Expression analysis of heat-shock protein gene Hsp845 in the Antarctic psychrotrophic bacterium Psychrobacter sp. G under temperature and salinity stress
2016-02-01CHEShuaiZHANGLiangZHANGZhengLINXuezheng
CHE Shuai, ZHANG Liang, ZHANG Zheng & LIN Xuezheng*
1 The First Institute of Oceanography, SOA, Qingdao 266061, China;
2 Key Lab of Marine Bioactive Substances, SOA, Qingdao 266061, China
1 Introduction
Heat shock response, which involves the synthesis of heat shock proteins (Hsps), plays a vital role in the defense of bacterial cells against temperature-induced damage. Hsps are highly conserved and exist in almost all species[1]. GrpE,a member of the Hsp family, is transiently induced under various stress conditions[2]. GrpE is a known nucleotideexchange factor for the molecular chaperone DnaK, which functions co-translationally and post-translationally to promote protein folding and disaggregation in cells[3-6]. In this molecular chaperone system, DnaK (an Hsp70 homolog)acts in concert with GrpE and its co-chaperone DnaJ, an Hsp40 homolog[7]. DnaJ stimulates the hydrolysis of DnaK-bound ATP and converts DnaK from a low-affinity state to a high-affinity state for substrates, whereas GrpE facilitates ADP/ATP exchange and restores DnaK to the low-affinity state[8-10]. GrpE gene expression is regulated by σ70 and σ32 promoters[11], thereby allowing rapid and transient induction in response to elevated temperatures[12-13]. Previous studies have also shown that this molecular chaperone system is essential to normal cell growth[14-16], cell division[16],membrane translocation[17], chromosome segregation, and maintenance of low-copy-number plasmids[3]. As Hsps, the expressions of DnaK, DnaJ, and GrpE are transiently induced under different stress conditions, such as heat[18], salinity[19],ethanol treatment, viral infection, and exposure to heavy metals[14,20].
Sea ice in the Southern Ocean is dominated by strong gradients of salinity and drastic ぼuctuations of temperature,space, and light[21]. Temperature and salinity are the two most important factors affecting the survival of microorganisms living in sea ice. These organisms have accordingly evolved adaptive mechanisms ranging from molecular to whole cell levels. The freezing and thawing of ice during different seasons in Antarctic seawater inhabited by Psychrobacter sp.affects the salinity of offshore seawater[22].
Although the heat-shock response of GrpE is well documented at the molecular level in mesophilic microorganisms such as Escherichia coli[12,14], Synechocystis sp.[15], and Bacillus subtilis[23]and in thermophilic microorganisms such as Thermus thermophilus[24]and Thermosynechococcus elongates[15], limited studies have been performed in cold-adapted microorganisms. Moreover,most of the earlier studies focused on response to only one stress factor, such as temperature. In fact, organisms in nature seldom undergo only one stress at a time[1]. To our knowledge,researchers have rarely investigated the mechanism of GrpE in response to salinity and to combinations of multiple stresses at both transcriptional and translational levels simultaneously.According to our previous study[2], the Hsp845 gene is 615 bp long and encodes a protein consisting of 204 amino acid residues with a molecular mass of 22.7 kDa and a calculated isoelectric point of 4.54. A BLAST analysis revealed that the deduced amino acid sequence of Hsp845 was a close homolog to amino acid sequences of other known cytosolic GrpEs. Among the four Hsp genes we investigated, Hsp845 was found to be strongly sensitive at the transcriptional level to temperature and salinity stress[2].In the present study, we further investigated the expression of the Hsp845 gene under different conditions (temperature,salinity, and combinations of temperature and salinity) at both transcriptional and translational levels by quantitative realtime PCR (qRT-PCR) and western blotting, respectively.
2 Materials and methods
2.1 Bacteria, medium, and growth conditions
The Antarctic psychrotrophic bacterium Psychrobacter sp.G was isolated from Antarctic seawater and maintained in our laboratory[25]. Psychrobacter sp. G was cultured at 20°C with rotation of 150 r∙min–1in medium consisting of 5 g tryptone, 1 g yeast extract and 14 g NaCl in 1 L seawater. The composition of the medium used for salinity stress treatments, which has been described previously, is summarized in Table 1[26]. Seawater (with an approximate salinity of 31‰) was taken from the coast at Qingdao(Shandong, China).
