Single-cell analyses of circulating tumor cells
2015-12-15XiXiChenFanBai
Xi-Xi Chen, Fan Bai
Biodynamic Optical Imaging Center, School of Life Science, Peking University, Beijing 100871, China
Introduction
Most cancer-related deaths are caused by metastasis.The presence of circulating tumor cells (CTCs) in peripheral blood was first observed in 18691; this phenomenon is an important“intermediate step” in cancer metastasis1.Studies on CTCs not only reveal the critical biological processes involved in cancer metastasis but also provide a non-invasive method for cancer detection, diagnosis, prognosis, and test of drug response.Research on CTCs has increased in recent years because of the vital role of CTCs in cancer metastasis and significant potential in clinical applications.In this review, we present a summary of these studies, with specific emphasis on single-cell analyses of CTCs.
CTCs and metastasis
A century has passed since the first observation of CTCs in cancer patients’ blood.The characterization of cancer metastasis has greatly advanced over time.The key steps can be summarized as intravasation, migration, and extravasation (Figure 1).
When metastasis initiates, the tumor cells in the primary site proliferate, thereby attracting blood vessels to provide nutrients and oxygen for tumor growth while giving the tumor cells a chance to invade the blood vessels2.Epithelial to mesenchymal transition (EMT) is speculated to play a critical role in this intravasation process.To intrude blood vessels,epithelium-originating tumor cells should lose cell polarity and tight junctions among different cells, thus forming spindlelike, mesenchymal-shaped cells and acquiring a higher motility power to break the defense line of the basement membrane and vascular wall3-5.Many cell surface markers, including the most commonly applied E-cadherin, vimentin, and fibronectin, are used to characterize the EMT process.Whether EMT is required in metastasis remains unclear.Several researchers reported that tumor cells cooperate with mesenchymal cells rather than transit into them to complete the metastasis process6.CTC is an ideal location to provide understanding on the issue, and direct investigations on CTCs mostly support the existence of this transition process7-11.Researchers identified epithelial and mesenchymal markers on CTCs, thereby suggesting that some CTCs express both markers, and they are in a partial EMT state12.
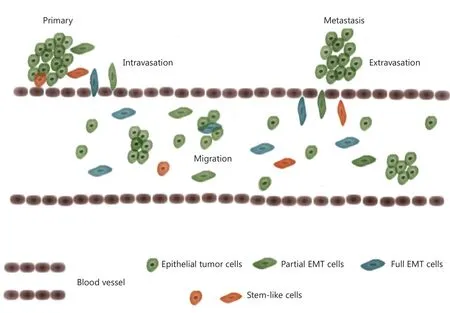
Figure 1 CTCs and metastasis.Tumor cells may transform to mesenchymal-like cells and intrude the blood vessel wall, entering blood streams.Tumor cells migrate as single or in microemboli.When located in appropriate distal sites, cells escape from blood streams (mesenchymal to epithelial transition maybe involved) and seed new tumor lesions.The population of CTCs includes both epithelial and mesenchymal-like cells,non-stem and stem-like cells.
Studies on CTCs can also provide clues on tumor initiation.CTCs represent a population capable of immigration and tumor initiation.A stem-like subpopulation exists in the tumor tissue,which possesses high ability of tumorigenesis13-15(Figure 1).For CTCs to seed metastasis, the entire process should have extremely low efficiency16.Even if CTCs succeed in intravasation,most of them cannot survive the tough environment in the blood stream and eventually die from anoikis.One of the main goals of CTC research is to identify the stem-like subpopulation within CTCs, which has real tumorigenesis ability.A successful characterization of these cells will provide significant clinical values.
CTCs sometimes aggregate to form microemboli (circulating tumor microemboli, CTM).In the early 1970s, researchers proved that injections of tumor cells in the form of microemboli into mouse significantly increase the efficiency of tumor initiation17,18.CTM may endow tumor cells advantages in survival and help to avoid anoikis.Meanwhile, the participation of mesenchymal cells may provide a “moving niche”, which gives CTCs strong viability and moving ability12.Tumor cells have the most effective initiation ability when they cooperate with mesenchymal cells19.Previous studies showed that when a combination of two tumor cell lines with clearly different metastatic abilities is injected into a mouse, approximately 90%of the metastatic tumors are multiclonal, whereas a cell line with low-metastatic ability can only form less than 10% of the metastasis when injected alone12,20.This result suggests that CTM formation could even increase the metastasis of weak metastatic tumor cells.
For several decades, strategies for metastasis research are mainly based on mouse models or cell lines.The lack of proper objects for research extensively limited further investigation.To date, studies on CTCs have created a new opportunity; CTCs are the true link between primary and metastatic tumors.These cells may carry valuable information of both primary and metastatic sites, as well as specific details of intravasation, migration, and extravasation.
