Herbivore defense responses and associated herbivore defense mechanism as revealed by comparing a resistant wild soybean with a susceptible cultivar
2015-12-21XioyiWngHifengChenZhihuiShnQingnnHoChnjunZhngZhongluYngXiojunZhngSongliYunDezhenQiuShuilinChenYongqinJioXinZhou
Xioyi Wng,Hifeng Chen,Zhihui Shn,Qingnn Ho,Chnjun Zhng, Zhonglu Yng,Xiojun Zhng,Songli Yun,Dezhen Qiu,Shuilin Chen, Yongqin Jio,Xin'n Zhou,*
aKey Laboratory of OilCrop Biology of the Ministry of Agriculture,Oil Crops Research Institute ofthe Chinese Academy of AgriculturalSciences, Wuhan 430062,China
bGraduate Schoolof the Chinese Academy of Agriculture Sciences,Beijing 100081,China
Herbivore defense responses and associated herbivore defense mechanism as revealed by comparing a resistant wild soybean with a susceptible cultivar
Xiaoyi Wanga,b,Haifeng Chena,Zhihui Shana,Qingnan Haoa,Chanjuan Zhanga, Zhonglu Yanga,Xiaojuan Zhanga,Songli Yuana,Dezhen Qiua,Shuilian Chena, Yongqin Jiaoa,Xin'an Zhoua,*
aKey Laboratory of OilCrop Biology of the Ministry of Agriculture,Oil Crops Research Institute ofthe Chinese Academy of AgriculturalSciences, Wuhan 430062,China
bGraduate Schoolof the Chinese Academy of Agriculture Sciences,Beijing 100081,China
A R T I C L E I N F O
Article history:
Accepted 8 July 2015
Available online 16 August 2015
Soybean H.armigera Morphology Gene expression level polymorphism(ELP) Detoxification and toxin-tolerance gene
Plants have evolved sophisticated defense mechanisms against herbivores to help them adapt to the environment.Understanding the defense mechanisms in plants can help us control insects in a more effective manner.In this study,we found that compared with Tianlong 2(a cultivated soybean with insect susceptibility),ED059(a wild soybean line with insect resistance) contains sharper pubescence tips,as well as lower transcript levels of wound-induced protein kinase(WIPK)and salicylic acid-induced protein kinase(SIPK),which are important mitogen-activated protein kinases involved in early defense response to herbivores.The observed lower transcript levels of WIPK and SIPK induced higher levels ofjasmonic acid(JA),JA biosynthesis enzymes(AOC3)and some secondary metabolites in ED059.Functionalanalysis of the KTI1 gene via Agrobacterium-mediated transformation in Arabidopsis thaliana indicated thatit plays an important role in herbivore defense in ED059.We further investigated the molecular response of third-instar Helicoverpa armigera(Hübner)larvae to Tianlong 2 and ED059.We found apoptotic cells only in the midguts of larvae that fed on ED059.Compared with larvae reared on the susceptible cultivar Tianlong 2,transcript levels of catalase(CAT)and glutathione S-transferase(GST)were up-regulated,whereas those of CAR,CHSB,and TRY were down-regulated in larvae that fed on the highly resistant variety ED059.We propose that these differences underlie the different herbivore defense responses of ED059 and Tianlong 2.
©2015 Crop Science Society of China and Institute of Crop Science,CAAS.Production and hosting by Elsevier B.V.This is an open access article under the CC BY-NC-ND license
(http://creativecommons.org/licenses/by-nc-nd/4.0/).
1.Introduction
Soybean,the main oil crop of China,has a long history of providing the country with plant protein and oil.However, various diseases and damage due to insect pests have led to huge yield losses in recent years.Pest control in China has relied mainly on chemical pesticides.However,use of such pesticides causes serious environmental pollution andincreases pest resistance to pesticides.Breeding and deployment of resistant varieties is a new way of pest management that is crucial for agricultural sustainable development and environmental protection.
Plants have evolved a diverse set of defense responses to herbivores.They use both constitutive and induced defenses to combat challenges posed by pests[1].Constitutive defenses include morphological and biochemical features such as sharp and thick trichomes[2,3],heavy epicuticular waxes[4],and secondary compounds,which are constitutively present in the plant.Induced defense is a complex physiological and biochemical process that can be enhanced upon recognition of herbivore attack and includes several signal transduction pathways(the mitogen-activated protein kinase(MAPK)signaling pathway,jasmonic acid(JA)pathway,salicylic acid(SA) pathway,and ethylene(ET)pathway,among others)[1].These signaling pathways produce many anti-herbivore compounds [5],including secondary metabolites such as proteinase inhibitors(PIs)and nicotine,which exert direct effects on herbivores by interfering with their physiology[6,7].Other compounds, such as volatile organic compounds,act by interfering with the behavior of herbivores[8].However,very little is known about the herbivore defense mechanisms in soybean.
Plants can also distinguish damages from herbivores and mechanical wounding.Some genes and proteins are activated both by mechanicalwounding and by herbivores[9].However, other studies indicate that very different transcript profiles are induced by mechanical wounding and insect attack[10-12]. Studies of the mechanism by which insects induce plant response have led to the discovery of fatty acid-amino acid conjugates(FACs)in herbivore oral secretions that elicit herbivory-specific responses,such as JA and ET bursts,that are greater than those elicited by mechanical wounding;high levels of trypsin proteinase inhibitor(TPI);and the release of volatile organic compounds(VOCs)[13].However,whether there is any difference in the early steps and the final defense metabolites between soybean response to herbivore attack and that to wounding is unknown.
When a plant is attacked by herbivores,the induced defense reaction notonly is activated in the localleafbeing attacked,but also is transferred to other leaves through a series of signal transduction events[14].Ultimately,the whole plant shows enhanced resistance against herbivores.In tomato,PIs induced by herbivores provide an ideal model for elucidating the molecular basis of systemic resistance signaling in plants[15]. A recent study showed that a signaling molecule that undergoes long-distance transport is JA,which induces expression of PIs in leaves throughout the plant[16].Systemin,a polypeptide signaling molecule,regulates the biosynthesis of JA [17].Systemin and JA collaborate to induce the plant to produce defense compounds after attack by herbivores.However, whether there is systemic resistance in soybean following local insult by herbivores remains unknown.
Differences in phenotypic characteristics can be ultimately attributed to genetic differences.In recent years,along with the growing number of completed genome sequences and transcript profiles of organisms,large-scale gene expression analyses have become more easily.A large number of studies have focused on gene expression-level polymorphisms(ELPs) in different individuals to analyze diverse phenotypic traits [18],such as rust resistance[19],herbivore-induced early signaling events,and secondary metabolite production in insect resistance[20].ELPs are of great help for understanding the complete metabolic,regulatory,and developmental pathways that underlie different phenotypic traits.
In a previous study,we found that the wild soybean line ED059(Glycine soja Sieb.et Zucc.)was highly resistant to the cotton bollworm Helicoverpa armigera(Hübner),and thata cultivar Tianlong 2[Glycine max(L.)Merr.]was highly susceptible to H. armigera[3].In the present study,we investigated differences in morphology,early response to stresses,and anti-herbivore metabolites between ED059 and Tianlong 2 after attack by H. armigera.Our results showed that ED059 not only had physical barriers against herbivores,but also exhibited an induced herbivore defense reaction.Our results reveallarge differences in phenotype and molecular response to herbivores in ED059 and Tianlong 2 and provide important new insights into the metabolic,regulatory and developmental pathways that underlie the resistance of wild soybean variety ED059 to herbivores.
2.Materials and methods
2.1.Plant materials
The cultivated soybean(Tianlong 2)and the wild soybean (ED059)used in this study were obtained from the Institute of Oil Crops Research,Chinese Academy of Agricultural Sciences,Wuhan,China.Tianlong 2 is susceptible and ED059 resistant to H.armigera.
Seeds were pre-germinated on moistened filter paper in a plant growth chamber at 27°C,85%ambient humidity,and 16:8 light:dark photoperiod for 3 to 4 days.The seedlings were transferred into 18 cm×18 cm individual plastic pots containing 2:3 Pinnstrup Substrate(Pinnstrup Substrate, Denmark):vermiculite at 27°C under 16 h of light.All plants used in the experiments had three fully expanded trifoliates.
2.2.Trichome density and the shape of pubescence tip on ED059 and Tianlong 2
Tianlong 2 and ED059 were observed for pubescence tip morphology and density using 10 randomly selected 4-week-old plants(plants in the V3 stage)from each variety. First,soybean leaves were cut into small pieces and fixed in phosphate buffer with 4%glutaraldehyde for 24 h.The leaves were then cleaned with phosphate buffer and dehydrated in an ethanol concentration series.Lastly,the leaves were sprayed with metal film and their pubescence tips were photographed under a scanning electron microscope(Hitachi SU8010)to classify the trichome tips as either sharp or blunt[21].
Using a punch with a diameter of 5 mm,5 samples from each leaf were obtained for examination.Then,the trichome density was quantified for both Tianlong 2 and ED059 by measurement of the number of trichomes in 1 mm2of the upper and under leaf surfaces of 10 plants of each variety using a fluorescence microscope(OLYMPUS IX71)at 20×magnification.The trichome density was represented as the number of trichomes per square millimeter(mm2).
2.3.Scanning and transmission electron microscopy analysis of wax on the leaf surface and cuticle thickness of Tianlong 2 and ED059
Treatment of materials for observation of wax on the surface of leaves was the same as that for observation of pubescence tip shape.All materials were observed under the scanning electron microscope.
For transmission electron microscopy(TEM),sections of the leaves of Tianlong 2 and ED059 were collected and fixed in FAA solution for 24 h.The leaves were then successively dehydrated with ethanolat 30%,50%,70%,80%,90%,and 100% concentrations and xylene was used as transpirant.Paraffin was infiltrated at 52°C to 54°C.Change ofparaffin was done 4 times every 2 h.Leaves were then embedded in a paraffin block and cut into 4-μm slices using a rotary microtome.The slices were placed on cover slips and washed with xylene before coating with gold film.The thickness of the cuticle was then measured using a transmission electron microscope (Hitachi H-7500)[22].