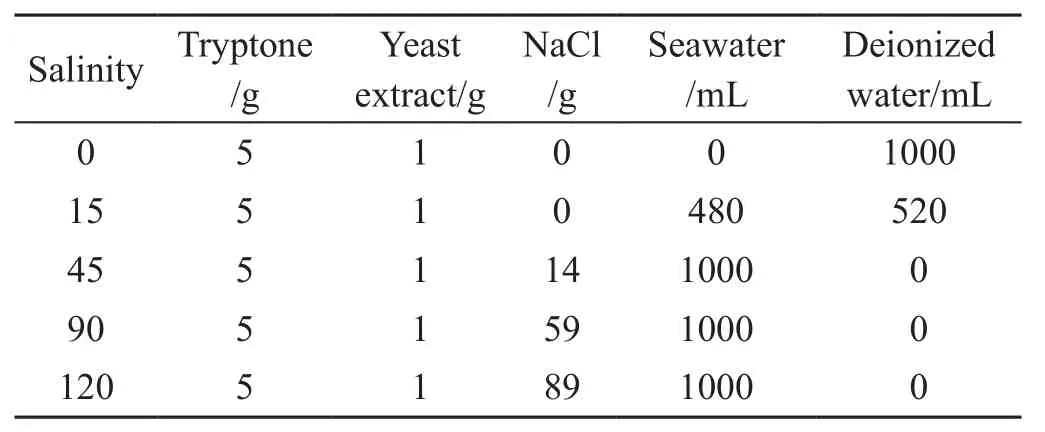
Table 1 Components of culture medium having different salinities
2.2 Recombinant expression and purification of Hsp845 protein
Primers Hsp845F (5′-GGAATTCCATATGAGCGAGCA AAATAACCAC-3′) and Hsp845R (5′-CCGCTCGAGCTGA CCGACACGTACCATCG-3′), respectively containing NdeI and XhoI restriction enzyme sites, were designed to amplify the open reading frame of the Hsp845 gene. After digestion with restriction enzymes NdeI and XhoI, the target PCR product was subcloned into plasmid pET22b digested with the same restriction enzymes. The recombinant plasmid was then transformed into E. coli BL21 (DE3) competent cells for recombinant protein expression. Positive clones were screened by PCR with primers T7F and T7R and then confirmed by nucleotide sequencing. Each positive clone was incubated in 250 mL Luria-Bertani medium(containing 100 µg∙mL–1ampicillin) at 37°C with shaking at 150 r∙min–1. When the absorbance of the culture at 600 nm (OD600) reached 0.6–0.8, the cells were incubated for an additional 4 h in the presence of the inducer isopropyl-β-D-thiogalactopyranoside (IPTG) added to afinal concentration of 1 mM. The cells were then collected by centrifugation at 10000×g for 5 min and resuspended in 5 mL of 50 mM Tris-HCl buffer (pH 8.0). After lysis of the cells with a one-shot cell disrupter (UL61010A-1, Constant Systems Ltd., Daventry, UK), the lysates were centrifuged at 10000×g for 20 min at 4°C. The recombinant protein was purified on a Ni Sepharose 6 Fast Flow column (11-0008-86 AE, GE Healthcare, Uppsala, Sweden) according to the manufacturer’s instructions. The antiserum was prepared and validated by GenScript (Nanjing, China; order ID:168267-1).
2.3 Stress treatments
Psychrobacter sp. G was initially cultured at its optimal growth temperature (20°C) and salinity (45‰)[26]. When the OD600of the culture reached approximately 0.5, temperature,salinity, and combined stress treatments were carried out as follows. For temperature stress treatments, cultures were kept at different temperatures (0°C, 10°C, or 30°C) for different durations (2 h, 6 h, or 12 h). For salinity stress treatments, the cultures were centrifuged at 8000 ×g for 5 min at 20°C. After thoroughly suspending the pellets with an equal volume of medium to a final salinity of 0‰, 15‰, 90‰, or 120‰, the suspensions were maintained at 20°C for 2 h, 6 h, or 12 h.In the combined stress treatments, cells were cultured under the following conditions: a final salinity of 15‰ at 0°C or 30°C or a final salinity of 90‰ at 0°C or 30°C. A strain cultured continuously under optimal conditions was used as the control for qRT-PCR and western blot analysis. Three biological replicates of each control and stress treatment were carried out.
