Endophytic Fungi from Nicotiana tabacum L.and Their Antibacterial Activity
2015-05-17ZHOUKaiyiWANGWeixuanPENGYuYURuitingYUEYangLAIDaowanZHOULigangCollegeofAgronomyandBiotechnologyChinaAgriculturalUniversityBeijing0093ChinaTechnicalCenterofHunanTobaccoIndustryCoLtdChangsha4004China
ZHOU Kai-yi,WANGWei-xuan,PENG Yu,YU Rui-ting,YUE Yang,LAIDao-wan,ZHOU Li-gang* College of Agronomy and Biotechnology,China Agricultural University,Beijing 0093,China; Technical Center of Hunan Tobacco Industry Co.Ltd.,Changsha 4004,China
周开谊1,王伟轩1,彭 宇2,于瑞婷1,岳 阳1,赖道万1,周立刚1*
1中国农业大学农学与生物技术学院,北京100193;2湖南中烟工业有限责任公司技术中心,长沙410014
Introduction
Plant endophytic fungi are microorganisms that live within plant tissues without causing symptoms of disease[1,2].Endophytic fungi are rich of valuable bioactive metabolites with antioxidant,anti-viral,insecticid-
al,anti-tumor and antimicrobial activities[3-5].
Tobacco(Nicotiana tabacum L.),a member of the family Solanaceae,is one of themost important research model plants,and of high agricultural and economic value worldwide[6].Some endophytic fungi have been obtained from N.tabacum.Among them,Alternaria,Fusarium and Chaetomium species were dominant fungi[7,8].To the best of our knowledge,there was no report about antimicrobial activity screening of the endophytic fungi isolated from N.tabacum.This study aimed to further investigate the endophytic fungi from N.taba-cum,as well as to detect antimicrobial activity of the crude extracts from the fungi on pathogenic bacteria in order to provide the support data for investigating antimicrobialmetabolites aswell as for developing biocontrol agents.
M aterials and M ethods
Plantmaterials
The fresh and whole plants of three years old N.tabacum(cv.Xiangyan No.3)were collected from the experimental field of China Agricultural University in July 2013.The healthy plant samples(stems and roots)were carefully excised from the host and stored in the sealed plastic bags at4℃for processingwithin 24 h of collection.
Isolation of the endophytic fungi
The stem and root explants of N.tabacum were firstly rinsed with tap water to remove the attached microbes and particles,and were then cut into 5.0 cm length.They were surface-sterilized with 75%ethanol for 2 min and resurface-sterilized with 2%sodium hypochlorite(NaClO)for 20 min.Finally,these surface-sterilized samples were rinsed five times with sterile distilled water and placed on sterile filter paper.After the dried explantswere cut into small pieces of0.5 cm ×0.5 cm,they were placed on potato dextrose agar(PDA)plates containing 500μg/mL of streptomycin sulfate and incubated at25℃in darkness to eliminate any bacterial growth until themycelia were apparent at the edge of the explants.The pure cultureswere finally isolated by the hyphal tip isolation,and cultured on PDA plateswithout antibiotics.
In order to prove that the obtained fungi were isolated from the inside segments(not isolated from the surface),the last rinsed sterile distilled water from the explants was spread on PDA plates,and no fungus was found to emerge after 48 h cultivation.
M orphological characterization
Themorphological characterization of the isolated fungi were observed and described based on the method of Ainsworth et al.[9]and other references[10-14],including colony texture and color,type of conidiophores and mycelia,growth rate.
DNA extraction,ITS-rDNA am plification and sequence analysis
Analysis of the ITSsequences of rDNA regionswere also used to identify endophytic fungi.The total genomic DNA extraction of the isolated fungi was based on the previousmethods[15,16].The ITS region with the primer pair ITS1(5'-TCCGTAGGTGAACCTGCGG-3')and ITS4(5'-TCCTCCGCTTATTGATATGC-3')was amplified by the polymerase chain reaction(PCR).To identification,the PCR productswere purified using the QIA quick gel purification kit(Hilden,Qiagen,Germany)as described by the protocol ofmanufacturer and sequenced using the primer pair ITS1 and ITS4 on the ABI PRISM 3730 sequencer(Applied Biosystem,USA).The sequences of the endophytic fungal isolates were run by BLAST program against the database(National Center for Biotechnology Information website:http://www.ncbi.nlm.nih.gov),and then they were submitted to GenBank database to obtain the accession numbers.