2.4.Plant treatment and qRT-PCR analyses
All the plants used in the experiment had 3 fully expanded trifoliates.The second fully expanded leaf was used for mechanical treatment and was wounded 6 times on each side of the midvein every 20 min using pliers to mimic the bite marks of insects.For H.armigera treatment,the second fully expanded leaf was infested with 3 third-instar larvae of H. armigera.After specific time periods,the wounded localleaves and the first fully expanded unwounded systemic leaves were harvested,flash frozen in liquid nitrogen,and stored at-80°C until use for qRT-PCR analyses and phytohormone quantification.Samples from untreated plants were used as controls.
Total RNA was extracted using Trizol reagent(Sigma,USA) and 2μg was reverse-transcribed to cDNA using Superscript II reverse transcriptase(Invitrogen).Quantitative Real-time PCR analyses were performed with SuperReal PreMix(SYBR Green, Tiangen)on a Rotorgene Q(Qiagen)real-time PCR system. Samples were harvested from three biological replicates for analysis.Quantification of the relative expression levels ofthe genes was performed by the 2-ΔΔCTmethod.The soybean HDC gene was used as an internal control for normalization of the expression levels of the selected genes.The primers used for SYBR Green-based analysis are listed in Table S1.
2.5.Phytohormone extraction and quantification
JA and SA were analyzed by HPLC-ESI-MS/MS as previously described[23].Plant treatment was the same as in the preceding method.Approximately 0.1 g of the treated leaves was collected at indicated times,immediately frozen in liquid nitrogen,and stored at-80°C until analysis.
About0.1 g ofthe treated leaves was ground into powder.The powder was extracted with 0.5 mL 1-propanol-H2O-concentrated HCl(2:1:0.002,v/v/v)containing 0.4μg each of[2H2]-JA and [2H4]-SA as internal standards.After centrifugation,the bottom organic phase was transferred to 10-mL tubes and evaporated in a nitrogen stream.Each sample was redissolved in 80%methanol and then extracted using a C18 solid-phase extraction(SPE) cartridge(CNWBOND HC-C18).Eluates were analyzed by HPLC-ESI-MS/MS(Agilent 1200,Agilent Technologies,CA,USA) and then quantified using a hybrid triple quadrupole/linear ion trap mass spectrometer(ABI 4000 Q-Trap,Applied Biosystems, Foster City,CA,USA)in multiple reaction monitoring(MRM)and information-dependent acquisition(IDA)mode.Standard curves for SA and JA quantification were generated using a series of SA and JA dilutions.These experiments were all performed with three biologicalreplicates and each sample was measured three times.
2.6.Changes in midgut cells and the molecular dynamics of detoxification and toxin tolerance genes of H.armigera after feeding on Tianlong 2 and ED059
Newly hatched larvae of H.armigera were fed on artificial diet until the third instar and then on ED059 for 24 h before being starved for the next 24 h.Larvae that fed on Tianlong 2 were collected as a control.
2.6.1.Paraffin wax
Five H.armigera individuals feeding on Tianlong 2 and ED059 plants were immediately fixed in 4%paraformaldehyde. Fixation was performed at room temperature for 24 h.The fixed tissues were subsequently dehydrated by passing through an ascending concentration series of ethanol(30%, 50%,70%,90%,and 100%)and then infiltrated with wax at 57°C.The tissues were then embedded in paraffin and sections of midgut of 4μm thickness were cut with a rotary microtome(Leica,Germany).
2.6.2.TUNEL and DAPIstaining
The paraffin wax sections were hydrated by first immersing in xylene,then passing through a descending concentration series ofethanol(100%,95%,90%,80%,70%)and finally through water. An in situ cell death detection kit(terminal deoxynucleotidyl transferase dUTP nick end labeling,TUNEL,11684817910,Roche, Germany)was used to detect apoptotic cells.Apoptotic cells were marked with a green fluorescent FITC fluorochrome.In this system,biotinylated nucleotides were briefly incorporated at the 3′-OH ends of DNAy using terminal deoxynucleotidyl transferase.Horseradish peroxidase-labeled streptavidin was then bound to these biotinylated nucleotides,which were detected using peroxidase and diaminobenzidine(DAB)stains and methyl green as counterstain after the completion of immunohistochemistry[24].At the same time,samples were incubated in 1%Triton X-100 for 8 min and washed with PBS (pH 7.4).The samples were then stained with 4,6-diamidino-2-phenyl-indole(DAPI,sc-3598,Santa Cruz Biotechnology,USA)for 20 min and examined under a fluorescence microscope.These experiments were allperformed with three biologicalreplicates.
2.6.3.RT analysis
After H.armigera larvae were fed on Tianlong 2 and ED059 for 24 h,they were collected and dissected for extraction of midgut tissues.The midguts were placed into PBS(pH 7.2)for cleaning and were immediately frozen in liquid nitrogen for RNA extraction.Insects feeding on Tianlong 2 were used as controls.Total RNA was extracted using Trizol reagent (Sigma)and 2μg was reverse-transcribed to cDNA usingSuperscript II reverse transcriptase(Invitrogen).Quantitative real-time PCR analyses were performed using SuperReal PreMix(SYBR Green,TianGen)on the Rotorgene Q(Qiagen) Real-Time PCR system.Target genes were amplified using gene-specific primers(Table S2).For each condition,three biological replicates were used.Quantification of relative expression levels of the genes was performed by the 2-ΔΔCT method.The 18 s gene in H.armigera was used as an internal control for normalization of the expression levels of other genes.
2.7.KTI1 overexpression and test of insect defense against H.armigera
Gene overexpression based on Agrobacterium-mediated transformation of Arabidopsis thaliana by the floral dip method was used to stably overexpress the gene KTI1 as described previously[24].The full-length cDNA of the gene were amplified by PCR using the specific primers listed in Table S1.The PCR products were digested with XcmΙand inserted into the plasmid PCX-DG as described previously[25].Control plants were transformed with empty vector(EV).Seeds from T0 transgenic A.thaliana plants were selected on MS agar plates containing 50 mg L-1hygromycin.Seeds from T1 plants were individually collected and selected on MS agar plates containing 50 mg L-1hygromycin with 3:1(number of positive plants:number of negative plants)segregation ratio.Selected T2 plants were propagated and homozygous overexpression lines were confirmed by RT-PCR analysis.
The insect defense test was performed in a growth chamber set at 27°C,85%ambient humidity,and 16:8(L:D) photoperiod.Eggs of H.armigera were hatched at 27°C.One freshly hatched larva of H.armigera was placed in a Petri dish (100×25 mm)and starved for 12 h before use in the test.Five mature rosette leaves from 4-week-old plants of either genotype were placed in each dish.The leaves were changed every 2 days,and the test was stopped after 6 d of infestation. The weight of larvae in each Petri dish(for each genotype) was recorded 2,4,and 6 days after feeding.The dish experiment was conducted in a randomized complete block design with 10 replications.
2.8.Data analysis
All data were analyzed using Statistica software(Statistica, SAS Institute Inc.,USA).
3.Results
3.1.The morphology oftrichome and cuticular wax of Tianlong 2 and ED059
The shape of the pubescence tip was significantly different between Tianlong 2 and ED059(Fig.1-a)and the shape of the pubescence tip could be easily identified without exception within a variety.ED059 leaves had sharp pubescence tips, whereas Tianlong 2 leaves had blunt tips.We collected the first fully expanded trifoliate of each plant for observation of trichome density.ED059 displayed a significantly higher trichome density on both the adaxial and the abaxial leaf surface than Tianlong 2(Fig.1-b).
SEMwas used to assess wax on the leaf surfaces of the two lines.The leaf surfaces of ED059 were clearly seen to be more sparsely covered by wax crystals than those of Tianlong 2 (Fig.1-c).The structures found on the ED059 and Tianlong 2 leaves were all irregular lamellar crystals(Fig.1-c).
TEM was used to observe the thickness of the cuticle. ED059 leaves had thicker cuticles than did Tianlong 2(Fig.1-d and e).
3.2.Comparison of gene expression-level polymorphisms (ELPs)in Tianlong 2 and ED059
3.2.1.Different transcript levels of protein kinase genes in Tianlong 2 and ED059
Wounding or herbivore attack activates a large number ofprotein kinases,such as wound-induced(WIPK),mitogen-activated (MAPK),and receptor-like(RLK)protein kinases.Many of these protein kinases have been implicated in the defense response of plants[26].
To investigate whether protein kinases are expressed at different levels between the two varieties after wounding or being attacked by an herbivore,we used H.armigera to infect the plants and pliers to make wounds that mimic insect bite marks every 20 min.Samples were collected at multiple time points.We chose several protein kinase genes known to participate in plant early response to biotic and abiotic stresses and measured their transcript levels in Tianlong 2 and ED059 using qRT-PCR.
Mitogen-activated protein kinase 2(MAPK2)and mitogenactivated protein kinase 1(MAPK1),which are orthologs of tobacco SIPK and WIPK,respectively,can be activated by pathogens[27].The levels of MAPK2 and MAPK1 changed more in Tianlong 2 than in ED059 after H.armigera treatment. Wounding treatment greatly induced MAPK2 and MAPK1 expression in both Tianlong 2 and ED059(Fig.2-a and b). MAPK3,which is a WIPK ortholog,showed a trend similar to that of MAPK1,suggesting similarities in their functions (Fig.2-c).