2.4 Total RNA isolation and cDNA synthesis
Total RNA was extracted using an RNAprep Pure Cell/Bacteria kit (DP430, Tiangen Biotech, Beijing, China)according to the manufacturer’s protocol. The purity of prepared RNA was judged by the A260/A280ratio. A PrimeScript RT Reagent kit (DRR037A, Takara, Dalian,China) was used for the reverse transcription of RNA into cDNA. The cDNA for qRT-PCR was prepared using 500 ng of total RNA. Primers Q-F (5′-TGGTGGATAACTTGGA-3′)and Q-R (5′-ACCGACCGCTTCAT-3′) for qRT-PCR were designed with the software program Primer 5.0. The GAPDH gene encoding glyceraldehyde-3-phosphate dehydrogenase was used as an internal reference[2,26].
2.5 Expression analysis of the Hsp845 gene by qRTPCR
SYBR Premix Ex Taq II (DRR820A, Takara) was used for the qRT-PCR analysis. qRT-PCR amplification was carried out on a Stratagene Mx3005P qPCR System for 40 cycles (95°C for 1 min, 45°C for 30 s, and 72°C for 20 s).All reactions were performed in triplicate. Quantification of mRNA was based on threshold cycle (Ct) values. After normalizing the Ctvalue of Hsp845 using the Ctvalue corresponding to the selected reference gene, the efficiency of each qRT-PCR amplification was calculated. Data analysis was based on the comparative Ct(2–ΔΔCt) method[27]. Data obtained from qRT-PCR analysis were subjected to analysis of variance to identify significant differences between mean values among treatments. Differences were considered to be statistically significant at P < 0.05 and highly significant at P< 0.01.
2.6 Western blot analysis of Hsp845 protein expression in Psychrobacter sp. G
Western blot analysis was carried out to investigate the expression of Hsp845 at the translational level. Total protein of Psychrobacter sp. G was dissolved in 50 mM Tris-HCl buffer (pH 8.0) and then centrifuged at 10000×g for 10 min at 4°C to collect the supernatant. Protein concentration was then assayed. Each sample (approximately 60 mg protein) was analyzed by 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The proteins were then electroblotted onto Immobilon-P transfer membranes (IPVH00011, SolarBio,Beijing, China). To prevent nonspecific antibody binding,membranes were first blocked with 5% non-fat milk in TBST(10 mM Tris-HCl [pH 7.5], 150 mM NaCl, and 0.1% Tween-20) accompanied by shaking for 3 h at room temperature and then incubated with the 1:1000 diluted antiserum to Hsp845 in TBST overnight. After three 10-min washes with TBST and one 10-min wash with TBS (10 mM Tris-HCl[pH 7.5], 150 mM NaCl), each membrane was incubated in peroxidase-conjugated goat anti-rabbit IgG (1:2000 in block reagents as indicated above) for 2 h and then washed three times with TBST (10 min per wash) followed by a 10-min wash in TBS. Finally, the membrane was soaked in a reaction mixture (1 mL of 6 mg∙mL–14-chloro-1-naphthol in methanol, 10 mL TBS, and 6 µL H2O2) in darkness for 10 min and the reaction was blocked in distilled water.Antibody binding was visualized through a peroxidasecatalyzed stain reaction[28].
3 Results
3.1 Expression and purification of recombinant Hsp845 protein
Recombinant Hsp845 protein was expressed in E. coli BL21(DE3) induced by IPTG (Figure 1, lanes 2 and 3). Expressed recombinant Hsp845 protein carrying a 1.08-kDa Histag was detected in the lysate supernatant by Coomassie Bright Blue staining and found to have a molecular mass of approximately 24 kDa, consistent with the expected value. The recombinant Hsp845 protein was purified by Ni Sepharose 6 Fast Flow chromatography (Figure 1, lane 4),which revealed that the purity of the obtained protein was higher than the threshold criterion of the antibody test (85%).The recombinant protein was sent to GenScript to revalidate its purity and obtain effective antigen protein.

Figure 1 Sodium dodecyl sulfate-polyacrylamide gel electrophoretic analysis of recombinant expression of Hsp845 protein. Lane 1, protein marker; lane 2, lysate of Escherichia coli with pET22b-Hsp845 without induction; lane 3, lysate of E. coli with pET22b-Hsp845 induced by isopropyl-β-D-thiogalactopyranoside; lane 4, recombinant Hsp845 protein purified by Ni Sepharose 6 Fast Flow chromatography.
