PCR-DGGE Analysis of Bacterial Communities Structure in Babylonia areolata Culture Systems of The Subtidal Zone and The Pond Mulched Plastic Film and Sand in Bottom
2015-12-14ShufangLIDequanQIUJidongZHANGShipingYANGMingshengQIU
Shufang LI, Dequan QIU, Jidong ZHANG*, Shiping YANG, Mingsheng QIU
1. Agricultural College of Guangdong Ocean University, Zhanjiang 524088, China;
2. Fisheries College of Guangdong Ocean University, Zhanjiang 524088, China
Babylonia areolate is of gastropoda, neogastropoda, buccinacea, and radula. It grows fast and is resistant to diseases. The flesh of Babylonia areolate is very tender and is of high economical and nutritional value. Artificial cultivation of Babylonia areolate has been in China for nearly 30 years, and Babylonia areolate mostly suffers from shell-flesh separating disease and proboscis edema-disease. Once the disease happens to Babylonia areolate, the chance of being cured is slim, which brings about grave economic loss to farmers. The causes and mechanism of such disease are unknown[1].In China,the ways to raise Babylonia areolate include intertidal perse seine enclosure culture, shallow sea cage cul-ture,pond cultivation and onshore cement-pond culture[2-4], with each culture showing its own ecological features[2,5].From 2011 to 2012,we monitored the bacterial communities in Babylonia areolata culture systems of the sub-tidal zone and the pond mulched plastic film and sand in bottom,and found that the microorganism ecological level in the Babylonia areolata culture systems of the pond mulched plastic film and sand in bottom could be similar to the sub-tidal zone culture systems through changing the pond seawater and monitoring the microbial population[6]. However,the several kinds of vibrio isolated from Lyme disease spirochete and the photobacterium beijerinck fail to cause diseases to Babylonia areolata, which suggested that although those vibrio are not the direct pathogenic bacteria to the shell-flesh separating disease and proboscis edema-disease, the structure and number of bacterial communities in the ecological environment of culture system are closely related to the health of Babylonia areolata. The community structure of microorganism is an essential part of the ecological structure of Babylonia areolata, which not only affects the circulation and transformation of mass and energy in the ecological environment, but also influences the formation and function of normal fungi.Therefore,the policies to observe the Babylonia areolata ecological environment are of great significance to the ecological safety to adjust the cultivation system. In order to know the bacterial communities structure in Babylonia areolata culture systems and to research and optimize the management pattern of Babylonia areolata culture systems of the pond mulched plastic film and sand in bottom, we collected samples from a Babylonia areolata cultivation center in Zhanjiang in the middle ten days of May in 2013, and studied characteristics of the bacterial communities in Babylonia areolata culture systems of the sub-tidal zone and the pond mulched plastic film and sand in bottom based on DGGE (denaturing gradient gel electrophoresis) technology,which lay firm foundations for the studies of Babylonia areolata culture systems.
Materials and Methods
Sample collection and treatment
Collection of the pond mulched plastic film and sand samples The culture system of pond mulched plastic film and sand is to put the mulched plastic film into the pond based on the features of Babylonia areolata, and then to cover the 10 cm sterilized sand over the plastic film. Before raising Babylonia areolata,we disinfected and sunbathed the pond. Then, we filtered the seawater and put them into the pond to establish a pond cultivation system. The water was between 80 and 100 cm deep, and we changed 50%to 100%of water every two week.The samples for this experiment were collected from the pond where Babylonia areolata had been raised for about six months. We used sterilized water sampler to collect the surface water in the pond.The collected samples were saved for preservation and sent to the lab for tests.
Collection of samples in the subtidal zone The sub-tidal zone of Babylonia areolata lies in Zhanjiang City, Guangdong Province. There is about 1 square kilometer of land for raising Babylonia areolata, and the surroundings live other marine animals.We collected water and the sediment as 1.1.1 suggested.The collected sand samples in the bottom of the ocean are fine sand, with little mud in it. The collected samples were preserved at low temperature and sent for experiments.