3.2.2.Different transcript levels of WRKY transcription factors between Tianlong 2 and ED059
Transcription factors play critical roles in mediating gene expression by binding to the promoter region to activate or inhibit target gene transcription in eukaryotes.WRKYs,which are unique transcription factors identified in plants in recent years,are involved in various aspects of plant physiological processes including plant response to biotic and abiotic stresses [28].WRKYs play important roles in signaling pathways underlying response to stress and not only target upstream MAPKs but also regulate downstream target genes[29,30].To observe whether any ELPs are present in WRKY genes in ED059 and Tianlong 2 after wounding and H.armigera treatment,we evaluated the transcript levels oftwo WRKY genes:WRKY 6 and WRKY20.The WRKY6 gene has been reported to be induced by O3response in soybean leaves.Its orthologous gene AtWRKY6 in Arabidopsis has been shown to be a part of the signal transduction pathway underlying the response of Arabidopsis to pathogens[31].WRKY20 expression is induced by manyabiotic stresses,such as high salt,drought,low temperature and ABA[32].
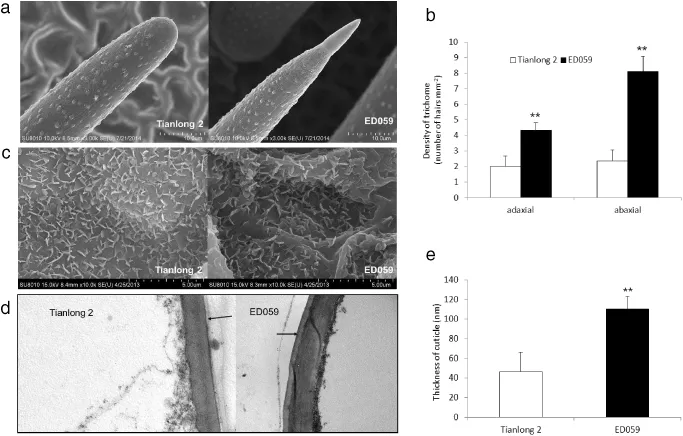
Fig.1–Morphology of trichome and cuticular wax of Tianlong 2 and ED059.(a)Scanning electron micrographs(SEM)ofthe trichome tips of soybean showing two distinct phenotypes:the blunt tips of Tianlong 2 and the sharp tips of ED059.(b)Trichome density on adaxialand abaxialsurfaces of the leaves using TEManalysis.Each value represents mean±SE ofthree independent measurements.(c)SEM images ofthe epicuticular wax crystals on the leaf surface.(d)TEMimages of cuticle membranes ofthe leaves.(e)Analysis of leaf cuticle thickness by TEM.Each value represents the mean±SE of three independent measurements.
When untreated,ED059 plants had higher transcript levels of WRKY6 than Tianlong 2.After 0.5 h,the transcript levels of WRKY6 reached a maximum value,increasing twofold and 24-fold after H.armigera and wounding treatment,respectively,in ED059.Although a similar induction pattern of WRKY6 transcript levels was observed in ED059 for the two treatment conditions,much higher transcript levels of WRKY6 were induced by wounding than by H. armigera treatment(Fig.3-a).
Prior to any treatment,WRKY20 transcripts were present at similar levels in both accessions.Both treatment conditions induced increased levels of WRKY20.H.armigera induced higher levels of WRKY20 in both accessions than wounding treatment.However,H.armigera treatment induced higher levels of WRKY20 in Tianlong 2 than in ED059 after 0.5 h.After 1 h of H.armigera treatment,the levels of WRKY 20 declined more quickly in Tianlong 2 than in ED059(Fig.3-b).
At present,very little is known about the biochemical and physiological functions of most WRKYs,in particular about the function of WRKYs in insect defense in soybean.In this study,we investigated the expression levels of several WRKYs with functions in biotic and abiotic defenses.We found that these transcription factors were expressed at different levels between ED059 and Tianlong 2 plants.Our results suggest that ELP in these transcription factors leads to different downstream gene transcript levels in the two plants.
3.2.3.Different levels of herbivore-induced JA and SA between Tianlong 2 and ED059
Induced herbivore defense responses are controlled largely by phytohormones such as jasmonates(JA and its derivatives) and salicylic acid(SA).Previous studies have shown that SIPK, WIPK,and other early signal transduction components play important roles in regulating the biosynthesis of phytohormones[33].To investigate whether ELPs of these early signaling genes affect ED059 and Tianlong 2,we measured JA and SA levels induced by wounding and H.armigera infection. In addition,we evaluated the gene transcript levels of some JA-and SA-biosynthesis related genes.
Biosynthesis of JA is essential for the production of induced defense responses to herbivores in many plants[20]. In the absence of treatment,ED059 showed significantly lower levels of JA than did Tianlong 2(Fig.4-a).In ED059,both treatments greatly induced high levels of JA;in contrast,in a previous study we found that H.armigera treatment induced lower levels of JA and AOC3 in Tianlong 2 than in ED059[34]. The levels of JA also declined after wounding treatment (Fig.4-a).Allene oxide cyclase 3(AOC3)is an important enzyme involved in JA biosynthesis[20].In the absence of treatment,the transcript levels of AOC3 showed nearly no difference between ED059 and Tianlong 2.Both wounding and H.armigera treatment induced higher AOC3 production in ED059 than in Tianlong 2.ED059 exhibited high transcript levels of AOC3 at 1 and 2 h after H.armigera and wounding treatments,respectively(Fig.4-a)[35].
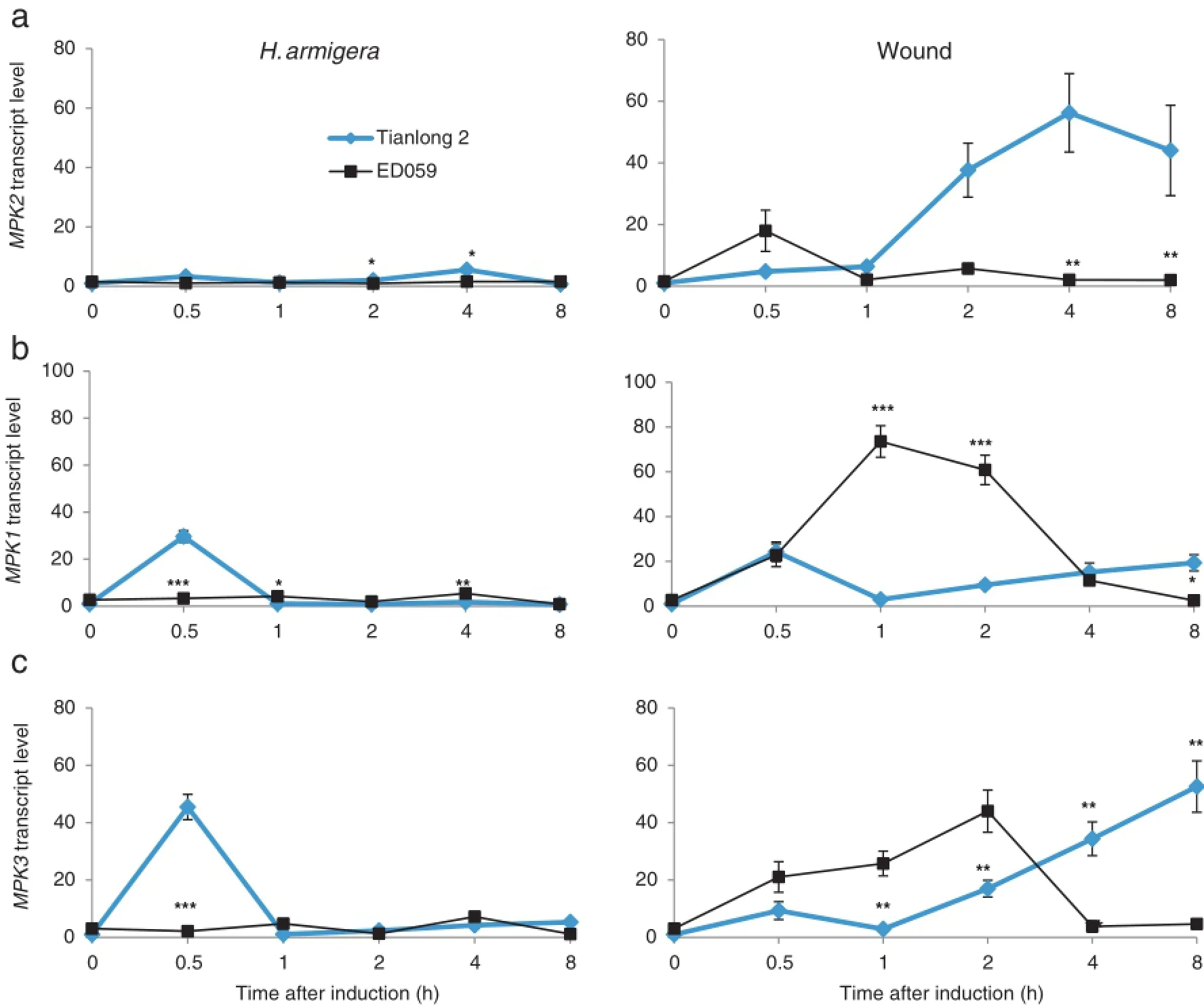
Fig.2–Different transcript levels of MAPK in Tianlong 2 and ED059.Tianlong 2 and ED059 plants were grown under identical conditions.The second fully expanded leaves were wounded with pliers or infested with 3 third-instar larvae of H.armigera. Individualleaves fromthe three replicate plants were harvested after treatmentatthe indicated times.The mean(±SE)expression levels of MPK2(a),MPK1(b),and MPK3(c)were quantified by qRT-PCR.Asterisks indicate significantly different transcript levels between Tianlong 2 and ED059 at the indicated times(unpaired t-test;*,P<0.05;**,P<0.01;***,P<0.001;n=3).