3.2 Expression characteristics of Hsp845 in response to temperature stress
qRT-PCR analysis revealed that Hsp845 gene expression was significantly inhibited by low temperatures (0°C and 10°C) and enhanced immediately after exposure to high temperature (30°C). Under 30°C conditions, Hsp845 gene expression reached a maximum after 2 h of treatment, with a 5.3-fold increased compared with the control (20°C), and then decreased during the next 10 h (Figure 2). Western blot analysis indicated that the expression of Hsp845 protein was also enhanced by high temperature (30°C) and inhibited by low temperatures (0°C and 10°C). Expression levels increased continuously at 30°C and reached a 2.2-fold maximum compared with the control (20°C) at 12 h (Figure 3), a trend not entirely consistent with the qRT-PCR results.
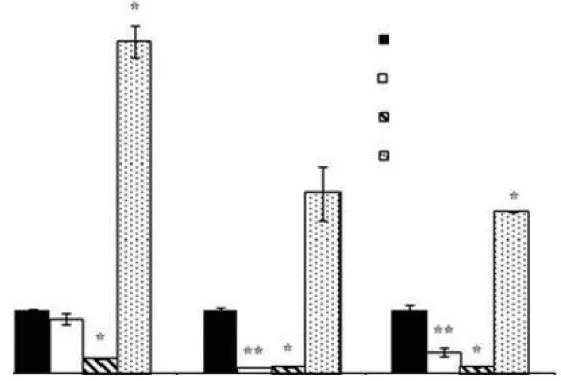
Figure 2 qRT-PCR analysis of Hsp845 gene expression in response to temperature stress. Standard error bars are shown: * P< 0.05; ** P < 0.01.
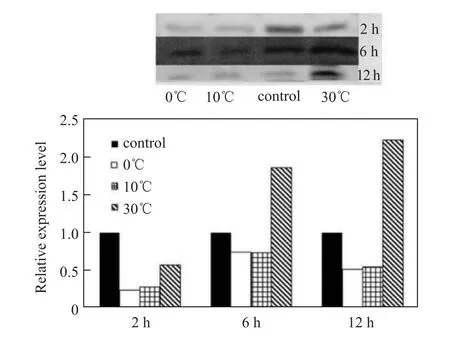
Figure 3 Western blot analysis of Hsp845 protein expression in response to temperature stress.
3.3 Expression characteristics of Hsp845 in response to salinity stress
With regard to salinity stress, Hsp845 gene expression was found to be immediately and significantly enhanced by both low salinity (0‰) and high salinity (90‰ and 120‰) during the first 2 h, but was not sensitive to 15‰ salinity (Figure 4). Over the next 10 h, Hsp845 expression decreased under exposure to salinity levels of 0‰ and 90‰. At a salinity level of 120‰, however, Hsp845 gene expression reached a maximum 44.8-fold increase compared with the control (45‰salinity) at 6 h and then exhibited a 9.8-fold decrease over the next 6 h relative to the control. Hsp845 expression was also enhanced by high salinity (90‰ and 120‰) and reached maximum relative expression increases of 1.9-fold and 1.7-fold after 12 h, respectively. In the absence of salt, Hsp845 protein expression was significantly inhibited, contrary to the qRT-PCR results. Similarly, Hsp845 expression was not sensitive to a salinity level of 15‰ (Figure 5).
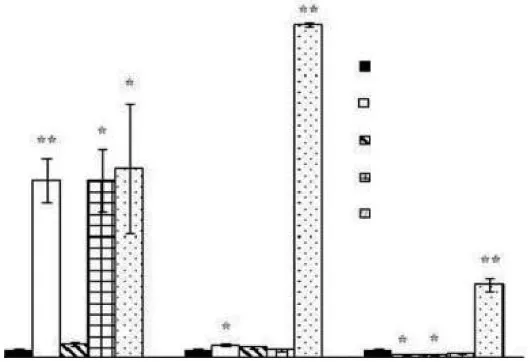
Figure 4 qRT-PCR analysis of Hsp845 gene expression in response to salinity stress. Standard error bars are shown:* P <0.05; **P < 0.01.