M ycelia suspension culture and extract preparation Fourmycelia plugs from the edge of the actively growing fungal colony were inoculated into 300 mL Erlenmeyer flasks containing 100 mL potato dextrose broth(PDB).The PDB cultures were incubated at 150 rpm on a rotary shaker at 25 ℃ for 20 days.After suspension culture,the fermented broth was filtrated under vacuum to afford the filtrate and mycelia.The filtrate was extracted thrice with an equal volume of ethyl acetate(1∶1,v/v).The mycelia were lyophilized and powdered,followed by extraction with ultrasound in methanol(0.1∶5,g/mL)for three times.The crude extracts from themycelia and filtrate were obtained by evaporation under vacuum,respectively.
Detection of antibacterial activity of the crude extract
The thin layer chromatography(TLC)-bioautography assay was used to detect the antibacterial activity of the ethyl acetate or methanol extracts[17].Six bacterial strains including Agrobacterium tumefaciens(G-),Bacillus subtilis(G+),Pseudomonas lachrymans(G-),Ralstonia solanacearum(G-),Staphylococcus haemolyticus(G+),and Xanthomonas vesicatoria(G-)from Department of Plant Pathology of China Agricultural U-niversity,were selected for antibacterial assay.The developed TLC platewhich was covered with the testbacteria,was incubated at28 ℃ for12 h,then 0.5mg/mL of3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide(MTT,purchased from Amresco,USA)was equably sprayed on the TLC plate,after that the TLC plate was incubated successively for another 2 min.The antibacterial activity of the extractswas determined by the formation ofwell-defined inhibition zones which wasmade visible by spraying MTT thatwas converted to the formazan dye by the livingmicroorganism[18].Antibacterial activity was detected as the white inhibition zones against a purple background,and the length of each antibacterial area wasmeasured in order to calculate its Rf value:Rf=D1/D2,where,D1is the distance(mm)between the antimicrobial area and initial sample point,and D2is the distance(mm)between the developing solvent front and initial sample point on a TLC plate.The diameter(mm)of the antibacterial area was alsomeasured[13].
Results and Discussion
Identification of the endophytic fungi
A total of 38 endophytic fungal isolates were obtained from the roots and stems of N.tabacum.According to theirmorphological features,23 representative fungal isolates were selected for further identification.Both morphological traits and ITS-rDNA gene sequence analysiswere used to identify these isolates.Based on the results of macro and microscopic identification,they were identified as 18 genera,namely Acremonium(isolates Nitaf01 and Nitaf02),Stephanonectria(isolate Nitaf03),Clonostachys(isolate Nitaf04),Colletotrichum(isolate Nitaf05),Ilyonectria(isolate Nitaf06),Fusarium(isolates Nitaf07,Nitaf08 and Nitaf09),Petriella(isolate Nitaf10),Plectosphaerella(isolates Nitaf11 and Nitaf12),Purpureocillium (isolate Nitaf13),Chaetomium(isolate Nitaf14),Podospora(isolate Nitaf15),Thielavia(isolate Nitaf16),Myriodontium(isolate Nitaf17),Penicillium (isolates Nitaf18 and Nitaf20),Cladosporium(isolate Nitaf19),Alternaria(isolate Nitaf21),Rhizopycnis(isolate Nitaf22)and Mortierella(isolate Nitaf23)(Table 1).Among them,Acremonium,Fusarium,Penicillium and Plectosphaerella were dominant genera.To our knowledge,13 genera(Acremonium,Cladosporium,Clonostachys,Ilyonectria,Mortierella,Myriodontium,Petriella,Plectosphaerella,Podospora,Purpureocillium,Rhizopycnis,Stephanonectria and Thielavia)were isolated from N.tabacum for the first time.
The 23 different ITS1-5.8S-ITS4 partial sequences of the fungal isolates were submitted to the GenBank to get their accession numbers(i.e.KM514480,KM095507-KM095528).According to BLAST analysis,the closest related species were obtained(Table 1).Except for the fungi Nitaf16 and Nitaf17,other isolated endophytic fungi had more than 99%similarity with their closest relative species.The molecular characters of the fungal isolates were basically consistent with theirmorphological ones.