Methods for CTC detection and enrichment
The main challenge in CTC research is their detection and enrichment, which requires the ability to detect one CTC out of almost 109normal blood cells21.Based on the known properties of tumor cells, numerous platforms for CTC detection and enrichment were developed and can be classified into two major categories: (I) immunochemistry-based methods, including positive selection (enrichment of epithelial marker-positive cells)and negative selection (depletion of CD45-positive blood cells);and (II) physical property-based methods (selection by cell size or electrical charge) (Figure 2).
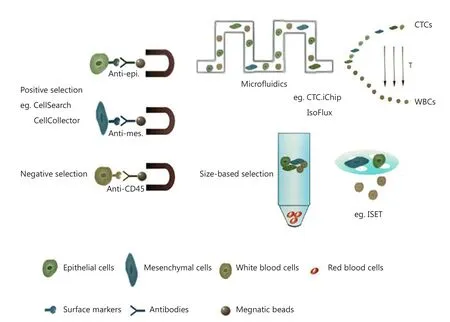
Figure 2 Methods for CTCs enrichment.CTCs enrichment can be based on their biological properties (e.g., surface markers) or physical properties (e.g., size).Surface marker-based methods include positive selection and negative selection.For CTCs detection, most applied marker in positive selection is EpCAM, and in negative selection is CD45.Subsequent enrichment can be managed by centrifugation or size-based filtration.Microfluidic-based enrichment can help to improve the sensitivity and recovery of target cells and can combine selection methods using both biological and physical properties.
Epithelial-marker based approaches are the most widely applied strategies for CTC detection.Among all the platforms,CellSearch from Janssen Diagnostics is considered the most successful.CellSearch is the only FDA-approved platform for CTC detection in clinical practice on patients with breast,prostate, and colorectal cancers.The practicability of CellSearch has been widely verified by many studies and referred as the“golden standard” for evaluating newly developed approaches22.CellSearch uses magnetic beads coupled with anti-EpCAM antibodies at the initial step to enrich EpCAM-positive epithelial cells and later stain the enriched cells for Cytokeratin (CK),CD45, and nucleus (DAPI) to capture CTCs (CK+CD45-DAPI+) and remove leukocytes (CK-CD45+DAPI+).
Currently, no other epithelial marker-based approach has performed better than CellSearch, but several platforms that implement microfludics have great potential.IsoFlux is a commercially availabe platform for CTC detection.This platform also relies on EpCAM-coupled-magnetic beads to capture CTCs, but the capture step is conducted in a microfluidic chip that can increase the chance to arrest CTCs with lower EpCAM expression.Both EpCAMlow(MDA-MB-231) and EpCAMhigh(SKBR3)-expressing tumor cells can be recovered with a successful rate of 74% and 85%, respectively23.Moreover,application of a combination of different epithelial markers (for example, EpCAM and MUC1) may also help to recover more epithelium-originating tumor cells24.The limitation of blood volume can also be a contributing factor to the low yield of CTC capture.CellCollector was developed to resolve this problem.This platform uses an EpCAM antibody-coated wire to capture CTCs in vivo25.However, metastasis often involves EMT; thus,epithelial marker-dependent approaches may miss numerous CTCs that have low or absent epithelial marker expression.Considering this phenomenon, the depletion of blood cells may be a supplementary method.
A chip-based platform CTC-iChip shows a good example of a combination of size-based selection and label-dependent enrichment26.This platform has positive (posCTC-iChip) and negative selection modes (negCTC-iChip).CTC-iChip initially applies a micro-column device to remove erythrocytes and cell fragments.Subsequently, the remaining mononuclear cells go through a second inertial focusing microfluidic device for further separation of EpCAM+ and CD45+ cells.Authors emphasized that even for several low-EpCAM-expressing cell lines,negCTC-iChip still achieved a high recovery.ISET27is another widely accepted size-based approach.This platform applies a specific membrane filter for tumor cell selection because tumor cells are often larger and stiffer than blood cells.Other size-based platforms include the ScreenCell and CanPatrol28.The main advantage for using membrane filter is that CTM can be retained for further investigation.Nonetheless, the sizes of tumor cells may vary significantly and not be always larger than blood cells because of the broad heterogeneity among tumor cells.Therefore, these size-based platforms need careful clinical validations.Furthermore, cells sticking on the filter are sometimes inconvenient for further manipulation; new devices were invented to address this problem, such as the Parsotix29and JETTA30systems.