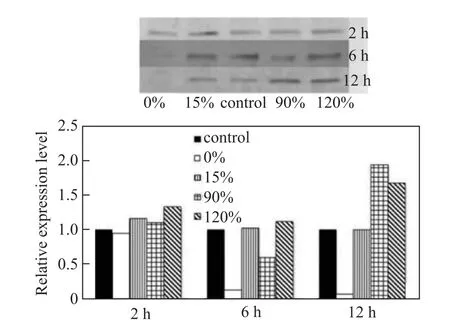
Figure 5 Western blot analysis of Hsp845 protein expression in response to salinity stress.
3.4 Expression characteristics of Hsp845 in response to combined temperature and salinity stress
qRT-PCR analysis revealed that Hsp845 gene expression was enhanced by heat treatment at 30°C at a salinity of 15‰ or 90‰, but was inhibited at 0°C at those salinity levels. After exposure to 30°C, the transcriptional expression of Hsp845 reached a maximum level that was 3.2-fold (under 15‰salinity) and 4.0-fold (90‰ salinity) higher at 6 h compared with the control (20°C, 45‰ salinity), and then decreased during the next 6 h (Figure 6). The expression characteristics of Hsp845 in response to combined temperature and salinity stress were similar to those detected by qRT-PCR. High temperature enhanced Hsp845 protein expression under both low (15‰) and high (90‰) salinity, with maximum increases of 1.7-fold and 1.8-fold respectively observed at 12 h compared with the control (20°C, 45‰ salinity). By contrast, treatment at 0°C with a salinity of 15‰ or 90‰inhibited Hsp845 expression (Figure 7).
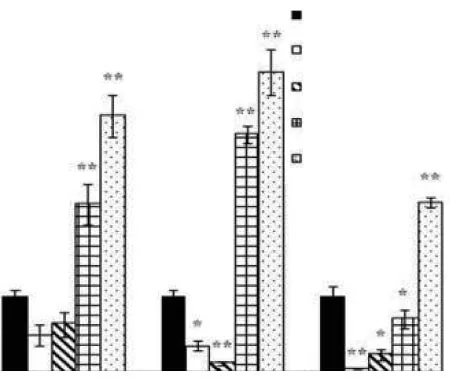
Figure 6 qRT-PCR analysis of Hsp845 gene expression in response to combined temperature and salinity stress. Standard error bars are shown: * P < 0.05; ** P < 0.01.
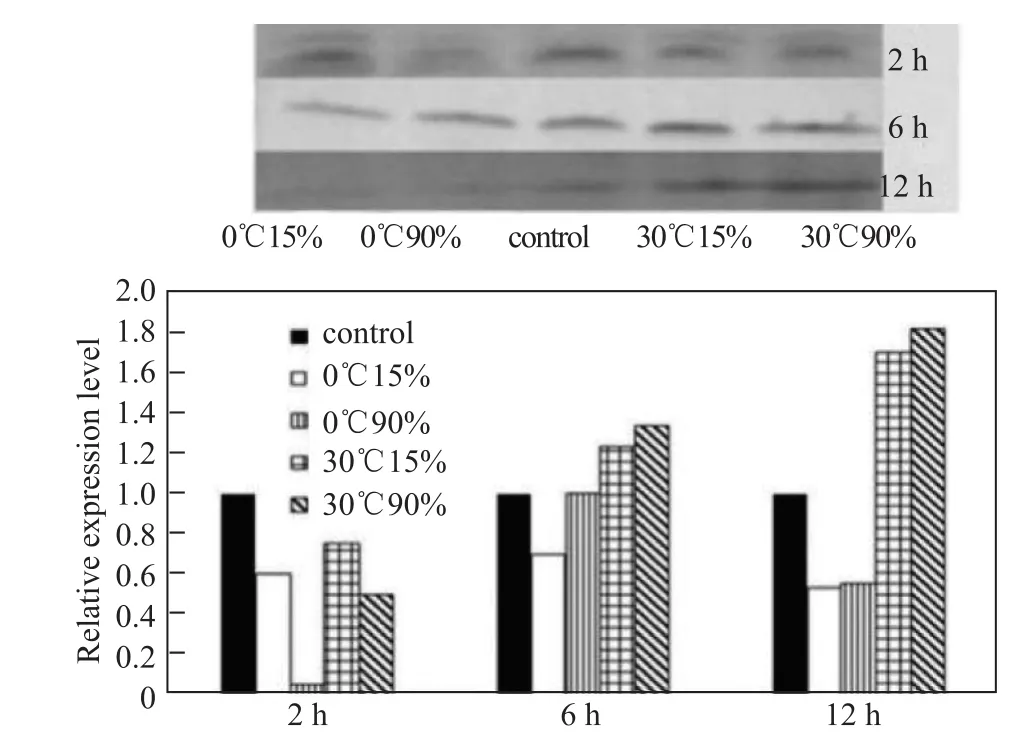
Figure 7 Western blot analysis of Hsp845 protein expression in response to combined temperature and salinity stress.
