolation by distance,positive correlation should be identified between genetic distance and geographic distance when populations reach equilibrium between gene drift and gene flow, and if disequilibrium,either gene flow or gene drift would be a dominant factor to the population genetic differentiation[51-52].In accordance with the theory, the gene flow inO.acuminatavar.crispawas very low (Nm=0.314 8) with no evident correlation between genetic distance and geographic distance, suggesting that gene drift might role the population structure. According to the present study,the population LD and LW didn’t lie in a close distance, but these two populations located not too far and connected by the water drift.Meanwhile, the populations LD,NSW and LW were basically located in a water drift according to geographic feature,and the population NSW didn’t clustered with LD and LW, seeds disposal often disturbed by algae and geographic features of the lake.As stated above, each population retained a unique micro ecological,population NSW located in a bay confined by the peninsula,for this reason it was reasonable to combine the two populations and separated from others based on assuming hydrochory influenced the gene flow ofO.acuminatavar.crispa.A similar study was documented in Sparganium emersum and Silene tatarica[53-54],of which with a high population similarities and no significant correlation between genetic and geographical distance.
Implications for conservation and priorities
It has been reported that genetic diversity was a potential evolutionary dynamic to sustained the ever-changing environment[55-56].The result in present study clearly depicted a pattern of genetic data and shed light on the conservation forO.acuminatavar.crispa. Accordingly,due to the low genetic diversity and absent of gene flow,in-situ conservation strategy should be complemented in order to enhance the population size and slowed down the rapid genetic erosion. At the same time,due to the environment fluctuation and the deterioration of water quality[57-58],ex-situ conservation should be implemented too.On account of STRUCTURE division showed that 11 populations was portioned in to 2 groups,two management units of this species was suggested for conservation. For the populations in the group A,NSW,DJC and CH should be the priority targets of protection because of their relatively higher genetic diversity than others.Besides,populations LD and LW,which maintained a great distinction to other populations,should be also considered as the principal conjoined populations for conservation.Considering translocation can enlarged gene flow and weaken the genetic variance among populations,translocation of individuals between different groups is not recommended.
Additionally,for ex-situ conservation, extensively sampled strategy should be implemented according to the genetic data in order to obtained sufficient genotypes and different populations may preserved separately while individuals from different patches needed to be keep in the same ponds accordingly.
Acknowledgments
We thank Cai-Zhen Gao and Wen-Juan Cao of Yunnan Normal University for their help in collecting plant materials ofO.acuminatavar.crispa.Also,we thanks Ms.Zhang jingxian for her helpful manuscript.This work was supported by the National Natural Science Foundation of China (Grant No.31460050, 31360049) and Financial Fund for Yunnan Biodiversity Protection:Hydro-ecology Investigation and Assessment for Three Plateau lakes in Yunnan Province-Lugu Lake,Erhai Lake and Fuxian Lake.
参考文献:
[1]Alan R.Templeton.Introduction to Conservation Genetics[J].Cambridge University Pr,2010(01):56.
[2]Yuan J.Evaluation on the Biological Resources of Provincial Nature Reserve Lugu Lake in Yunnan[J].Shaanxi Forest Science & Technology,2010(02):10.
[3]Ningyun L I,Tian K,Chen Y,et al.Changes of Ottelia acuminata communities in the lakes of northwestern Yunnan Plateau over the past three decades[J].Journal of Lake Sciences,2015,27(03):401-406.
[4]Li H, Hsu T Z.The Geobotanical Expedition on Lake Luguhu[J].Acta Botanica Yunnanica,1979(01):011.
[5]Chen Y,Li X,Yin L,et al.Genetic diversity and migration patterns of the aquatic macrophyte Potamogeton malaianus in a potamo-lacustrine system[J].Freshwater Biology,2009,54(06):1178-1188.
[6]Xia J,Lu J,Wang Z X,et al.Pollen limitation and Allee effect related to population size and sex ratio in the endangered Ottelia acuminata(Hydrocharitaceae):implications for conservation and reintroduction[J].Plant Biology, 2013,15(02):376-383.