Current clinical applications of CTCs
To date, CTC enumeration has been widely used as a prognostic marker for patients’ overall survival rate.CellSearch is the only FDA-approved system for CTC detection applicable in clinical practice.A cut-off value of ≥5 (breast and prostate cancers) or≥3 (colorectal cancer) in 7.5 mL blood is highly significant in predicting worse prognosis (Figure 3) (modified from Janssen Diagnostics, LLC, 2015)31-33.Counting the number of CTCs also shows predictive power in lung cancer34, melanoma35, head,and neck carcinoma36, as well as pancreatic cancer37.Even for several early-stage cancers, such as colorectal and breast cancers,CTC numeration also displays potential value in prognostic prediction38,39.
In addition to its role in prognosis prediction, changes in CTC number during medical treatment can also deliver valuable information of therapy response.Patients with a sharp decrease in CTC count after treatment often show better outcomes33,40.Accordingly, CTC enumeration can also help guide drug development41,42.
Single-cell analyses of CTCs
Besides enumeration, CTC characterization at the molecular level is also in progress.Key genetic variation events that are welldemonstrated in solid tumors are also assessed in CTC analysis.For example, in FISH and quantitative PCR implementation,researchers evaluate the TMPRSS2-ERG fusion, AR gain,and PTEN loss in CTCs, primary tumor, and metastatic site of prostate cancer patients43; the fusion event of TMPRSS2-ERG in CTCs is also used as a biomarker for sensitivity of abiraterone acetate treatment in castration-resistant prostate cancer patients44.Furthermore, the HER2 overexpression,which is an important determinant of therapy, in breast cancer can also be evaluated in CTCs at certain time points45.Another study conducted on lung cancer patients focused on the mutational status of the drug resistance-related gene EGFR in CTCs.Researchers collected CTCs from 12 patients, and 11 of them had activated mutations on EGFR in CTCs.Some of the mutations in CTCs seemed to be de novo, indicating that the mutations could not be detected in the primary tumor46.These studies demonstrate the potential use of CTCs as a realtime monitoring of response to therapy and guide in therapeutic planning.
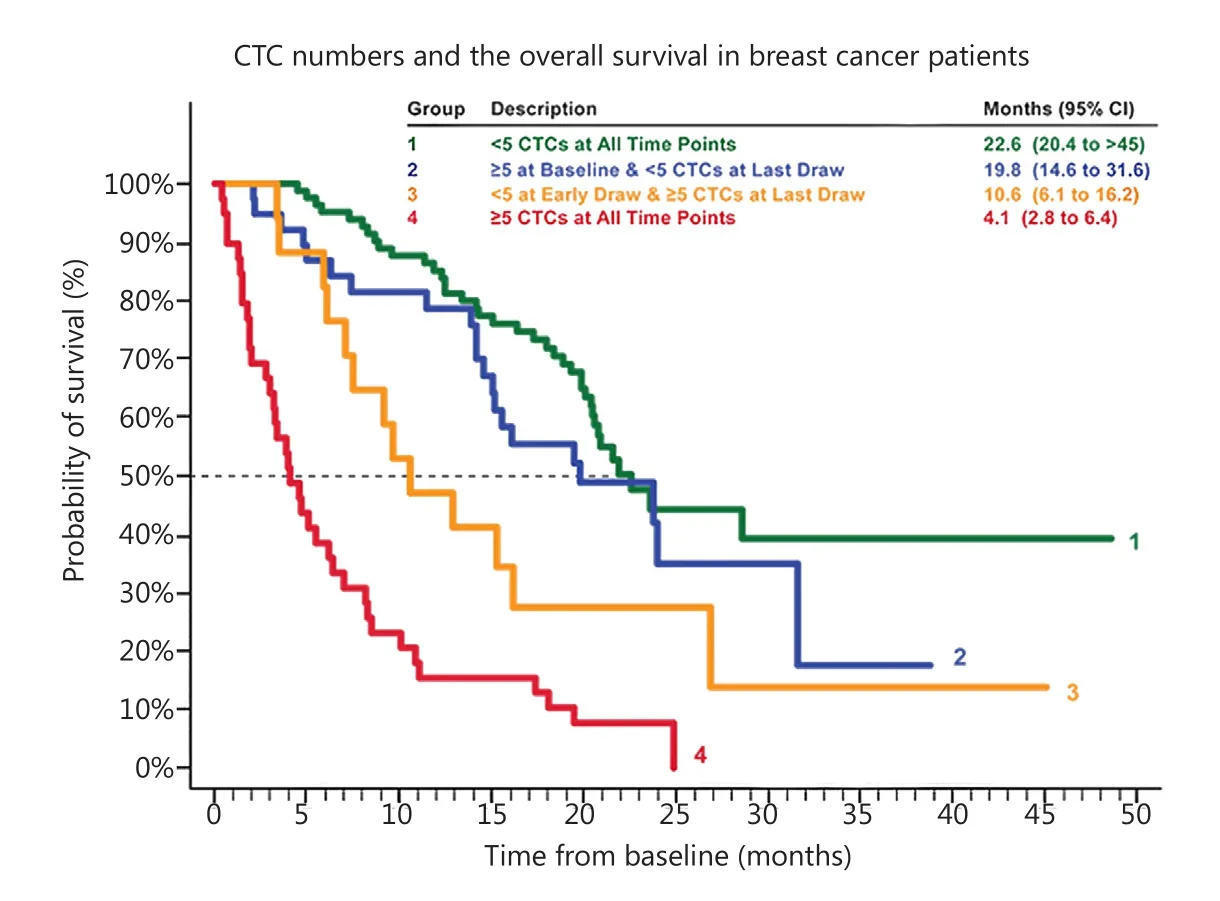
Figure 3 CTCs and prognostic prediction.The number of CTCs detected per 7.5 mL blood shows a strong correlation with the survival rate in breast cancer.If more than 5 CTCs detected, the survival rate is lowered to <50%.Similar results have been obtained in prostate cancer,colorectal cancer and lung cancer.(Modified from Janssen Diagnostics, LLC, 2015).