The biosynthesis and perception of JA-IIe are also important for induced herbivore defense response in plants[35,36]. Threonine deaminase(TD)supplies IIe to JA for JA-IIe biosynthesis and JA-IIe induces the defense response to herbivores through a COI1-dependent pathway[37].We investigated the ELP of TD and COI1,which are involved in JA-IIe biosynthesis and the JA-IIe-dependent signaling pathway.ED059 and Tianlong 2 exhibited almost the same basal transcript levels of TD and COI1.The transcript levels of TD were greatly induced in ED059 after 2 and 1 h of H.armigera and wounding treatments,respectively.The transcript levels of TD remained almost the same in Tianlong 2 during the whole process of H.armigera treatment.In contrast,TD transcript levels increased gradually with wounding treatment(Fig.4-b).In both accessions,the transcript levels of COI1 increased to high values at 0.5 h after H.armigera and wounding treatment(Fig.4-c).
Without treatment,SA levels were not significantly different between Tianlong 2 and ED059.In ED059,H.armigera treatment decreased SA levels,whereas wounding treatment increased them.In contrast,SA levels were slightly increased in Tianlong 2 after both treatments(Fig.5-a).Phenylalanine ammonia lyase 1(PAL1)has been identified as the key enzyme involved in biosynthesis of SA[1].We measured the transcript levels of PAL1 in ED059 and Tianlong 2.After 0.5 h, H.armigera treatment induced higher PAL1 transcript levels in Tianlong 2 than in ED059.In contrast,wounding treatment induced much higher levels of PAL1 in ED059 than in Tianlong 2 after 0.5 h(Fig.5-b).
These results showed that wounding and H.armigera induced different levels of changes in phytohormones such as JA and SA between Tianlong 2 and ED059.ELPs of these genes,which are involved in the biosynthesis of phytohormones,contribute to the phenotypic differences between Tianlong 2 and ED059.
3.2.4.Different transcript levels of wounding and H.armigerainduced antiherbivory secondary metabolites between Tianlong 2 and ED059

Fig.3–WRKYgenes in Tianlong 2 and ED059 showdifferent levels oftranscript accumulation.Tianlong 2 and ED059 plants were grown under the same conditions.The second fully expanded leaves were wounded with pliers or infested with 3 third-instar larvae of H.armigera.Individualleaves from three replicate plants were harvested after the treatment at the indicated times.Mean (±SE)expression levels of WRKY6(a)and WRKY20(b)were quantified by qRT-PCR.Asterisks represent significantly different transcript levels between Tianlong 2 and ED059 at the indicated times(unpaired t-test;*,P<0.05;**,P<0.01;***,P<0.001;n=3).
Wounding and H.armigera treatment caused the transcript levels of some protein kinases,transcription factors,and genes involved in phytohormone biosynthesis to change between ED059 and Tianlong 2.These changes likely increase the production of secondary metabolites that are toxic and antinutritive to herbivores[38-40].In soybean,these herbivoreinduced metabolites include PIs,polyphenoloxidase(PPO),and flavonoids,among others[41-46].We studied the transcript levels of severalsecondary metabolites that may contribute to soybean defense against herbivory.They are as follows:Bowman-Birk proteinase inhibitor(BBI),Kunitz trypsin inhibitor 1(KTI1), polyphenol oxidase(PPOJH2),nerol synthase(NES),flavonoid 3′-hydroxylase(SF3'H1),and1-aminocyclopropane-1-carboxylate oxidase(ACO)[43].
The Bowman-Birk type proteinase inhibitor(BBI)is expressed in a wide variety of plants and contributes to plant defense against insects[38,41].However,the constitutive transcriptlevels of BBI in untreated control plants were the same for both accessions.H.armigera treatment induced significantly higher BBI transcript levels than wounding treatment in both Tianlong 2 and ED059.After the treatment,however,ED059 showed higher BBI transcriptlevels than Tianlong 2.The transcriptlevels of BBI in systemic leaves of ED059 increased in both treatments,whereas the transcript levels of BBI were almost unchanged in systemic leaves of Tianlong 2(Fig.6-a).
Kunitz trypsin inhibitor 1(KTI1)has been proposed to defend plants against insects by inhibiting the activity of serine proteases[44].In digital gene expression profiling analysis,we found that a KTI1 gene was highly induced in ED059 after H.armigera treatment(unpublished data).Under normal conditions,ED059 has the same KTI1 transcript levels as Tianlong 2.H.armigera and wounding treatment both significantly increased the transcript levels of KTI1 in localbut not systemic leaves of ED059.H.armigera treatment induced no increase in KTI1 levels in local or systemic leaves of Tianlong 2,but wounding treatment induced an increase in both(Fig.6-b).
Polyphenol oxidase(PPOJH2)has been reported to function in nematode resistance[45,47].In our study,the transcript levels of PPOJH2 increased to high values in both ED059 and Tianlong 2;however,after 48 h of H.armigera treatment,local leaves of ED059 contained 50%more PPOJH2 than those of Tianlong 2.In contrast,PPOJH2 levels did not change significantly with wounding treatment or H.armigera treatment in Tianlong 2(Fig.6c).
Flavonoid 3′-hydroxylase(SF3′H1)and 1-aminocyclopropane-1-carboxylate oxidase(ACO)are just two of the key enzymes in the falvonoid biosynthesis pathway[48].Flavonoids can enhance the ability of plants to defend themselves against herbivores[49]. After 8 h of H.armigera treatment,the transcript levels of SF3′H1 and ACO increased in the local leaves of both Tianlong 2 and ED059,with higher levels of SF3′H1 and ACO found in ED059.After wounding treatment,local leaves of Tianlong 2 and ED059 showed no significant change in transcript levels of SF3′H1. However,after wounding treatment,transcript levels of ACO increased in local leaves of both Tianlong 2 and ED059.In contrast,after H.armigera and wounding treatment,transcript levels of ACO remained the same in systemic leaves of Tianlong 2 and ED059.However,after 0.5 h ofboth treatments,the transcript levels of SF3'H1 increased fourfold in systemic leaves of ED059 (Fig.6-d and e).

Fig.4–Tianlong 2 and ED059 show different levels of wound-and H.armigera-elicited JA expression.Tianlong 2 and ED059 plants were grown under the same conditions.The second fully expanded leaves were wounded with pliers or infested with 3 third-instar larvae of H.armigera.Individual leaves from three replicate plants were harvested at the indicated times after treatment.(a)Mean(±SE)JA concentrations in leaves harvested at the indicated times were measured with HPLC-MS/MS.The mean(±SE)transcript levels of AOC3 were quantified by qRT-PCR.(b)Mean(±SE)transcript levels of TD were quantified by qRT-PCR.Asterisks represent significantly different transcript levels between Tianlong 2 and ED059 at the indicated times (unpaired t-test;*,P<0.05;**,P<0.01;***,P<0.001;n=3).(c)The mean(±SE)transcript levels of COI1 were quantified by qRT-PCR.Asterisks indicate significantly different transcript levels between Tianlong 2 and ED059 at the indicated times (unpaired t-test;*,P<0.05;**,P<0.01;***,P<0.001;n=3).
Aside from directly defending themselves against herbivores,plants also produce volatile organic compounds such as terpenoids.In addition to serving in plant defense against herbivores,terpenoids can also mediate indirect defense by attracting the predators of herbivores[50]and can send insect defense signals from an attacked leaf to an unwounded leaf or to other nearby plants[46].NES is the most important enzyme for the biosynthesis of monoterpene,which is one of the most important indirect defense compounds induced by herbivores [1].We found that after 48 h,H.armigera treatment increased the transcript levels of NES in local leaves of ED059 by nearly 17-fold.After 24 h,wounding treatment also enhanced the transcript levels of NES in local leaves of ED059.However, neither H.armigera nor wounding treatment induced any increase in transcript levels of NES in systemic leaves of ED059 (Fig.6-f).
3.3.Functional characterization of Kunitz trypsin inhibitor 1 (KTI1)by transgenic technology
We used a method based on Agrobacterium-mediated stable transformation of A.thaliana to generate transgenic plants in order to investigate whether the gene KTI1 plays a role in insect resistance.In two homozygous independently transformedlines of KTI1,transcript levels increased in comparison with those in wild-type plants(Fig.7-a).
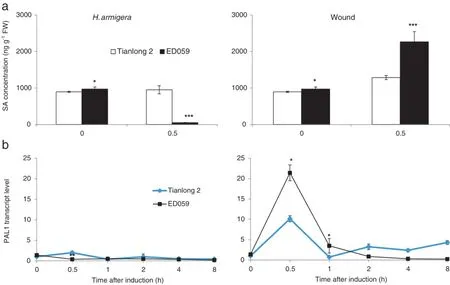
Fig.5–Tianlong 2 and ED059 have differentlevels ofwounding-and H.armigera-elicited SA expression.Tianlong 2 and ED059 plants were grown under identicalconditions.The second fully expanded leaves were wounded with pliers or infested with 3 third-instar larvae of H.armigera.Individual leaves from three replicate plants were harvested at the indicated times after treatment.(a)Mean(±SE)SA concentrations in leaves harvested at indicated times were measured by HPLC-MS/MS.Asterisks indicate significantly different transcript levels between Tianlong 2 and ED059 at the indicated times(unpaired t-test;*,P<0.05;**,P<0.01;***,P<0.001;n=3).(b)The mean(±SE)transcript levels of PAL1 were quantified by qRT-PCR.Asterisks indicate significantly different transcript levels between Tianlong 2 and ED059 at the indicated times(unpaired t-test;*,P<0.05;**,P<0.01;***,P<0.001;n=3).
The masses of newly hatched H.armigera larvae placed on plants overexpressing KTI1,EV and wild-type plants were determined 2,4,and 6 days after the start of the experiment. The results showed that H.armigera larvae feeding on plants overexpressing KTI1 gained less mass after 6 days of feeding than those grown on EV and wild-type plants(Fig.7-b and c). These results suggest that KTI1 plays an important role in defense against H.armigera.