Sample treatment and preservation The collected water and sediment samples were mixed sufficiently in the container, and numbered respectively.The sediment samples from the pond mulched plastic film and sand were S1, and the sediment samples from the sub-tidal zone were S2. The samples in the pond were S3,and the water samples in the sub-tidal zone were S4.Those samples were conserved in the fridge at-20 ℃.
Extraction of total DNA in the bacteria
The extraction of total DNA in the sediment samples (S1 and S2) was carried out according to the rapid extraction agent of DNA in soil samples(Sangon Biotech). The total DNA extraction of water samples (S3 and S4)was based on Yi Qi’s methods[7]. The obtained water samples were filtered by 10 μm filter membrane to remove impurities, and then we shredded the membrane with scissors into pieces before putting them into the EP tube.Ultraviolet spectrophotometer was applied to determine the concentration and purity of DNA, which was conserved at-20 ℃.
Bacteria 16SrDNA (V6 -V8 zone)amplification
According to Luo Pengs’ approach, bacteria 16rDNA(V6-V8 variation zone) gene amplification universal primer 968F/1401R 5’ end connected a GC (synthesized by Sangon Biotech). We considered the bacteria total DNA as the model, and we used 968F/1401R to amplify 16SrDNA. The total volume of PCR reaction system was 50 ml, 10×Buffer (including 2.0 mM MgCl2) 5 μl, dNTP (10 mM) 1 ml,968F-GC (10 mM) 1 ml, 1401 R(10 mM)1 ml, Taq enzyme (5 U/ml) 0.25 ml, model DNA 0.5 ml, adding ddH2O to 50 μl. The PCR reaction software was set at 94 ℃for 4 minutes, 94 ℃for 0.5 minutes, 56 ℃for 1 minutes,and 72 ℃for 0.5 minutes. There were 30 circulations,and the duration for 72℃lasted to 7 minutes. PCR produce electronic swim in 1.5%of agarose gel for 30 minutes. We used UVI gel image system to analyze the results(Gene Genius company).
Denaturing gradient gel electrophoresis and DGGE belt reclaiming
We collected the 16SrDNA (V6-V8 zone) PCR produce from each sample, and used D-Code general variation to detect Bio-rad for DGGE electrophoresis. The concentration of polyacrylamide gel was 8%(N-hydroxymethyl acrylamide(NMA)∶methylenebisacrylamide=37.5∶1).The denaturant concentration ranged from 30% to 60% (100%denaturant concentration was made up of 7 mol/L urea and 40%formamide). The electrophoresis at a stable temperature of 60 ℃under 180 V lasted for 4 hours. After dying decoloration,and the gel imaging,we analyzed the diversity and similarity of fungi in the samples by Quantity One software.
DGGE band cutting, clone and sequencing
We carefully cut down the bandfrom DGGE gel, and used UNIQ-10 high DNA gel extraction kid (Sangon Biotech), and used primers 968F/1401R for 16SrDNA amplification. We connected PCR to pUCm-T, and transformed DH5a cell over the X-gal/IPTG platform. We detected positive clones and white fungi by PCR, and sent them to Sangon Biotech.
16SrDNA sequence analysis
The obtained sequence was compared on the website http: // rdp.cme.msu.edu/seqmatch/seqmatch_intro.jsp, and obtained the most similar sequence as the sample 16SrDNA.We constructed the phylogenetic tree with 16SrDNA sequence.
Results and Analyses
The bacteria community DGGE fingerprint spectrum of two culture systems of Babylonia areolata and the diversity and similarity analysis of fungi
Fig.1 shows the DGGE fingerprint spectrum of bacteria community of the sediment samples from the culture system of the sub-tidal zone and the culture system of the pond mulched plastic film and sand (S1 and S2), and water samples (S3 and S4). We analyzed DGGE spectrum by Quantity one,and obtained the number of special belts in the samples, Shannonwiener index, Simpson index and Shannon concentration index (Table 1).Results suggested that the bacteria diversity of sediment samples from the pond mulched plastic film and sand(S1) was higher than that from the sub-tidal zone (S2). The bacteria diversity of both water samples was similar. The Shannon concentration index of the bacteria community of four samples all was 1, which showed the even distribution of bacteria communities of Babylonia areolata in two different culture systems.