The rapid development of next-generation sequencing is accompanied by progress in genomic and transcriptomic characterization of CTCs to obtain a comprehensive understanding of a specific population of tumor cells.Meanwhile, sequencingbased analyses can identify novel genetic variation events during tumor development.Yu et al.8conducted the first transcriptome sequencing of isolated CTCs and identified some of the EMT-related genes with abnormal expression, including TGF-β and FOXC1, thereby suggesting the dynamic change of epithelial and mesenchymal composition.These results are in accordance with the observed RNA-FISH and immunofluorescence results.
CTCs are generally rare in patients’ blood; one of the main challenges for sequencing-based analyses is to exclude the contamination from white blood cells (WBCs).CTCs can be separated from WBCs according to surface markers by using flow cytometry or micromanipulation.However, this phenomenon brings another critical problem, that is, the remaining CTCs are insufficient to provide DNA or RNA for the next generation sequencing.Given the progress in developing whole genome amplification and whole transcriptome library construction,sequencing-based analyses for CTCs can be scaled down to single-cell level.In 2009, Tang et al.47improved the protocol for mRNA library construction to enable the mRNA-Seq of one single cell.Since the study was conducted, single-cell transcriptome analysis has been widely used.
The first single-cell transcriptome analysis of CTCs was conducted with the Smart-Seq method for full-length mRNASeq48.Researchers compared the libraries generated from six CTCs from a melanoma patient with those of other melanocyteoriginating cells (including primary melanocytes and melanoma cell lines), as well as blood, immune, and embryonic stem cells.The results proved the melanoma origin of the identified NG2+ CTCs.Moreover, the results identified important plasma membrane proteins with significant gain or loss of expression,including CDH1 and HLA1.
Whole genome amplification has always been a challenging step for any genomic analysis at the single-cell level.To date,the dominant method for single-cell genome amplification is multiple displacement amplification (MDA)49,50, which uses phi-29 polymerase, producing long amplicons in a strand-displacing method at 30 ℃.MDA provides relatively high fidelity in amplified products and is widely applied to the mutation detection of single cells.Nevertheless, the amplification bias of MDA is severe, which may cause distortion of copy number determination and allele dropout.In 2012, Sunny Xie’s group51developed multiple annealing and looping-based amplification cycles (MALBAC).This method significantly reduces the amplification bias and provides a high coverage and evenness across the whole genome.
The first whole-genome analysis for CTCs was published in 2012.In this work, Ni et al.52applied MALBAC for the wholegenome amplification of single CTCs.In this pioneering work,CTCs from seven lung adenocarcinoma patients (including a patient with a combination of adenocarcinoma and small cell lung cancer, and a patient with phenotypic transition from adenocarcinoma to small cell lung cancer) and four patients with small cell lung cancer carcinoma were isolated by CellSearch System.Subsequently, whole exome sequencing was performed on each single CTC to detect single nucleotide variations (SNVs), and low-depth whole-genome sequencing(~0.1×) was used to detect the copy number variations(CNVs).Ni et al.52reported that SNVs detected in single CTCs are highly heterogeneous.They also reported that the important mutations associated with drug target (EGFR),drug resistance (PIK3CA), and phenotypic transition (TP53,RB1) can all be detected in CTCs, thus implying the significant clinical value of CTC genomic analyses.Moreover, Ni et al.52found that regardless of the extensive heterogeneity in SNVs,almost all the CTCs from the same patient represented a reproducible CNV pattern.In lung adenocarcinoma patients, similarity in CNV pattern among different patients is also higher than expected.However, patients with SCLC exhibited distinct CNV pattern in their CTCs (Figure 4).Ni et al.52also reported that reproducibility in the CNV patterns of CTCs remains stable at different time points during treatment in one SCLC patient.