3.4.Response of H.armigera larvae fed with Tianlong 2 and ED059
Previous studies have shown that toxic secondary metabolites such as protease inhibitors,nicotine,and polyphenol oxidase exist widely in plants and influence the physiology of insects' midgut,thereby affecting nutrient digestion and absorption [20].When larvae of H.armigera fed on ED059,transcript levels of BBI,KTI1,PPOJH2,NES,SF3′H1 and ACO increased.Based on this result,we studied the midguts of H.armigera larvae that fed on Tianlong 2 and ED059.
We performed TUNEL and DAPI staining to identify the presence of apoptotic cells in larvae midguts.Midgut cells of H.armigera larvae that had fed on ED059 showed green fluorescent signal,indicated apoptotic cells,whereas H. armigera larvae that had fed on Tianlong 2 showed no such signal(Fig.8-a and b).
In a previous study,larvae of H.armigera that fed on ED059 produced significantly less frass than those that fed on Tianlong 2[51,52].Interestingly,after 24 h of hunger,the midguts of larvae that fed on Tianlong 2 had almost no frass, whereas the midguts of larvae that fed on ED059 had large amounts of frass(Fig.8-a and 8-b).These results suggest that some antinutritive factors hindered nutrient absorption, utilization,and digestion in H.armigera larvae that had fed on ED059.
Interactions between insect and plants are complex;on one hand,plants can generate antiherbivore secondary metabolites,while on the other hand,insects can adapt and be tolerant to these secondary metabolites,by regulating expression of genes coding for acetylcholinesterase(ACE), carboxylesterase(CAR),glutathione S-transferase(GST),catalase(CAT),trypsin,and chitin synthase(CHS)[1,53].
We performed RT-PCR to obtain the expression profiles of ACE,CAR,GST,CAT,CHSB and trypsin in H.armigera larvae that had fed on ED059.We used the transcript levels of the genes expressed in H.armigera that fed on Tianlong 2 as the control.The transcript levels of CAT and GST were up-regulated(Fig.9-a and b),whereas those of CAR,CHSB,and trypsin were down-regulated(Fig.9-c,d, and e).H.armigera larvae that had fed on Tianlong 2 and ED059 showed no difference in transcript levels of ACE (Fig.9-f).
4.Discussion
We compared gene expression in two soybean accessions, Tianlong 2 and ED059,in response to an herbivore.We measured (1)morphological characteristics,such as trichomes and waxes on the leaf surface;(2)gene expression-levelpolymorphism(ELP) in early herbivore-induced response genes(protein kinases, transcription factors,phytohormone biosynthesis enzymes, and phytohormone signal transduction pathway)and anti-herbivore metabolites;(3)herbivore induced phytohormones,namely JA and SA;and(4)the molecular response of H.armigera larvae after being fed on ED059.
4.1.Difference in morphology between Tianlong 2 and ED059
Plant trichome and cuticular wax are the first lines of plant defense against environmental threats such as mechanical injury and pathogen and herbivore attacks[3,54,55].A sharp pubescence tip,denser trichomes,and large amounts of wax on leaves improve soybean insect resistance[56-58].In contrast,less wax on the leaf surface reduces insect feeding. For example,sorghum,cabbage,and Cercis canadensis var. mexicana(L.)have less wax but higher resistance to Phaonia soccata(W.),Brevicoryne brassica(L.),and Megachile sp.than plants that have more wax[32].
We found that ED059 has a sharp pubescence tip,dense trichomes,less crystal,and thick cuticle on the leaf surface compared to Tianlong 2.These morphological differences may underlie differences in insect resistance between Tianlong 2 and ED059.
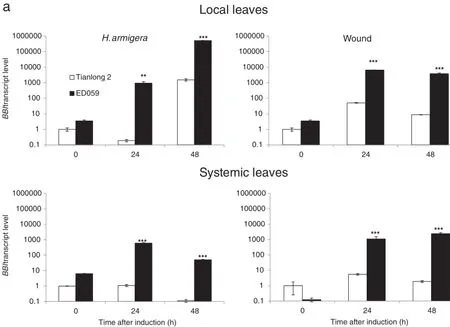
Fig.6–Anti-herbivory secondary metabolite genes in Tianlong 2 and ED059 show different levels of transcript accumulation in localand systemic leaves.Tianlong 2 and ED059 plants were grown under the same conditions.The second fully expanded leaves were wounded with pliers or infested with 3 third-instar larvae of H.armigera.Individualtreated leaves and third fully expanded untreated leaves fromthree replicate plants were harvested atthe indicated times after treatment.Mean(±SE)expression levels of BBI(a),KTI1(b),PPOJH2(c),SF3′H1(d),ACO(e),and NES(f)were quantified by qRT-PCR.The upper picture displays the expression levelof a gene in localleaves and the bottom picture displays the expression levelofthe same gene in systemic leaves.Asterisks indicate significantly different transcript levels between Tianlong 2 and ED059 at the indicated times(unpaired t-test;*,P<0.05;**,P<0.01;***,P<0.001;n=3).
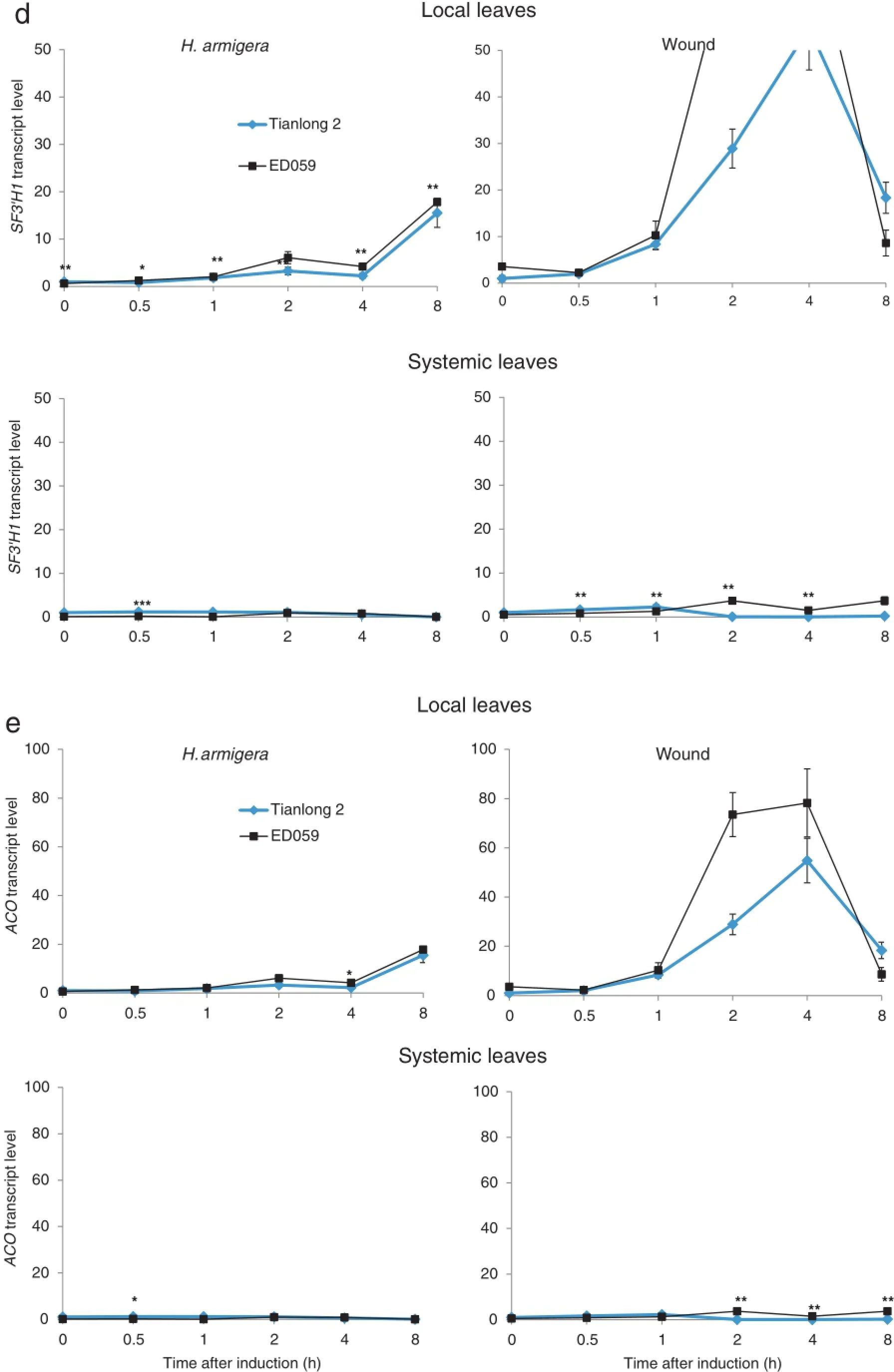
Fig.6(continued).