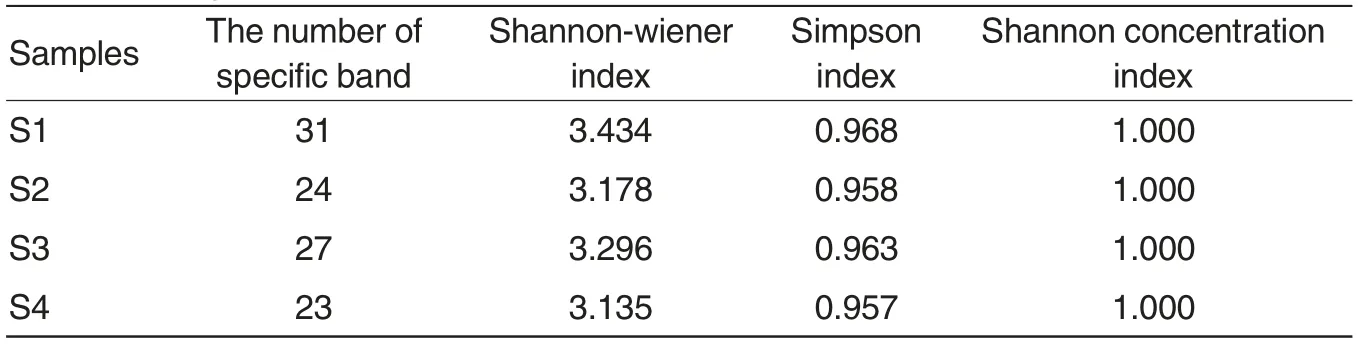
Table 1 The diversity and concentration of bacteria community of culture systems of Babylonia areolata
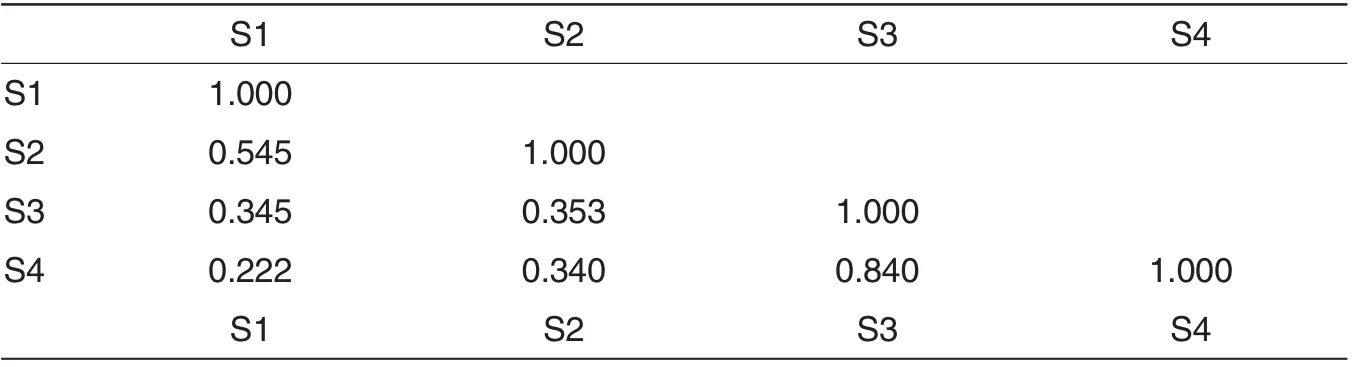
Table 2 Similarity of bacteria community structure of samples %
The microorganisms in the samples were treated with cluster analysis based on the unweight pair group method with arithmetic averages. Results (Fig.2 and Table 2) suggested that the four samples were classified into two groups. The sediment samples S1 and S2 were one group whose similarity was 54.5%, while the water samples S3 and S4 were one group whose similarity was 84.0%.The bacteria community structure of sediment samples and water samples of the same culture system were not similar,while the similarity of sediment sample S1 from the pond mulched plastic film and sand samples was 34.5% similar to the water sample S3, and the similarity of sediment sample S2 from the sub-tidal zone was 34.0% similar to the water sample S4.