Similar works are also conducted on colorectal cancer53.Researchers isolated 37 intact CTCs from six patients with stage IV colorectal carcinoma by using CellSearch System; and a limited MDA was applied to amplify the whole genome.The amplified DNA was then subjected to comparative genome hybridization microarray for copy number evaluation, as well as massive parallel sequencing of a panel of 68 colorectalassociated genes for mutation detection.Many important genetic variation events previously reported in colorectal carcinoma are also detected in CTCs, including mutations in APC, KRAS,and PIK3CA.Previous studies also verified by targeted deep sequencing that most of the seemingly exclusive mutations in CTCs are also present in primary tumor at a relatively low frequency in minor subclones.Steinert et al.54also found that immune escape pathways are up-regulated in CTCs of colorectal cancer patients, thereby implying a mechanism for CTC survival in the bloodstream.

Figure 4 Genome features of single CTCs from lung cancer patients.CTCs were isolated from different lung cancer patients and the whole genome was amplified for sequencing.(A) Exome sequencing results suggested heterogeneity in SNVs of single CTCs from an individual patient.(B,C) Critical tumor mutations related to drug target, drug resistance, phenotypic transition can be detected in CTCs, including mutations on EGFR, PIK3CA, RB1, and TP53.(D) Differed from SNVs, the global CNV pattern is highly reproducible among CTCs from the same individual.(E) Even from different patients, CTCs share a relatively similar global CNV pattern.(Modified from Ni et al.52).
Lohr et al.55recently proposed a validated pipeline for whole exome sequencing of single CTCs.Isolated CTCs were amplified for the genome, and libraries were then constructed.Using low-depth sequencing, some poorly qualified libraries were eliminated; finally, by combining different whole exome libraries in one patient, they obtained a relatively accurate spectrum of mutations.However, this combination of libraries ignored the heterogeneity among different CTCs in an individual patient,which may also play important roles in metastasis.
Compared with blood samples collected directly from cancer patients, mouse models may be a better alternative for the biological characterization of CTCs.In certain cancer types, CTCs are difficult to isolate, which makes mouse model advantageous in these types of cancer.Ting et al.56applied their developed CTC-iChip device on an engineered pancreatic cancer mouse model (KPC) to isolate 168 single CTCs from five mice and managed to perform RNA-seq on 45% of the single CTCs.They found that CTCs show high level of multiple gene transcripts in the ECM pathway, which is normally expressed in reactive stromal cells rather than in epithelial cancer cells,including Dcn and Sparc.Sparc is a well-known ECM protein found in the stroma of primary human pancreatic ductal adenocarcinoma (PDAC).Ting et al.56also verified that SPARC overexpression may enhance the metastasis in pancreatic cancer by using shRNA-mediated knockdown test in two human PDAC cell lines with relatively high SPARC expression (PDAC2 and PDAC3).
Many factors may influence the single CTC analysis results.For example, in choosing platforms for CTC enrichment,EpCAM-based platform usually provide the highest capture purity, which would lead to minor blood cells contamination.But they are only suitable with those high-epithelial-markerexpressing CTCs.Negative selection or size-based selection may provide a broader range of CTCs detection, thus would be more proper when heterogeneity among different CTCs is emphasized.However, negative selection is often accompanied by higher rate of normal cells contamination, thus it requires further careful identification of “real” CTCs.Microfluidic devices may also help to increase the sensitivity of low-epithelialmarker-expressing CTCs detection.For transcriptome analysis,it requires cells to be intact, thus platforms causing much damage to the cells are not suitable.Besides enrichment methods,whole genome amplification methods or library construction methods should also be carefully selected according to different purpose of research.Taking the whole genome amplification as an example, MALBAC performs better for CNV capture, while MDA is more accurate for SNV detection.
Research on CTCs at single cell level holds much promise because of the heterogeneity among CTCs.Polzer et al.57reported that there are pre-existing CTCs resistant to ERBB2-targeted therapies, whose exploration may guide personalized treatment decisions.Yet currently the application of single CTC analysis in clinic is far from developed, mainly due to the limitation of CTC enrichment and isolation.Rare platforms could adapt to all types of cancer and the situations among different patients are highly diverse, making it hard to generally evaluate the efficiency or accuracy of single CTC analysis in clinic for now.