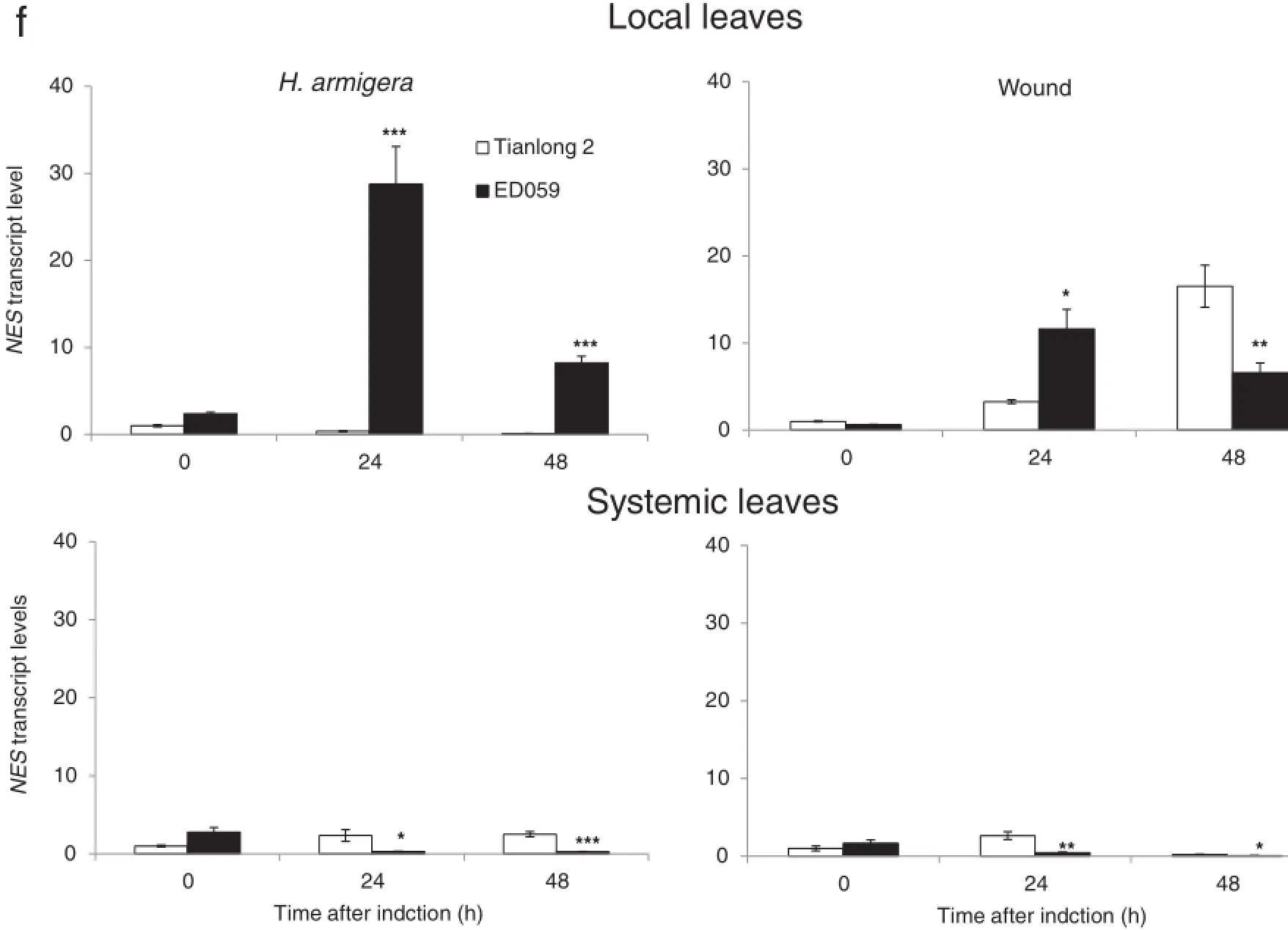
Fig.6(continued).
Pubescence on the surface of soybean plants is a highly important factor for insect resistance.For this reason,a useful avenue of research would be detection of QTLs for pubescence tip shape and density in ED059.The relationship between antixenosis resistance and pubescence characteristics should also be investigated,with comparison of the positions and effects of the corresponding QTLs.If the relationship between antixenosis resistance and pubescence characteristics is confirmed,insect-resistant lines can be selected based on their pubescence characteristics.
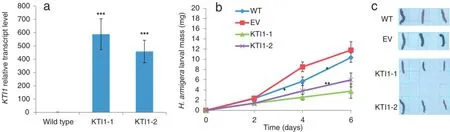
Fig.7–H.armigera larval performance on Arabidopsis thaliana plants overexpressing the KTI1 gene.(a)Mean(±SE)of KTI1 transcriptlevels in leaves oftransgenic plants.Numbers following the gene name represent different homozygous independently transformed lines.Asterisks indicate significantdifferences in KTI1 transcriptlevels between wild-type and KTI1 transgenic plants (unpaired t-test;***,P<0.001;n=3;three independent biologicalreplicates were used per genotype).(b)Mean(±SE)of H.armigera larvalmass after 2,4,and 6 days of feeding on wild-type(WT)plants,plants transformed with the empty vector(EV)and Arabidopsis thaliana plants overexpressing KTI1.Asterisks indicate significant differences between larvae feeding on wild-type plants and those feeding on different transgenic plants(unpaired t-test;*,P<0.05;n=10;10 independent biological replicates were used).(c)Example of H.armigera larvae at 6 days after feeding on WT plants,EV plants,and Arabidopsis thaliana plants overexpressing KTI1.
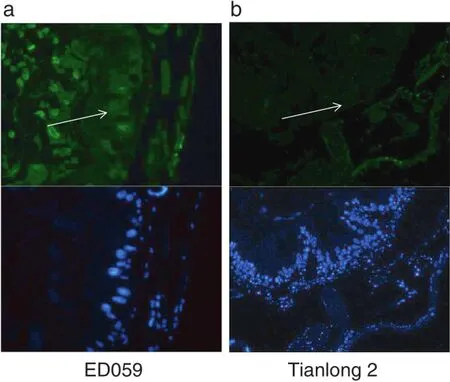
Fig.8–Detecting apoptosis in the midguts of H.armigera larvae feeding on Tianlong 2 and ED059.Newly hatched larvae were fed on Tianlong 2 and ED059 until the third-instar stage and then starved for 24 h.(a)Section of midgut 24 h after H.armigera feeding on ED059.(b)Section of midgut 24 h after H.armigera feeding on Tianlong 2.Arrows indicate apoptotic midgut cells. Green signal indicates DAPI and blue signal indicates TUNEL staining.
4.2.ELPs of genes underlying herbivore-induced responses in Tianlong 2 and ED059
Arabidopsis and tobacco are model plants and numerous in-depth studies of Arabidopsis/tobacco-insect interaction have recently been published.However,few reports on soybeaninsect interaction have been published.For this reason,we chose to focus on orthologs in Arabidopsis and tobacco in the current study.
MAPKsignaling is an early-acting signalin mediating plant resistance to herbivores,especially WIPK and SIPK[32]. MAPK1,MAPK3,and MAPK2 can be activated by pathogens and are orthologs of tobacco SIPK and WIPK,which regulate herbivore-induced JA biosynthesis[59].In the present study, the transcript levels of the three genes were evaluated in both accessions,and higher levels of gene transcripts were detected in Tianlong 2 than in ED059.Still,H.armigera subsequently induced higher levels of JA and several genes encoding transcription factors,JA biosynthesis enzymes (LOX3,AOC3,TD)and some secondary metabolites(BBI,KTI1, PPOJH2,NES,SF3′H1,and ACO)in ED059 than in Tianlong 2.In addition,fewer larvae fed on ED059 than on Tianlong 2, suggesting that although both accessions are equally able to perceive herbivores,ED059 needs only a few stimulito initiate herbivory-induced signaling pathways.
As a core signaling molecule,JA acts with other hormone pathways that integrate the information perceived atthe plantinsect interface into broad-spectrum defense responses[60].In the present study,H.armigera induced higher levels of JA,JA biosynthesis enzymes(AOC3),and the signaltransduction gene Cornatine insensitive 1(COI1)in ED059 than in Tianlong2. However,after H.armigera treatment,levels of SAand transcript levels of SA biosynthesis enzymes(PAL1)decreased in ED059. The function of SA in defense mechanisms of plants against herbivory remains unclear.In a previous study,Manduca sexta induced only minor accumulations of SA in N.attenuate.This finding suggests that SA plays only a minor role in plants' resistance to chewing insects.In comparison,phloem-feeding insects such as aphids and silverleaf whiteflies induced SA-dependent responses[61].In the present study,the herbivore defense in ED059 may be attributed to activation of the JA-dependent response pathway.
Analysis of the KTI1 gene by Agrobacterium-mediated stable transformation of A.thaliana identified KTI1,which participated in resistance to H.armigera.Newly hatched larvae of H. armigera fed for 6 d with plants overexpressed in KTI1 showed more than twofold lower mass than larvae grown on WT and EV controlplants(Fig.7).In this study,although we found that the KTI1 gene has a function in insect resistance,we used the heterologous plant Arabidopsis rather than soybean as a host plant to test the function of the gene KTI1.The results imply that the same function is conserved in Arabidopsis and soybean,and thus provides clues to its biological role in soybean.However,the results could also be the result of fortuitous interaction with a new set of partners present in Arabidopsis but not in soybean,and the results cannot explainthe defense mechanism of KTI1 gene in soybean.A thorough future study should reveal how the KTI1 gene functions in insect resistance in soybean and Arabidopsis.
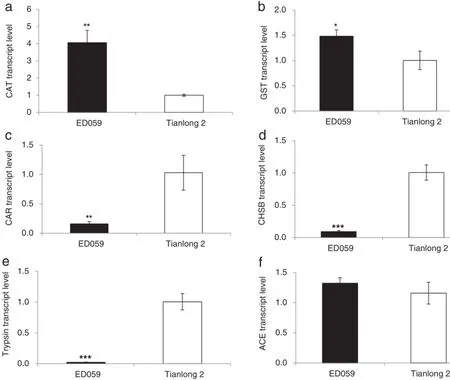
Fig.9–qRT-PCR analysis ofthird-instar larvae of H.armigera for transcriptlevels of ACE,CAR,GST,trypsin,and CHS.Newly hatched larvae were fed on Tianlong 2 and ED059 untilthe third-instar stage and then starved for 24 h.The mean(±SE)expression levels of CAT(a),GST(b),CAR(c),CHSB(d),trypsin(e),and ACE(f)were quantified by qRT-PCR.Asterisks indicate significantly different transcript levels between Tianlong 2 and ED059 at the indicated times(unpaired t-test;*,P<0.05;**,P<0.01;***,P<0.001;n=3).