The clone, sequencing and sequence analysis
The cloned and measured sequence of representative band was checked on the website http://rdp.cme.msu.edu/seqmatch/seqmatch_intro.jsp to get the sequence which was the most similar one to cutting gel. We set up the 16SrDNA(V6-V8 zone)sequence growth tree of the pond mulched plastic film and sand (Fig.3)and the 16SrDNA(V6-V8 zone)of the sub-tidal culture system(Fig.4).
As is shown in the phylogenetic tree of bacteria from the culture system of the pond mulched plastic film and sand (Fig.3), the bacteria community from the culture system of the pond mulched plastic film and sand included Proteobacteria, Chloroflexi,Actinobacteria and Cyanobacteria.Proteobacteria had the largest number of bacteria. Gammaproteobacteria included Vibrionaceae (band 17)and Photobacterium (band 27), Oceanospirillaceae (band 33), Francisellaceae (band 3), and Alteromonadaceae (band 30). Besides, there were several unclassified_Gammaproteobacteria (band 2, 22, 28, 34 and 36). Epsilonproteobacteria included Helicobacteraceae (band 4,9,12 and 13). Alphaproteobacteria involved Rhodobacteraceae (band 19).Deltaproteobacteria involved Desulfobacteraceae (band 6 and 18).Chloroflexi included Anaerolineaceae(band 37, 1 and 5). Cyanobacteria involved band 11 and 14. In addition,Actinobacteria included Nocardioidaceae(band 10).
According to the phylogenetic tree of bacteria from the sub-tidal culture system, the bacteria community of the system mainly included Proteobacteria, Chloroflexi, and Cyanobacteria.The proteobacteria involved some unclassified gammaproteobacteria (band 2, 22, 36, 28 and 34), Chromatiaceae and Photobacterium(band 17 and 27).The alphaproteobacteria included phodobacteraceae (band 19). The deltaproteobacteria included desulfobacteraceae (band 6). The chloroflexi mainly involved Anaerolineaceae (band 16 and 37). The Cyanobacteria largely included band 11,20 and 21.
Discussions
Because merely 1% microorganisms in the nature can be developed through normal approaches[9], DGGE technology modified in the corresponding denaturant concentration through different sequencing DNA images,which resulted in the decease in the electrophoresis speed, and stayed stable at last at the corresponding denaturant gradient for rapid and accurate identification of microorganisms in the nature or the artificial surroundings[10].Therefore,such technology has been widely used in studies on soil, mud, plants root, animals digestion, ocean and lake, etc.[11-14]This paper optimized the management pattern of Babylonia areolata culture systems of the pond mulched plastic film and sand in bottom by adopting the denaturing gradient gel electrophoresis (DGGE). The results indicated that the dominant bacterial communities in Babylonia areolata culture systems of the sub-tidal zone and the pond mulched plastic film and sand in bottom, which were built on the basis of the seawater in East-island of Zhanjiang, included Proteobacteria, Chloroflexi, Cyanobacteria and Actinobacteria.The dominant bacterial groups in the above pond culture system were Gammaproteobacteria, Alphaproteobacteria, Deltaproteobacteria, Epsilonproteobacteria, Anaerolineae, Cyanobacteria and Actinobacteria. The dominant bacterial communities in the subt-idal zone culture system were Gammaproteobacteria, Alphaproteobacteria, Deltaproteobacteria, Anaerolineae and Cyanobacteria, and there were less Epsilonproteobacteria and Actinobacteria in the culture system.The higher diversity was detected in the above two culture systems. The results of unweight pair group method with arithmetic average (UPGMA)showed that the bacterial communities of the sediment samples S1 and S2 in the above two culture systems were acluster,the similarity of bacterial communities was 54.5% . The bacterial communities of seawater samples S3 and S4 in the above culture systems were in clusters, and the similarity of the bacterial communities was 84.0%.The results showed that the microorganism ecological level in the Babylonia areolata culture systems of the pond mulched plastic film and sand in bottom could be similar to the sub-tidal zone culture systems through changing the pond seawater and monitoring the microbial population.