4 Discussion
In this study, we investigated the expression characteristics of the Hsp845 gene at both transcriptional and translational levels in the Antarctic psychrotrophic bacterium Psychrobacter sp. G under various stress treatments using qRT-PCR and western blotting, respectively.
Hsp845, which is affiliated with GrpE, is the cochaperone of Hsp70[33]. In this study, high temperature(30°C) significantly enhanced Hsp845 expression at both transcriptional and translational levels. Previous studies have also indicated that the Hsp70 chaperone is generally induced in response to an external temperature rise of 5°C–10°C above the optimal growth temperature of an organism[29]and that GrpE expression is rapidly and transiently induced in response to elevated temperature[13].
Clark et al.[30]cloned Hsp70 family genes Hsc70, Hsp70,and Grp78 from Antarctic mollusks Laternula elliptica and Nacella concinna and studied the expression characteristics of the Hsp845 gene under high temperatures (15°C and 20°C) by qRT-PCR. When exposed to high temperatures, the expression of Hsp70 was found to be significantly induced, indicating that this gene plays an important role in the high temperature adaptation of Antarctic mollusks. Park et al.[31]also studied the expression characteristics of Hsp70 under high temperature conditions in the Antarctic clam Laternula elliptica. They observed that high temperature (10°C) significantly increased Hsp70 gene expression in gills and digestive gland tissue. The expression of Hsp70 reached a maximum (4.6-fold greater compared with the control) at 12 h in gill tissue, whereas maximum expression (3.6-fold greater compared with the control) in digestive gland tissue was observed at 24 h. Liu et al.[22]cloned the 2232-bp full-length sequence of the Hsp70 gene (CiHsp70) of the Antarctic ice alga Chlamydomonas sp. ICE-L from a cDNA library. qRT-PCR showed that both cold and heat shock treatments induced CiHsp70 expression.Under high salinity (62‰) stress, CiHsp70 expression was induced 3.0-fold compared with the control (31‰).Ultraviolet radiation induced expression of CiHsp70 gene expression as well. Briefly, CiHsp70, as a chaperone, may play an important role in Chlamydomonas sp. ICE-L adaption to the polar environment.
Hsp845 protein expression at the translational level lagged behind transcriptional-level expression and reached a maximum at 12 h after exposure to 30°C. The highest relative expression level, 2.2-fold greater than that of the control (20°C), was much lower than that of transcriptionallevel expression (5.3-fold greater compared with the control).The same tendency was also observed in the other two stress treatments, indicating that regulation may occur between transcriptional and translational levels.
Freezing and thawing of sea ice during different seasons in Antarctica, the habitat of Psychrobacter sp. G, affects the salinity of offshore seawater. Kilstrup et al.[19]have reported the Hsp70 chaperone is induced during salinity stress in Lactococcus lactis, while Nath et al.[32]have additionally discovered that hypersalinity creates a more general stress response than hyposaline conditions. The former authors observed that the Hsp70 chaperone had a transient response after a shift to low or high salinity during adaptation to a particular salinity. This finding may explain why Hsp845 gene expression was immediately and significantly enhanced at the transcriptional level by both low (0‰) and high (90‰ and 120‰) salinity during the first 2 h of treatment. The effects of high salinity (120‰) on Hsp845 gene expression were longer lasting, as the maximum relative expression level was reached at 6 h. In our study, however, we found that the Hsp845 protein was over-expressed at the transcriptional level after exposure to a salinity of 0‰, but was obviously inhibited at the translational level. The functional contribution of Hsps to salinity stress adaptation is still not clear. One possibility is that Hsps are induced as a consequence of the accumulation of denatured proteins resulting from lowered water activity during plasmolysis of salinity-shocked cells[33].