Conclusion and perspectives
Although CTC research is still in its infancy, it is a promising field of study because of its scientific significance in elucidating cancer metastasis and clinical value in non-invasive cancer detection, prognosis, and diagnosis.
Combining the advanced sequencing methods, CTC analyses show an apparent advantage over tumor tissues.The access to CTCs is much easier and less invasive than that to tumor tissues,which can only be obtained from surgery or biopsy samples.However, the current detection and isolation of CTCs are the bottleneck for CTC analyses.More works are needed to validate the efficiency of the newly developed platforms.Over the past decade, significant improvements have been made.Thus, we are highly confident that the sensitivity and efficiency of CTC detection will be improved in the near future to provide a solid base for the downstream genomic and functional analyses.
Conflict of interest statement
No potential conflicts of interest are disclosed
1.Ashworth TR.A case of cancer in which cells similar to those in the tumors were seen in the blood after death.Aus Med J 1869;14:146-149.
2.Fidler IJ.The pathogenesis of cancer metastasis: the ‘seed and soil’hypothesis revisited.Nat Rev Cancer 2003;3:453-458.
3.Thiery JP.Epithelial-mesenchymal transitions in tumour progression.Nat Rev Cancer 2002;2:442-454.
4. Thiery JP, Lim CT.Tumor dissemination: an EMT affair.Cancer Cell 2013;23:272-273.
5.Kalluri R.EMT: when epithelial cells decide to become mesenchymal-like cells.J Clin Invest 2009;119:1417.
6.Tsuji T, Ibaragi S, Shima K, Hu MG, Katsurano M, Sasaki A, et al.Epithelial-mesenchymal transition induced by growth suppressor p12CDK2-AP1 promotes tumor cell local invasion but suppresses distant colony growth.Cancer Res 2008;68:10377-10386.
7.Kallergi G, Papadaki MA, Politaki E, Mavroudis D, Georgoulias V,Agelaki S.Epithelial to mesenchymal transition markers expressed in circulating tumour cells of early and metastatic breast cancer patients.Breast Cancer Res 2011;13:R59.
8.Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, et al.Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition.Science 2013;339:580-584.
9.Hou JM, Krebs M, Ward T, Sloane R, Priest L, Hughes A, et al.Circulating tumor cells as a window on metastasis biology in lung cancer.Am J Pathol 2011;178:989-996.
10.Lecharpentier A, Vielh P, Perez-Moreno P, Planchard D, Soria J, Farace F.Detection of circulating tumour cells with a hybrid(epithelial/mesenchymal) phenotype in patients with metastatic non-small cell lung cancer.Br J Cancer 2011;105:1338-1341.
11.Balasubramanian P, Lang JC, Jatana KR, Miller B, Ozer E, Old M, et al.Multiparameter analysis, including EMT markers, on negatively enriched blood samples from patients with squamous cell carcinoma of the head and neck.PLoS One 2012;7:e42048.
12.Krebs MG, Metcalf RL, Carter L, Brady G, Blackhall FH, Dive C.Molecular analysis of circulating tumour cells-biology and biomarkers.Nat Rev Clin Oncol 2014;11:129-144.
13.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF.Prospective identification of tumorigenic breast cancer cells.Proc Natl Acad Sci U S A 2003;100:3983-3988.
14.Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M,Peschle C, et al.Identification and expansion of human coloncancer-initiating cells.Nature 2007;445:111-115.
15.Stewart JM, Shaw PA, Gedye C, Bernardini MQ, Neel BG,Ailles LE.Phenotypic heterogeneity and instability of human ovarian tumor-initiating cells.Proc Natl Acad Sci U S A 2011;108:6468-6473.
16.Fidler IJ.Metastasis: Quantitative Analysis of Distribution and Fate of Tumor Emboli Labeled With 125I-5-Iodo-2’-deoxyuridine2,3.J Natl Cancer Inst 1970;45:773-782.
17.Fidler IJ.The relationship of embolic homogeneity, number, size and viability to the incidence of experimental metastasis.Eur J Cancer 1973;9:223-227.
18. Thompson SC.The colony forming efficiency of single cells and cell aggregates from a spontaneous mouse mammary tumour using the lung colony assay.Br J Cancer 1974;30:332.
19.Duda DG, Duyverman AM, Kohno M, Snuderl M, Steller EJ,Fukumura D, et al.Malignant cells facilitate lung metastasis by bringing their own soil.Proc Natl Acad Sci U S A 2010;107:21677-21682.
20.Küsters B, Kats G, Roodink I, Verrijp K, Wesseling P, Ruiter DJ, et al.Micronodular transformation as a novel mechanism of VEGFA-induced metastasis.Oncogene 2007;26:5808-5815.