The culture system of pond mulched plastic film and sand was a kind of economic culture method since the structure within the pond was simple and was based on the habits of Babylonia areolata. However, the Babylonia areolata grew slowly than the ones that were raised by other methods.Studies found that the distribution of Gammaproteobacteria and Actinobacteria in the pond mulched plastic film and sand culture system were higher than that in the sub-tidal culture system, and the Gammaproteobacteria mostly were Desulfobulbus, Nitrosomonas and Pseudomonassp,which played an essential role in the mass and energy circulation process[16].Many Actinobacteria in the ocean can restrain and sterilize many kinds of insects and some virus.Some Nocardiaceae can decompose nitrile compound, which is good for environment purification[17]. Therefore, the similarity of bacteria community of the pond mulched plastic film and sand culture system and the sub-tidal culture system was up to 84.0%. Therefore, the pond mulched plastic film and sand culture system can be an essential culture approach to replace sub-tidal culture system to raise Babylonia areolata, in order to protect the sustainable development of marine ecology.
Vibrio and photobacteria were found in both Babylonia areolata culture systems. Studies by Dempsey[18]suggested that vibrio and photobacteria are the primary bacteria in the intestine of shrimps.The certain amount of both bacteria can contribute to the carbon source metabolism and use of intestines[19]. However, some bacteria of vibrio and photobacteria were major causes of some diseases in marine animals. Huang Yucong[20]proved that the swelling of Babylonia areolata was related to vibrio harveyi infection. Although the influences of other vibrio and photobacteria on Babylonia areolata were not verified, vibrio cholera and vibrio parahaemolyticus are major causes for diseases to human beings.Therefore, it is of great significance to control the number of vibrio bacteria in the Babylonia areolata culture systems to ensure the food safety of Babylonia areolata.
[1]WANG JG(王建钢),QIAO ZG(乔振国).A preliminary study on the causes of separation between meat and shell of Babylonia areolata (斑东风螺肉壳分离病病因的初步研究)[J].Journal of Modern Fisheries Information (现代渔业信息),2011,26(10):16-18.
[2]YI SW (尹绍武), LIAO JQ (廖经球),HUANG H (黄海), et al. Research advancement on biology and cultural ecology in Babylonia sp.(东风螺生物学及养殖生态学研究进展)[J]. 水产科学,2007,26(11):632-636.
[3]ZHENG GX(郑冠雄),XING YY(邢贻远).Grow-out trails of maculated ivory shell babylonia areolate in cement cisterm(方斑东风螺水泥池养成试验)[J].Fisheries Science&Technology Information(水产科技情报),200,33(1):46-48.
[4]PEI Kun (裴琨). Key techniques of industrial culture of moculated ivory shell Babylonia areolate (方斑东风螺工厂化养殖的关键技术)[J].Fisheries Science&Technology Informaiton(水产科技情报),2006,33(3): 107-108.
[5]LUO J(罗杰),DU T(杜涛),LIANG FL(梁飞龙), et al. Preliminary study on the cultivation method of Babylonia areolate Lamarck(方斑东风螺养殖方式的初步研究)[J]. Marine Science (海洋科学),2004,28(7):39-43.
[6]LI SF (李淑芳), QIU DQ (邱德全),ZHANG JD(张继东),et al.Study on microflora of pathogenic and conditional pathogenic bacteria in babylonia areolate attacked by shell-flesh separating disease and proboscis edema-disease(脱壳病和吻肿病东风螺体内致病菌及条件致病菌菌相研究)[J]. Advances in Marine Science (海洋科学进展),2013,31(2):266-271.
[7]YI Q(尹琦),SHI XC(史晓翀),DX(董雪),et al. Comparison study on four methods for microbial DNA extraction from seawater(四种海水样品总基因组DNA提取方法的比较研究)[J].Marine Environmental Science (海洋环境科学),2013,23(3):448-450.
[8]LUO P(罗鹏), HU CQ(胡超群), ZHANG LP(张吕平),et al.PCR-DGGE analysis of bacterial communities in marine Litopenaeus vannamei culture system(凡纳滨对虾海水养殖系统内细菌群落的PCR-DGGE 分析) [J]. Journal of Fishery Sciences of China(中国水产科学),2009,16(1):31-38.