The known differences in insect resistance level between Tianlong 2 and ED059 are unlikely to result solely from differences in genes studied in this investigation.A large and perhaps unimaginable amountofgenetic variation may underlie the insect defense response of ED059 and insect susceptibility of Tianlong 2.A clearer study of different herbivore-induced early responses and final anti-herbivore metabolites,asking whether these different expressed genes are involved in bollworm resistance,should provide an understanding of phenotypic variation between the two soybean accessions with respect to their herbivore defense systems.
4.3.How H.armigera larvae react to ED059
Using TUNEL and DAPI staining,we found that apoptotic cells are present in the midguts only oflarvae thatfed on ED059.If an insect eats toxic secondary metabolites of plants,its free radical metabolism in vivo may become abnormal,in turn inducing cellular oxidative stress and damage[62,63].CAT and GST eliminate oxidative stress and maintain the normal physiological activity of insects[64].In this study,newly hatched larvae were fed on Tianlong 2 until the third instar and then changed to feed on ED059.Transcript levels of CAT and GST were higher in H.armigera larvae that fed on ED059,suggesting that these larvae underwent oxidative stress and damage.
Transcript levels of KTI1 were higher in ED059 than in Tianlong 2.Trypsin inhibitor leads to excessive secretion of trypsin and ultimately to decreased trypsin mRNA levels in the midguts of herbivores[65].In this study,transcript levels of trypsin in H.armigera larvae that fed on ED059 were lower than those that fed on Tianlong 2.In addition,TPI-overexpressing transgenic plants were more resistant to insects than wild-type plants.Thus,our results suggest that trypsin inhibitors have anti-herbivore effects on ED059.
CHSB is mainly responsible for the formation of the peritrophic membrane in herbivore[66].Our finding that transcript levels of CHSB were decreased in H.armigera larvae that fed on ED059 suggests that ED059 plants contain toxic metabolites that prevent the formation of the peritrophic membrane in H.armigera larvae.
The function of ACE is to decompose the neurotransmitter acetylcholine that terminates nerve impulse transmission in the nervous system.Inhibiting ACE activity leads to insect death[66].Transcript levels of ACE were unchanged in H. armigera larvae that fed on ED059,indicating that the toxic secondary metabolites of ED059 do not contain insect neurotoxins.Interestingly,CAR is one ofthe important detoxificationenzymes in insects that reduce the toxicity ofplant's secondary metabolites.However,transcript levels of CAR in the H.armigera larvae that fed on ED059 decreased.It is possible that the CAR gene did not act in H.armigera larvae in response to ED059.
In conclusion,recent studies on interactions between Arabidopsis and various insect populations have revealed a large number of differences at transcriptomic,proteomic,and metabolomic levels.We predict that thousands of genes and proteins show polymorphism at the transcript and sequence levelbetween Tianlong 2 and ED059.With advances in genomic and proteomic technologies,we expect to find variations in more genes and proteins,ultimately contributing to a more comprehensive understanding of the different levels of insect resistance between Tianlong 2 and ED059.
Acknowledgments
This work was funded by the China Agriculture Research System(CAAS-04-PS08),the National Transgenic Project of China(2014ZX08004-005),and the Agricultural Science and Technology Innovation Program of China.We thank Dr. Weihua Ma of Huazhong Agriculture University for supplying eggs of H.armigera.
Supplementary material
Supplementary material to this article can be found online at http://dx.doi.org/10.1016/j.cj.2015.07.001.
R E F E R E N C E S
[1]R.M.Van Poecke,Arabidopsis-insect interactions,The Arabidopsis book/American Society of Plant Biologists,52007 e0107,http://dx.doi.org/10.1199/tab.0107.
[2]R.Mauricio,Ontogenetics of QTL:the genetic architecture of trichome density over time in Arabidopsis thaliana,Genetica 123(2005)75-85.
[3]D.J.Hulburt,H.R.Boerma,J.N.All,Effect of pubescence tip on soybean resistance to lepidopteran insects,J.Econ.Entomol. 97(2004)621-627.
[4]M.A.Jenks,S.D.Eigenbrode,B.Lemieux,Cuticular waxes of Arabidopsis,The Arabidopsis Book/American Society of Plant Biologists,12002 e0016,http://dx.doi.org/10.1199/tab.0016.
[5]B.N.Kunkel,D.M.Brooks,Cross talk between signaling pathways in pathogen defense,Curr.Opin.Plant Biol.5(2002) 325-331.
[6]G.I.Arimura,C.Kost,W.Boland,Herbivore-induced,indirect plant defences,BBA-Mol.Cell Biol.Lipids 1734(2005)91-111. [7]R.M.P.van Poecke,Indirect defence of plants against herbivores:using Arabidopsis thaliana as a model plant,Plant Biol.6(2004)387-401.
[8]J.W.Stratmann,C.A.Ryan,Myelin basic protein kinase activity in tomato leaves is induced systemically by wounding and increases in response to systemin and oligosaccharide elicitors,Proc.Natl.Acad.Sci.U.S.A.94 (1997)11085-11089.
[9]P.Reymond,H.Weber,M.Damond,E.E.Farmer,Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis,Plant Cell12(2000)707-720.
[10]J.Kahl,D.H.Siemens,R.J.Aerts,R.Gäbler,F.Kühnemann,C.A. Preston,I.T.Baldwin,Herbivore-induced ethylene suppresses a direct defense but not a putative indirect defense against an adapted herbivore,Planta 210(2000)336-342.
[11]R.Halitschke,I.T.Baldwin,Antisense LOX expression increases herbivore performance by decreasing defense responses and inhibiting growth-related transcriptional reorganization in Nicotiana attenuata,Plant J.36(2003) 794-807.
[12]A.Kessler,I.T.Baldwin,Defensive function of herbivoreinduced plant volatile emissions in nature,Science 291(2001) 2141-2144.
[13]C.A.Ryan,The search for the proteinase inhibitor-inducing factor,PIIF,Plant Mol.Biol.19(1992)123-133.
[14]C.A.Ryan,G.Pearce,Systemin:a polypeptide signal for plant defensive genes,Annu.Rev.Cell Dev.Biol.14(1998)1-17.
[15]J.W.Stratmann,Long distance run in the wounding response-jasmonic acid is pulling ahead,Trends Plant Sci.8 (2003)247-250.
[16]C.A.Ryan,D.S.Moura,Systemic wounding signaling in plants:a new perception,Proc.Natl.Acad.Sci.U.S.A.99 (2002)6519-6520.
[17]R.W.Doerge,Mapping and analysis of quantitative trait loci in experimental populations,Nat.Rev.Genet.3(2002)43-52.
[18]T.E.Coram,M.L.Settles,M.Wang,X.Chen,Surveying expression levelpolymorphism and single-feature polymorphismin near-isogenic wheat lines differing for the Yr5 stripe rust resistance locus,Theor.Appl.Genet.117(2008) 401-411.
[19]J.Wu,C.Hettenhausen,M.C.Schuman,I.T.Baldwin,A comparison of two Nicotiana attenuata accessions reveals large differences in signaling induced by oral secretions of the specialist herbivore Manduca sexta,Plant Physiol.146 (2008)927-939.
[20]X.Wang,H.Chen,A.Sha,R.Zhou,Z.Shang,X.Zhang,C. Zhang,L.Chen,Q.Hao,Z.Yang,D.Qiu,S.Chen,X.Zhou, Laboratory testing and molecular analysis ofthe resistance of wild and cultivated soybeans to cotton bollworm,Helicoverpa armigera(Hübner),Crop J.3(2015)19-28.
[21]B.X.Luo X,X.Sun,D.Zhu,B.Liu,W.Ji,H.Cai,L.Cao,J.Wu,M. Hu,X.Liu,L.Tang,Y.Zhu,Expression of wild soybean WRKY20 in Arabidopsis enhances drought tolerance and regulates ABA signalling,J.Exp.Bot.64(2013)2155-2169.
[22]Y.H.Li,F.Wei,X.Y.Dong,J.H.Peng,S.Y.Liu,H.Chen, Simultaneous analysis of multiple endogenous plant hormones in leaf tissue of oilseed rape by solid-phase extraction coupled with high-performance liquid chromatography-electrospray ionisation tandem mass spectrometry,Phytochem.Anal.22(2011)442-449.
[23]G.E.Metz,M.S.Serena,M.M.Abeyá,A.B.Dulbecco,A. Massone,S.Díaz,M.G.Echeverría,Equine arteritis virus gP5 protein induces apoptosis in cultured insect cells,Virus Res. 183(2014)81-84.
[24]H.Chen,C.G.Wilkerson,J.A.Kuchar,B.S.Phinney,G.A.Howe, Jasmonate-inducible plant enzymes degrade essentialamino acids in the herbivore midgut,Proc.Natl.Acad.Sci.U.S.A. 102(2005)19237-19242.
[25]M.Heinrich,I.T.Baldwin,J.Q.Wu,Protein kinases in plant growth and defense,J.Endocytobiosis Cell Res.22(2012)48-51. [26]A.Daxberger,A.Nemak,A.Mithöfer,J.Fliegmann,W. Ligterink,H.Hirt,J.Ebel,Activation of members of a MAPK module inβ-glucan elicitor-mediated non-host resistance of soybean,Planta 225(2007)1559-1571.
[27]T.Eulgem,P.J.Rushton,S.Robatzek,I.E.Somssich,The WRKY superfamily of plant transcription factors,Trends Plant Sci.5 (2000)199-206.
[28]S.H.Yang,A.D.Sharrocks,A.J.Whitmarsh,Transcriptional regulation by the MAP kinase signaling cascades,Gene 320 (2003)3-21.
[29]J.C.Moon,W.C.Yim,S.D.Lim,K.Song,B.M.Lee,Differentially expressed genes and in silico analysis in response to ozone (O3)stress of soybean leaves,Aust.J.Crop.Sci.8(2014) 276-283.