21.Yu M, Stott S, Toner M, Maheswaran S, Haber DA.Circulating tumor cells: approaches to isolation and characterization.J Cell Biol 2011;192:373-382.
22.Riethdorf S, Fritsche H, Müller V, Rau T, Schindlbeck C, Rack B,et al.Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: a validation study of the CellSearch system.Clin Cancer Res 2007;13:920-928.
23.Harb W, Fan A, Tran T, Danila DC, Keys D, Schwartz M, et al.Mutational Analysis of Circulating Tumor Cells Using a Novel Microfluidic Collection Device and qPCR Assay.Transl Oncol 2013;6:528-538.
24.Thege FI, Lannin TB, Saha TN, Tsai S, Kochman ML,Hollingsworth MA, et al.Microfluidic immunocapture of circulating pancreatic cells using parallel EpCAM and MUC1 capture: characterization, optimization and downstream analysis.Lab Chip 2014;14:1775-1784.
25.Saucedo-Zeni N, Mewes S, Niestroj R, Gasiorowski L, Murawa D, Nowaczyk P, et al.A novel method for the in vivo isolation of circulating tumor cells from peripheral blood of cancer patients using a functionalized and structured medical wire.Int J Oncol 2012;41:1241-1250.
26.Ozkumur E, Shah AM, Ciciliano JC, Emmink BL, Miyamoto DT,Brachtel E, et al.Inertial focusing for tumor antigen-dependent and-independent sorting of rare circulating tumor cells.Sci Transl Med 2013;5:179ra47.
27.Vona G, Sabile A, Louha M, Sitruk V, Romana S, Schütze K,et al.Isolation by Size of Epithelial Tumor Cells.Am J Pathol 2000;156:57-63.
28.Wu S, Liu Z, Liu S, Lin L, Yang W, Xu J.Enrichment and enumeration of circulating tumor cells by efficient depletion of leukocyte fractions.Clin Chem Lab Med 2014;52:243-251.
29.Joosse SA, Gorges TM, Pantel K.Biology, detection, and clinical implications of circulating tumor cells.EMBO Mol Med 2015;7:1-11.
30.Riahi R, Gogoi P, Sepehri S, Zhou Y, Handique K, Godsey J, et al.A novel microchannel-based device to capture and analyze circulating tumor cells (CTCs) of breast cancer.Int J Oncol 2014;44:1870-1878.
31.Cristofanilli M, Hayes DF, Budd GT, Ellis MJ, Stopeck A,Reuben JM, et al.Circulating tumor cells: a novel prognostic factor for newly diagnosed metastatic breast cancer.J Clin Oncol 2005;23:1420-1430.
32.Cohen SJ, Punt CJ, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY, et al.Relationship of circulating tumor cells to tumor response,progression-free survival, and overall survival in patients with metastatic colorectal cancer.J Clin Oncol 2008;26:3213-3221.
33.de Bono JS, Scher HI, Montgomery RB, Parker C, Miller MC,Tissing H, et al.Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer.Clin Cancer Res 2008;14:6302-6309.
34.Krebs MG, Sloane R, Priest L, Lancashire L, Hou J-M, Greystoke A, et al.Evaluation and prognostic significance of circulating tumor cells in patients with non–small-cell lung cancer.J Clin Oncol 2011;29:1556-1563.
35.Rao C, Bui T, Connelly M, Doyle G, Karydis I, Middleton MR,et al.Circulating melanoma cells and survival in metastatic melanoma.Int J Oncol 2011;38:755-760.
36.Nichols AC, Lowes LE, Szeto CCT, Basmaji J, Dhaliwal S,Chapeskie C, et al.Detection of circulating tumor cells in advanced head and neck cancer using the CellSearch system.Head Neck 2012;34:1440-1444.
37.Han L, Chen W, Zhao Q.Prognostic value of circulating tumor cells in patients with pancreatic cancer: a meta-analysis.Tumour Biol 2014;35:2473-2480.
38.Iinuma H, Watanabe T, Mimori K.Clinical significance of circulating tumor cells, including cancer stem-like cells, in peripheral blood for recurrence and prognosis in patients with Dukes’ stage B and C colorectal cancer.J Clin Oncol 2011;29:1547-1555.
39.Rack B, Schindlbeck C, Jückstock J, Andergassen U, Hepp P,Zwingers T, et al.Circulating tumor cells predict survival in early average-to-high risk breast cancer patients.J Natl Cancer Inst 2014;106:2504-2511.
40.de Bono JS, Logothetis CJ, Molina A, Fizazi K, North S, Chu L, et al.Abiraterone and increased survival in metastatic prostate cancer.N Engl J Med 2011;364:1995-2005.