[9]Ward DM, Bateson MM, Weller R, et al.Ribosomal RNA analysis of microorganisms as they occur in nature[J].Adv Microb Ecol,1992,12:219-286.
[10]XING DF(邢德峰), REN NQ(任南琪).Common problems in the analysis of microbial community by denaturing gradient gel electrophoresis (DGGE)(应用DGGE 研究微生物群落时的常见问题分析)[J]. Acta Microbiologica Sinica(微生物学报),2006,46(2):331-335.
[11]ZHANG ZD(张振冬), WANG SF(王淑芬), CAO YF (曹玉峰). DGGE technique and its application in study on microbial diversity in marine environment (DGGE 技术及其在海洋环境微生物多样性研究中的应用)[J]. Marine Environmental Science (海洋环境科学),2008,27(3):297-300.
[12]MAO YZ (毛玉泽),YAN TR (颜婷茹),ZHU L (朱玲),et al.Effect of mariculture activities on bacterial community structure in the water of Sanggou Bay(桑沟湾不同养殖区水体微生物群落结构特征)[J]. Journal of Fishery Sciences of China(中国水产科学),2013,20(4):824-831.
[13]JIN H(金浩), LI BL(李柏林), OU J(欧杰),et al.Microbial population diversity of activated sludge for wastewater treatment(污水处理活性污泥微生物群落多样性研究)[J].Journal of Microbiology(微生物学杂志),2012,32(4):1-5.
[14]CHEN MX(陈明霞),LI HY(李和阳), LI G(李刚),et al.The study on the diversity and distribution of vibrios and the correlation between them and their ambient environmental factors in Shenzhen costal waters (深圳海域弧菌种类组成、数量分布及其与环境因子的关系研究)[J]. Acta Oceanologica Sinica (海洋学报), 2012, 32(5): 117-125.
[15]LIU L (刘璐). Studies on the microbial diversity of Babylonia areolata marine cultivation ecological system (东风螺海水养殖生态系统内微生物多样性研究)[D].Hainan University,2011:7-49.
[16]WANG Y(王莹),HU CS(胡春胜). Research advances on community structure and function of denetrifiers(环境中的反硝化微生物种群结构和功能研究进展)[J]. Chinese Journal of Eco-Agriculture (中国生态农业学报),2010,18(6):1378-1384.
[17]ZHANG Y(张媛),ZHANG YY(张媛媛),LI ZJ (李振军), et al. Research progress on Nocardia (诺卡氏菌研究进展)[J].Chinese Journal of Zoonoses(中国人兽共患病学报), 2012, 28(6):628-634.
[18]DEMPSEY A C, KITTING C T, ROSSON RA. Bacterial variability among individual Penaei dshrimp digestive tract[J].Crustaceana,1989,56(3):267-278.
[19]MA SS(马生生),YU MC(于明超),LI ZJ(李卓佳). A review of studies on the effects and influencing factors of microflora in the digestive tract of shrimp(虾类消化道菌群研究进展)[J].Periodical of Ocean University of China(中国海洋大学学报),2007,37(6):889-893.
[20]HUANG YC(黄郁葱),JIAN JC(简纪常),WU ZH (吴灶和), et al. Preliminary study on the pathogenic bacteria isolated from proboscis edema-diseased Babylonia areolata(方斑东风螺吻管水肿病病原菌的初步研究)[J]. Fishery Modernization(渔业现代化),2009,36(4):37-41.
猜你喜欢
杂志排行
Agricultural Science & Technology的其它文章
- The Scale of Constructing Mountainous Cities in Yunnan Province based on"Benchmark"Farmlands
- The Measurement and Grading of Pressure on Cultivated Land in City-level in Yunnan Province
- Exploration of Water and Soil Conservation’s Function in Construction of Eco-environment
- Shelf-cultivation of Strawberry with Mulch and Heat Preservation Technology with Natural Energy
- Screening and Taxonomic Status of a Highly Efficient Antifungal Strain against Cytospora chrysosperma
- Adsorption Kinetics of NH4+by Purple Soils with Different pH Values