[7]Xiaocheng Y.The Aquatic Vegetation of the Lugu Lake[J].Journal of Chongqing Teachers College(Natural Science Edition),1993(02):017.
[8]Baillie J,Hilton-Taylor C,Stuart S N.2004 IUCN red list of threatened species: a global species assessment[M].IUCN,2004.
[9]Elias M,Panaud O,Robert T.Assessment of genetic variability in a traditional cassava(Manihot esculenta Crantz)farming system, using AFLP markers[J].Heredity,2000,85(03):219-230.
[10]Schmidt K,Jensen K.Genetic structure and AFLP variation of remnant populations in the rare plant Pedicularis palustris(Scrophulariaceae)and its relation to population size and reproductive components[J].American Journal of Botany,2000,87(05):678-689.
[11]Arrigo N,Buerki S,Sarr A,et al.Phylogenetics and phylogeography of the monocot genus Baldellia(Alismataceae):Mediterranean refugia,suture zones and implications for conservation[J].Molecular phylogenetics and evolution,2011,58(01):33-42.
[12]Cieslak E,Cieslak J,Szelag Z,et al.Genetic structure of Galium cracoviense(Rubiaceae):a naturally rare species with an extremely small distribution range[J].Conservation Genetics,2015,16(04):929-938.
[13]Vos P,Hogers R,Bleeker M,et al.AFLP:a new technique for DNA fingerprinting[J].Nucleic acids research, 1995,23(21):4407-4414.
[14]Lauterbach D,Burkart M,Gemeinholzer B.Rapid genetic differentiation between ex situ and their in situ source populations:an example of the endangered Silene otites(Caryophyllaceae)[J].Botanical Journal of the Linnean Society,2012,168(01):64-75.
[15]Holland M M,Parson W.GeneMarker®HID:a reliable software tool for the analysis of forensic STR data[J].Journal of forensic sciences,2011,56(01):29-35.
[16]Yeh F C,Yang R C,Boyle T B J,et al.POPGENE,the user-friendly shareware for population genetic analysis[J].1997(01):31.
[17]Nei M,Li W H.Mathematical model for studying genetic variation in terms of restriction endonucleases[J].Proceedings of the National Academy of Sciences,1979,76(10):5269-5273.
[18]Slatkin M,Barton N H.A comparison of three indirect methods for estimating average levels of gene flow[J].Evolution,1989,43(07):1349-1368.
[19]Mantel N.The detection of disease clustering and a generalized regression approach[J].Cancer research,1967,27(2 Part 1):209-220.
[20]Peakall R O D,Smouse P E.GENALEX 6:genetic analysis in Excel. Population genetic software for teaching and research[J].Molecular ecology notes,2006,6(01):288-295.
[21]Meirmans P G.Using the AMOVA framework to estimate a standardized genetic differentiation measure[J].Evolution,2006,60(11):2399-2402.
[22]Jensen R J.Ntsys-Pc-numerical taxonomy and multivariate-analysis system-version 1.40[J].Quarterly Review of Biology,1989,64(02):250-252.
[23]Falush D,Stephens M,Pritchard J K.Inference of population structure using multilocus genotype data:dominant markers and null alleles[J].Molecular ecology notes,2007,7(04):574-578.
[24]Pritchard J K,Stephens M,Donnelly P.Inference of population structure using multilocus genotype data[J].Genetics,2000,155(02):945-959.
[25]Evanno G,Regnaut S,Goudet J.Detecting the number of clusters of individuals using the software STRUCTURE:a simulation study[J].Molecular ecology,2005,14(08):2611-2620.
[26]Falush D,Stephens M,Pritchard J K. Inference of population structure using multilocus genotype data:linked loci and correlated allele frequencies[J].Genetics,2003,164(04):1567-1587.
[27]Nei M.Genetic distance between populations[J].American naturalist,1972,106:283-292.