41.Yap TA, Olmos D, Brunetto AT, Tunariu N, Barriuso J, Riisnaes R, et al.Phase I trial of a selective c-MET inhibitor ARQ 197 incorporating proof of mechanism pharmacodynamic studies J Clin Oncol 2011;29:1271-1279.
42.Bianchini D, Omlin AG, Pezaro CJ, Mukherji D, Lorente Estelles D,Zivi A, et al.First-in-human phase I study of EZN-4176, a locked nucleic acid antisense oligonucleotide (LNA-ASO) to androgen receptor (AR) mRNA in patients with castration-resistant prostate cancer (CRPC).J Clin Oncol 2013;31:abstr 5052.
43.Attard G, Swennenhuis JF, Olmos D, Reid AH, Vickers E, A’Hern R, et al.Characterization of ERG, AR and PTEN gene status in circulating tumor cells from patients with castration-resistant prostate cancer.Cancer Res 2009;69:2912-2918.
44.Danila DC, Anand A, Sung CC, Heller G, Leversha MA, Cao L, et al.TMPRSS2-ERG Status in Circulating Tumor Cells as a Predictive Biomarker of Sensitivity in Castration-Resistant Prostate Cancer Patients Treated With Abiraterone Acetate.Eur Urol 2011;60:897-904.
45.Fehm T, Müller V, Aktas B, Janni W, Schneeweiss A, Stickeler E, et al.HER2 status of circulating tumor cells in patients with metastatic breast cancer: a prospective, multicenter trial.Breast Cancer Res Treat 2010;124:403-412.
46.Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B,Collura CV, et al.Detection of mutations in EGFR in circulating lung-cancer cells.N Engl J Med 2008;359:366-377.
47.Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, et al.mRNA-Seq whole-transcriptome analysis of a single cell.Nat Methods 2009;6:377-382.
48.Ramsköld D, Luo S, Wang YC, Li R, Deng Q, Faridani OR, et al.Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells.Nat Biotechnol 2012;30:777-782.
49.Dean FB, Nelson JR, Giesler TL, Lasken RS.Rapid Amplification of Plasmid and Phage DNA Using Phi29 DNA Polymerase and Multiply-Primed Rolling Circle Amplification.Genome Res 2001;11:1095-1099.
50.Dean FB, Hosono S, Fang L, Wu X, Faruqi AF, Bray-Ward P, et al.Comprehensive human genome amplification using multiple displacement amplification.Proc Natl Acad Sci U S A 2002;99:5261-5266.
51.Zong C, Lu S, Chapman AR, Xie XS.Genome-wide detection of single-nucleotide and copy-number variations of a single human cell.Science 2012;338:1622-1626.
52.Ni X, Zhuo M, Su Z, Duan J, Gao Y, Wang Z, et al.Reproducible copy number variation patterns among single circulating tumor cells of lung cancer patients.Proc Natl Acad Sci U S A 2013;110:21083-21088.
53.Heitzer E, Auer M, Gasch C, Pichler M, Ulz P, Hoffmann EM, et al.Complex tumor genomes inferred from single circulating tumor cells by array-CGH and next-generation sequencing.Cancer Res 2013;73:2965-2975.
54.Steinert G, Scholch S, Niemietz T, Iwata N, Garcia SA, Behrens B,et al.Immune escape and survival mechanisms in circulating tumor cells of colorectal cancer.Cancer Res 2014;74:1694-1704.
55.Lohr JG, Adalsteinsson VA, Cibulskis K, Choudhury AD,Rosenberg M, Cruz-Gordillo P, et al.Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer.Nat Biotechnol 2014;32:479-484.
56.Ting DT, Wittner BS, Ligorio M, Vincent Jordan N, Shah AM,Miyamoto DT, et al.Single-cell RNA sequencing identifies extracellular matrix gene expression by pancreatic circulating tumor cells.Cell Rep 2014;8:1905-1918.
57.Polzer B, Medoro G, Pasch S, Fontana F, Zorzino L, Pestka A, et al.Molecular profiling of single circulating tumor cells with diagnostic intention.EMBO Mol Med 2014;6:1371-1386.
杂志排行
Cancer Biology & Medicine的其它文章
- Acute coronary syndrome: a rare case of multiple endocrine neoplasia syndromes with pheochromocytoma and medullary thyroid carcinoma
- Quantitative proteomic analysis for high-throughput screening of differential glycoproteins in hepatocellular carcinoma serum
- Development of targeted therapies in treatment of glioblastoma
- Advances in immunotherapy for treatment of lung cancer
- Fueling the engine and releasing the break: combinational therapy of cancer vaccines and immune checkpoint inhibitors
- Reality of evidence-based practice in palliative care