Psychrobacter sp. G may simultaneously encounter multiple stress factors. For example, freezing is often accompanied by salinity stress caused by the absence of unfrozen water[21,34]. We therefore also investigated the combined effect of temperature and salinity on Hsp845 gene expression. qRT-PCR revealed that Hsp845 gene expression was enhanced by treatment at 30°C in combination with 15‰or 90‰ salinity, but was inhibited at 0°C under the same salinity levels. The results of western blotting also uncovered the same trend. Nonetheless, Hsp845 appeared to be more sensitive to high temperature than to salinity in the combined stress treatment at both transcriptional and translational levels,a observation inconsistent with the results of the single-factor experiment.
In this study, we found that Hsp845 was induced at both transcriptional and translational levels by high temperature(30°C). Hsp845 was induced at the transcriptional level by both low (0‰) and high (90‰ and 120‰) salinity, but was only induced at the translational level by high salinity (90‰and 120‰). In the single-factor experiment, the relative expression of Hsp845 under salinity stress was far higher than that observed under temperature stress. Under combined stress,however, Hsp845 was more sensitive to high temperature than to different salinities at both transcriptional and translational levels. The mechanism responsible for this inconsistent pattern needs further study. The maximum relative expression at the transcriptional level was higher than that at the translational level; moreover, the maximum expression of Hsp845 at the translational level lagged behind that at the transcriptional level. Finally, expression at the transcriptional level in the absence of salt seemed to conflict with results at the translational level; the underlying mechanism also needs to be studied further. Although the mechanism of GrpE proteins contributing to adaption to various stresses in microorganisms living in Antarctic seawater is still far from being understood, we believe that the present work is a good starting point for further investigations.
AcknowledgementsThis work was supported financially by the National Natural Science Foundation of China (Grant no. 41176174).
1 Feder M E, Hofmann G E. Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu Rev Physiol, 1999, 61(1): 243–282
2 Che S, Song W Z, Lin X Z. Response of heat-shock protein(HSP) genes to temperature and salinity stress in the Antarctic psychrotrophic bacterium Psychrobacter sp. G. Curr Microbiol, 2013,67(5): 601–608
3 Sugimoto S, Saruwatari K, Higashi C, et al. The proper ratio of GrpE to DnaK is important for protein quality control by the DnaK-DnaJGrpE chaperone system and for cell division. Microbiology, 2008,154(7): 1876–1885
4 McCarty J S, Buchberger A, Reinstein J, et al. The role of ATP in the functional cycle of the DnaK chaperone system. J Mol Biol, 1995,249(1): 126–137
5 Russell R, Jordan R, McMacken R. Kinetic characterization of the ATPase cycle of the DnaK molecular chaperone. Biochemistry, 1998,37(2): 596–607
6 Szabo A, Langer T, Schröder H, et al. The ATP hydrolysis-dependent reaction cycle of the Escherichia coli Hsp70 system DnaK, DnaJ, and GrpE. Proc Natl Acad Sci U S A, 1994, 91(22): 10345–10349
7 Liberek K, Marszalek J, Ang D, et al. Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc Natl Acad Sci U S A, 1991, 88(7): 2874–2878
8 Siegenthaler R K, Christen P. The importance of having thermosensor control in the DnaK chaperone system. J Biol Chem, 2005, 280(15):14395–14401
9 Brehmer D, Gässler C, Rist W, et al. Influence of GrpE on DnaK-substrate interactions. J Biol Chem, 2004, 279(27): 27957–27964
10 Zylicz M, Ang D, Georgopoulos C. The grpE protein of Escherichia coli. Purification and properties. J Biol Chem, 1987, 262(36): 17437–17442
11 Yura T, Nagai H, Mori H. Regulation of the heat-shock response in bacteria. Annu Rev Microbiol, 1993, 47(1): 321–350
12 Harrison C. Grp E, a nucleotide exchange factor for DnaK. Cell Stress Chaperones, 2003, 8(3): 218–224
13 Lipinska B, King J, Ang D, et al. Sequence analysis and transcriptional regulation of the Escherichia coli grpE gene, encoding a heat shock protein. Nucl Acids Res, 1988, 16(15): 7545–7562