[28]Long C,Jiang Z,Dao Z.Research Genetic diversity of Ottelia acuminata(Hydrocharitaceae)from the Eastern Himalayas,revealed by ISSR markers[J].Botanica Orientalis:Journal of Plant Science,2010(07):56-63.
[29]Zhang H Y,Tian K,Yu Y,et al.Genetic diversity among natural populations of Ottelia acuminata(Gaghep)Dandy revealed by ISSR[J].African Journal of Biotechnology,2009,8(22):6089-6093.
[30]Chen Y,Li X,Yin L,et al.Genetic diversity and migration patterns of the aquatic macrophyte Potamogeton malaianus in a potamo lacustrine system[J].Freshwater Biology,2009,54(06):1178-1188.
[31]Kang M,Ye Q,Huang H.Genetic consequence of restricted habitat and population decline in endangered Isoetes sinensis(Isoetaceae)[J].Annals of botany,2005,96(07):1265-1274.
[32]Hamrick J L,Godt M J W.Effects of life history traits on genetic diversity in plant species[J].Philosophical Transactions of the Royal Society of London B:Biological Sciences,1996,351(1345):1291-1298.
[33]Hamrick J L,Godt M J W,Sherman-Broyles S L.Factors influencing levels of genetic diversity in woody plant species[M].Population genetics of forest trees.Springer Netherlands,1992:95-124.
[34]Waller D M,O'malley D M,Gawler S C.Genetic variation in the extreme Endemic Pedicularis furbishiae,(Scrophulariaceae)[J].Conservation Biology,1987,1(04):335-340.
[35]Fér T,Hroudová Z.Genetic diversity and dispersal of Phragmites australis in a small river system[J].Aquatic Botany,2009,90(02):165-171.
[36]Les D H.Breeding systems,population structure, and evolution in hydrophilous angiosperms[J].Annals of the Missouri Botanical Garden,1988,75(03):819-835.
[37]Shibayama Y,Kadono Y.Genetic variations in the endangered aquatic plant Nymphoides coreana(Menyanthaceae)in south‐western Japan[J].Plant species biology,2008,23(03):212-216.
[38]Oudot-Canaff J,Bornette G,Vallier F,et al.Genetic temporal dynamics in restored wetlands:A case of a predominantly clonal species,Berula erecta(Huds)Coville[J].Aquatic Botany,2015,126:7-15.
[39]Wang X,Zhou W,Lu J,et al.Effects of population size on synchronous display of female and male flowers and reproductive output in two monoecious Sagittaria species[J].PloS one,2012,7(10):e48731.
[40]Cousens M I.Gametophyte ontogeny,sex expression,and genetic load as measures of population divergence in Blechnum spicant[J].American Journal of Botany,1979,66(02):116-132.
[41]Eckert C G,Samis K E,Lougheed S C.Genetic variation across species’ geographical ranges:the central-marginal hypothesis and beyond[J].Molecular ecology,2008,17(05):1170-1188.
[42]Eriksson O.Regional dynamics of plants: a review of evidence for remnant,source-sink and metapopulations[J].Oikos,1996,77(02):248-258.
[43]Holling C S.Resilience and stability of ecological systems[J].Annual review of ecology and systematics,1973,4(01):1-23.
[44]Li W.Environmental opportunities and constraints in the reproduction and dispersal of aquatic plants[J].Aquatic Botany,2014,118:62-70.
[45]Barrett S C H,Eckert C G,Husband B C.Evolutionary processes in aquatic plant populations[J].Aquatic Botany,1993,44(02):105-145.
[46]Llorens T M,Ayre D J,Whelan R J.Evidence for ancient genetic subdivision among recently fragmented populations of the endangered shrub Grevillea caleyi (Proteaceae)[J].Heredity,2004,92(06):519-526.
[47]Rottenberg A,Parker J S.Conservation of the critically endangered Rumex rothschildianus as implied from AFLP diversity[J].Biological conservation,2003,114(02):299-303.
[48]Wright S.The genetical structure of populations[J].Annals of eugenics,1949,15(01):323-354.