[30]S.Robatzek,I.E.Somssich,Targets of At WRKY6 regulation during plant senescence and pathogen defense,Genes Dev. 16(2002)1139-1149.
[31]X.Luo,X.Bai,X.Sun,D.Zhu,B.Liu,W.Ji,H.Cai,L.Cao,J.Wu, M.Hu,Expression of wild soybean WRKY20 in Arabidopsis enhances drought tolerance and regulates ABA signalling,J. Exp.Bot.64(2013)2155-2169.
[32]M.Kallenbach,F.Alagna,I.T.Baldwin,G.Bonaventure, Nicotiana attenuata SIPK,WIPK,NPR1,and fatty acid-amino acid conjugates participate in the induction of jasmonic acid biosynthesis by affecting early enzymatic steps in the pathway,Plant Physiol.152(2010)96-106.
[33]P.E.Staswick,I.Tiryaki,The oxylipin signaljasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis,Plant Cell 16(2004)2117-2127.
[34]Q.Wu,J.Wu,H.Sun,D.Zhang,D.Yu,Sequence and expression divergence of the AOC gene family in soybean: insights into functional diversity for stress responses, Biotechnol.Lett.33(2011)1351-1359.
[35]P.A.Gilardoni,C.Hettenhausen,I.T.Baldwin,G.Bonaventure, Nicotiana attenuata LECTIN RECEPTOR KINASE1 suppresses the insect-mediated inhibition of induced defense responses during Manduca sexta herbivory,Plant Cell23(2011)3512-3532.
[36]A.Paschold,R.Halitschke,I.T.Baldwin,Co(i)-ordinating defenses:NaCOI1 mediates herbivore-induced resistance in Nicotiana attenuata and reveals the role of herbivore movement in avoiding defenses,Plant J.51(2007)79-91.
[37]Z.Chen,Z.Zheng,J.Huang,Z.Lai,B.Fan,Biosynthesis of salicylic acid in plants,Plant Signal.Behav.4(2009)493-496. [38]O.L.Franco,S.C.Dias,C.P.Magalhaes,A.Monteiro,C.Bloch Jr., F.R.Melo,O.B.Oliveira-Neto,R.G.Monnerat,M.F.Grossi-de-Sa, Effects of soybean Kunitz trypsin inhibitor on the cotton boll weevil(Anthonomus grandis),Phytochemistry 65(2004)81-89.
[39]X.Y.Wang,L.H.Zhou,B.Xu,X.Xing,G.Q.Xu,Seasonal occurrence of Aphis glycines and physiological responses of soybean plants to its feeding,Insect Sci.21(2014)342-351.
[40]D.Sato,H.Akashi,M.Sugimoto,M.Tomita,T.Soga, Metabolomic profiling of the response of susceptible and resistant soybean strains to foxglove aphid,Aulacorthum solani Kaltenbach,J.Chromatogr.B Analyt.Technol.Biomed. Life Sci.925(2013)95-103.
[41]D.Kim,K.Lee,J.B.Kim,S.Kim,J.Song,Y.Seo,B.M.Lee,S.Y. Kang,Identification of Kunitz trypsin inhibitor mutations using SNAP markers in soybean mutant lines,Theor.Appl. Genet.121(2010)751-760.
[42]A.Nagamatsu,C.Masuta,M.Senda,H.Matsuura,A.Kasai,J.S. Hong,K.Kitamura,J.Abe,A.Kanazawa,Functional analysis of soybean genes involved in flavonoid biosynthesis by virus-induced gene silencing,Plant Biotechnol.J.5(2007) 778-790.
[43]J.M.Baek,S.Kim,Nucleotide sequence of a cDNA encoding soybean Bowman-Birk proteinase inhibitor,Plant Physiol. 102(1993)687.
[44]M.M.Shimizu,P.Mazzafera,Polyphenoloxidase is induced by methyljasmonate and Meloidogyne javanica insoybean roots but is not involved in resistance,Nematology 9(2007)625-632.
[45]S.Jain,D.K.Choudhary,Induced defense-related proteins in soybean plants by Carnobacterium sp.SJ-5 upon challenge inoculation of Fusarium oxysporum,Planta 239(2014)1027-1040.
[46]M.Zhang,J.Y.Liu,K.Li,D.Y.Yu,Identification and characterization of a novel monoterpene synthase from soybean restricted to neryl diphosphate precursor,PLoS One 8(2013),e75972.
[47]A.Nagamatsu,C.Masuta,H.Matsuura,K.Kitamura,J.Abe,A. Kanazawa,Down-regulation of flavonoid 3′-hydroxylase gene expression by virus-induced gene silencing in soybean reveals the presence of a threshold mRNA levelassociated with pigmentation in pubescence,J.Plant Physiol.166(2009)32-39.
[48]J.B.Harborne,R.J.Grayer,Flavonoids and insects,The Flavonoids,Springer,USA 1994,pp.589-618.
[49]N.Dudareva,E.Pichersky,J.Gershenzon,Biochemistry of plant volatiles,Plant Physiol.135(2004)1893-1902.
[50]I.Baldwin,J.C.Schultz,Talking trees,Science 221(1983)277-279.
[51]X.Ren,Z.Han,Y.Wang,Mechanisms of monocrotophos resistance in cotton bollworm,Helicoverpa armigera(Hübner), Arch.Insect Biochem.Physiol.51(2002)103-110.
[52]D.P.Bown,H.S.Wilkinson,J.A.Gatehouse,Differentially regulated inhibitor-sensitive and insensitive protease genes from the phytophagous insect pest,Helicoverpa armigera,are members of complex multigene families,Insect Biochem. Mol.Biol.27(1997)625-638.
[53]S.Eigenbrode,T.Castagnola,M.B.Roux,L.Steljes,Mobility of three generalist predators is greater on cabbage with glossy leaf wax than on cabbage with a wax bloom,Entomol.Exp. Appl.81(1996)335-343.
[54]H.Kanno,Role of leaf pubescence in soybean resistance to the false melon beetle,Atrachya menetriesi FALDERRMANN (Coleoptera:Chrysomelidae),Appl.Entomol.Zool.31(1996) 597-604.
[55]R.Bodnaryk,Leafepicuticular wax,an antixenotic factor in Brassicaceae thataffects the rate and pattern offeeding offlea beetles,Phyllotreta cruciferae(Goeze),Can.J.Plant Sci.72(1992) 1295-1303.
[56]S.D.Eigenbrode,K.E.Espelie,A.M.Shelton,Behavior of neonate diamondback moth larvae[Plutella xylostella(L.)]on leaves and on extracted leaf waxes of resistant and susceptible cabbages,J.Chem.Ecol.17(1991)1691-1704.
[57]S.D.Eigenbrode,S.K.Pillai,Neonate Plutella xylostella responses to surface wax components of a resistant cabbage (Brassica oleracea),J.Chem.Ecol.24(1998)1611-1627.
[58]S.Eigenbrode,C.White,M.Rohde,C.Simon,Behavior and effectiveness of adult Hippodamia convergens(Coleoptera: Coccinellidae)as a predator of Acyrthosiphon pisum (Homoptera:Aphididae)on a wax mutant of Pisum sativum, Environ.Entomol.27(1998)902-909.
[59]M.McConn,R.A.Creelman,E.Bell,J.E.Mullet,Jasmonate is essentialfor insect defense in Arabidopsis,Proc.Natl.Acad. Sci.U.S.A.94(1997)5473-5477.
[60]J.Wu,I.T.Baldwin,Herbivory-induced signalling in plants: perception and action,Plant CellEnviron.32(2009)1161-1174.
[61]A.Valavanidis,T.Vlahogianni,M.Dassenakis,M.Scoullos, Molecular biomarkers of oxidative stress in aquatic organisms in relation to toxic environmental pollutants, Ecotoxicol.Environ.Saf.64(2006)178-189.
[62]Z.Yang,F.Zhang,Q.He,G.He,Molecular dynamics of detoxification and toxin-tolerance genes in brown planthopper(Nilaparvata lugens Stål.,Homoptera: Delphacidae)feeding on resistant rice plants,Arch.Insect Biochem.Physiol.59(2005)59-66.
[63]R.E.Lynch,I.Fridovich,Autoinactivation ofxanthine oxidase: the role of superoxide radical and hydrogen peroxide, Biochim.Biophys.Acta 571(1979)195-200.
[64]C.A.Ryan,Protease inhibitors in plants:genes for improving defenses against insects and pathogens,Annu.Rev. Phytopathol.28(1990)425-449.
[65]X.Chen,H.Tian,L.Zou,B.Tang,J.Hu,W.Zhang,Disruption of Spodoptera exigua larvaldevelopment by silencing chitin synthase gene A with RNA interference,Bull.Entomol.Res. 98(2008)613-619.
[66]S.Keane,M.Ryan,Purification,characterisation,and inhibition by monoterpenes of acetylcholinesterase from the waxmoth,Galleria mellonella(L.),Insect Biochem.Mol.Biol.29 (1999)1097-1104.
8 March 2015
in revised form 3 July 2015
.
E-mail address:zhouxinan@caas.cn(X.Zhou).
Peer review under responsibility of Crop Science Society of China and Institute of Crop Science,CAAS
杂志排行
The Crop Journal的其它文章
- Effectofelevated[CO2]and nutrientmanagement on wetand dry season rice production in subtropicalIndia
- A nucleotide substitution at the 5′splice site of intron 1 of rice HEADING DATE 1(HD1)gene homolog in foxtail millet,broadly found in landraces from Europe and Asia
- Resistance to powdery mildew in the pea cultivar Xucai 1 is conferred by the gene er1
- Responses in gas exchange and water status between drought-tolerant and-susceptible soybean genotypes with ABA application
- Genetic gains in wheat in Turkey:Winter wheat for irrigated conditions
- Barnyard millet globalcore collection evaluation in the submontane Himalayan region of India using multivariate analysis