14 Wu B, Ang D, Snavely M, et al. Isolation and characterization of point mutations in the Escherichia coli grpE Heat Shock Gene. J Bacteriol,1994, 176(22): 6965–6973
15 Barthel S, Rupprecht E, Schneider D. Thermostability of two cyanobacterial GrpE thermosensors. Plant Cell Physiol, 2011, 52(10):1776–1785
16 Bukau B, Walker G C. Cellular defects caused by deletion of the Escherichia coli dnaK gene indicate roles for heat shock protein in normal metabolism. J Bacteriol, 1989, 171(5): 2337–2346
17 Hartl F U, Hayer-Hartl M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science, 2002, 295(5561): 1852–1858
18 Lindquist S, Craig E A. The heat-shock proteins. Annu Rev Genet,1988, 22(1): 631–677
19 Kilstrup M, Jacobsen S, Hammer K, et al. Induction of heat shock proteins DnaK, GroEL, and GroES by salt stress in Lactococcus lactis. Appl Environ Microbiol, 1997, 63(5): 1826–1837
20 Neidhardt F C, Van Bogelen R A. Heat shock response//Neidhardt F C.Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, DC: American Society for Microbiology, 1987:1334–1345
21 Thomas D N, Dieckmann G S. Antarctic Sea ice—a habitat for extremophiles. Science, 2002, 295(5555): 641–644
22 Liu S H, Zhang P Y, Cong B L, et al. Molecular cloning and expression analysis of a cytosolic Hsp70 gene from Antarctic ice algae Chlamydomonas sp. ICE-L. Extremophiles, 2010, 14(3): 329–337
23 Holtmann G, Brigulla M, Steil L, et al. RsbV-independent induction of the SigB-dependent general stress regulon of Bacillus subtilis during growth at high temperature. J Bacteriol, 2004, 186(18): 6150–6158
24 Grimshaw J P A, Jelesarov I, Siegenthaler R K, et al. Thermosensor action of GrpE: the DnaK chaperone system at heat shock temperatures. J Biol Chem, 2003, 278(21): 19048–19053
25 Lin X Z, Cui S S, Xu G Y, et al. Cloning and heterologous expression of two cold-active lipases from the Antarctic bacterium Psychrobacter sp. G. Polar Res, 2010, 29(3): 421–429
26 Song W Z, Lin X Z, Huang X H. Characterization and expression analysis of three cold shock protein (CSP) genes under different stress conditions in the Antarctic bacterium Psychrobacter sp. G. Polar Biol,2012, 35(10): 1515–1524
27 Livak K J, Schmittgen T D. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCTmethod. Methods,2001, 25(4): 402–408
28 Yu S S, Yang H, Chai Y M, et al. Molecular cloning and characterization of a C-type lectin in roughskin sculpin (Trachidermus fasciatus). Fish Shellfish Immunol, 2013, 34(2): 582–592
29 Lindquist S. The heat-shock response. Annu Rev Biochem, 1986,55(1): 1151–1159
30 Clark M S, Fraser K P P, Peck L S. Antarctic marine molluscs do have an HSP70 heat shock response. Cell Stress Chaperones, 2008, 13(1):39–49
31 Park H, Ahn I Y, Lee H E. Expression of heat shock protein 70 in the thermally stressed antarctic Clam Laternula elliptica. Cell Stress Chaperones, 2007, 12(3): 275–282
32 Nath I V A, Bharathi P A L. Diversity in transcripts and translational pattern of stress proteins in marine extremophiles. Extremophiles,2011, 15(2): 129–153
33 Overpeck J, Hughen K, Hardy D, et al. Arctic environmental change of the last four centuries. Science, 1997, 278(5341): 1251–1256
34 Pearce D A. Climate change and the microbiology of the Antarctic Peninsula region. Sci Prog, 2008, 91(2): 203–217
杂志排行
Advances in Polar Science的其它文章
- Norwegian contributions to Arctic environmental sciences from the 1880s to the third International Polar Year
- Meteorite classification for building the Chinese Antarctic Meteorite Depository—Introduction of the classification of 500 Grove Mountains meteorites
- Biodiversity and phylogenetic analysis of the gut microbiome of Euphausia superba Dana from the Rose Sea of the Antarctic Ocean
- Structure and seasonal variability of fronts in the Southeast Indian Ocean along sections from Fremantle, Australia to Antarctic Zhongshan Station
- Outreach channels for polar science: an expedition to Kerguelen Islands as a case study
- Potential methane production rates and its carbon isotopic composition from ornithogenic tundra soils in coastal Antarctic