[49]Kudoh H,Whigham D.Microgeographic genetic structure and gene flow in Hibiscus moscheutos(Malvaceae)populations[J].American Journal of Botany,1997,84(09):1285-1285.
[50]Pujolar J M,Lucarda A N,Simonato M,et al.Restricted gene flow at the micro-and macro-geographical scale in marble trout based on mtDNA and microsatellite polymorphism[J].Frontiers in zoology,2011,8(01):7.
[51]Hufford K M,Mazer S J.Plant ecotypes: genetic differentiation in the age of ecological restoration[J].Trends in Ecology & Evolution,2003,18(03):147-155.
[52]Petit R J,Excoffier L.Gene flow and species delimitation[J].Trends in Ecology & evolution,2009,24(07):386-393.
[53]Pillon Y,Qamaruz-Zaman F,Fay M F,et al.Genetic diversity and ecological differentiation in the endangered fen orchid(Liparis loeselii)[J].Conservation Genetics,2007,8(01):177-184.
[54]Tero N,Aspi J,Siikamäki P,et al.Genetic structure and gene flow in a metapopulation of an endangered plant species,Silene tatarica[J].Molecular Ecology,2003,12(08):2073-2085.
[55]Frankham R,Briscoe D A,Ballou J D.Introduction to conservation genetics[M].Cambridge:Cambridge University Press,2002.
[56]Pollux B J A,Jong M D E,Steegh A,et al.Reproductive strategy,clonal structure and genetic diversity in populations of the aquatic macrophyte Sparganium emersum in river systems[J].Molecular Ecology,2007,16(02): 313-325.
[57]Li H.The flourishing and declining of Ottelia acuminata in the lake Dian Chi[J].Journal of Yunnan University:Natural Sciences,1985(07):138-142.
[58]Penas J,Barrios S,Bobo-Pinilla J,et al.Designing conservation strategies to preserve the genetic diversity of Astragalus edulis Bunge,an endangered species from western Mediterranean region[J].PeerJ,2016(04):e1474.
收稿日期:2016-04-06基金项目:国家自然科学基金项目(31460050, 31360049);云南省生物多样性保护专项资金资助项目
作者简介:李一贤(1990—),男,山东日照人,在读硕士,研究方向为保护遗传学。张永洪为通信作者,
文章编号:1004-7999(2016)02-0139-10
DOI:10.13478/j.cnki.jasyu.2016.02.009
中图分类号:Q943.2
文献标识码:A
Genetic diversity of Ottelia acuminate var. crispa (Hydrocharitaceae):An endangered aquatic herb with extremely narrow distribution
LI Yixian1,YU Yanhong2, ZHANG Xiaoyun1,GUO Jianling1,SUN Wenguang1,ZHANG Yonghong1*
(1.LifeScienceofCollege,YunnanNormalUniversity,KunmingYunan650500,China2.YunnanInstituteofEnvironmentalScience,KunmingYunan650034,China)
Abstract:Ottelia acuminata var. crispa is an endangered aquatic herb which narrowly confined to Lugu Lake in southwest China.By using AFLP fingerprint, the genetic diversity and population structure of O. acuminata var. crispa and its conservation strategy were illustrated, and the pattern between spatial distribution and genetic variation was identified. The results showed that O. acuminata var. crispa retained a low level of genetic diversity (PPB=44.2%) with the mean Nei’s gene diversity index (HE=0.138 0±0.189 2) and mean Shannon’s information index(I=0.207 9±0.278 3).AMOVA revealed that approximately 64% genetic variation was due to between populations and 36% by within populations due to restricted gene flow (Nm=0.314 8). No correlation between genetic distance and geographic distance was found by Mantel test.Bayesian assignment suggested 2 main genetic groups that were according with the results of PCoA analysis.The results above indicated that the restriction of gene flow between populations of O. acuminata var. crispa might be a major cause of endangerment.
Key words:Ottelia acuminata var. crispa; genetic diversity; population structure; AFLP
yhzhang@mail.kib.ac.cn