Table 1 Closest relatives of the endophytic fungal isolates based on BLAST analysis and morphological identification
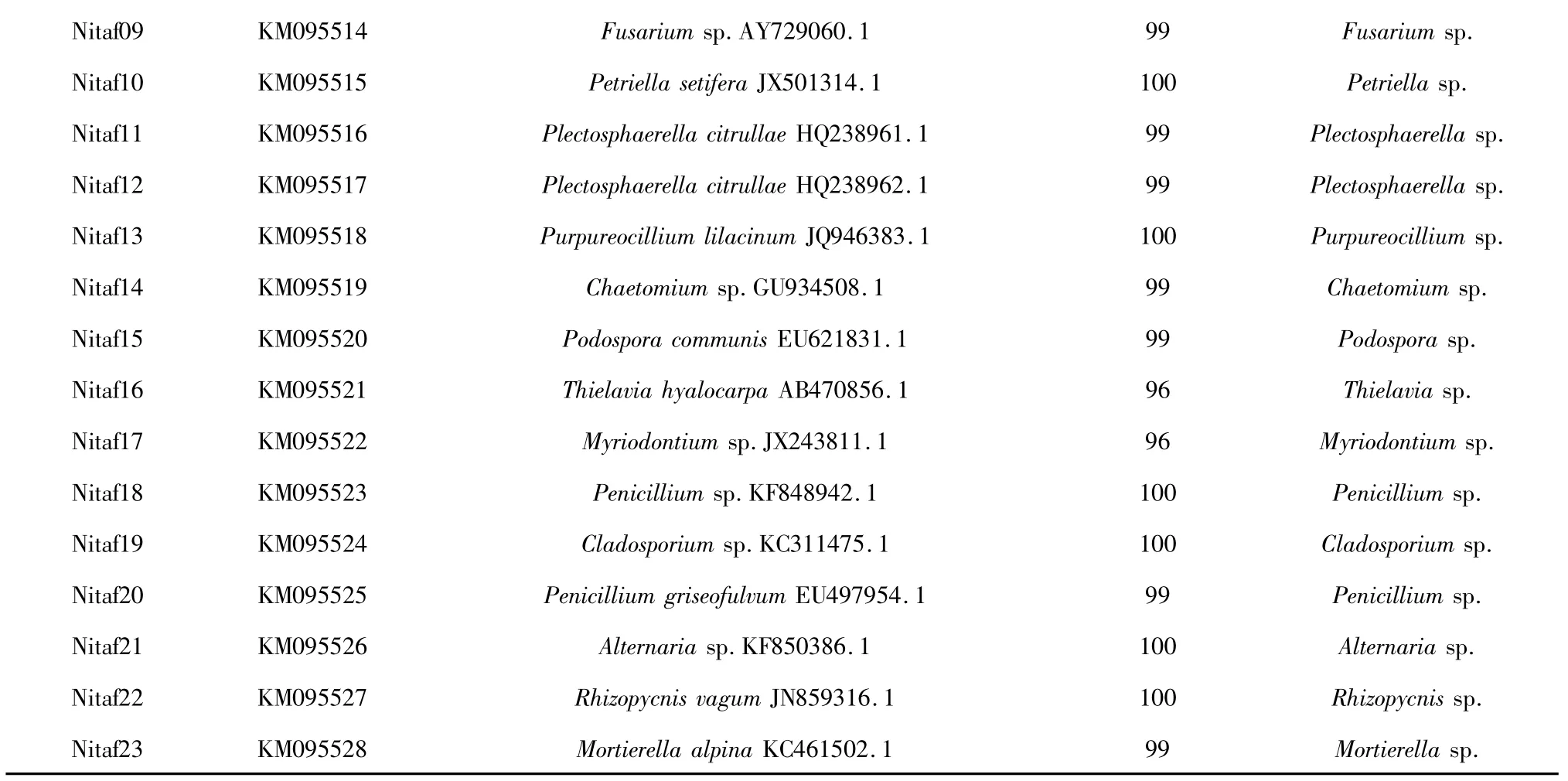
Nitaf09 KM095514 Fusarium sp.AY729060.1 99 Fusarium sp.Nitaf10 KM095515 Petriella setifera JX501314.1 100 Petriella sp.Nitaf11 KM095516 Plectosphaerella citrullae HQ238961.1 99 Plectosphaerella sp.Nitaf12 KM095517 Plectosphaerella citrullae HQ238962.1 99 Plectosphaerella sp.Nitaf13 KM095518 Purpureocillium lilacinum JQ946383.1 100 Purpureocillium sp.Nitaf14 KM095519 Chaetomium sp.GU934508.1 99 Chaetomium sp.Nitaf15 KM095520 Podospora communis EU621831.1 99 Podospora sp.Nitaf16 KM095521 Thielavia hyalocarpa AB470856.1 96 Thielavia sp.Nitaf17 KM095522 Myriodontium sp.JX243811.1 96 Myriodontium sp.Nitaf18 KM095523 Penicillium sp.KF848942.1 100 Penicillium sp.Nitaf19 KM095524 Cladosporium sp.KC311475.1 100 Cladosporium sp.Nitaf20 KM095525 Penicillium griseofulvum EU497954.1 99 Penicillium sp.Nitaf21 KM095526 Alternaria sp.KF850386.1 100 Alternaria sp.Nitaf22 KM095527 Rhizopycnis vagum JN859316.1 100 Rhizopycnis sp.Nitaf23 KM095528 Mortierella alpina KC461502.1 99 Mortierella sp.
Detection of antibacterial activity

Fig.1 Exam p les of antibacterial activity screening of the fungal extracts on Agrobacterium tumefaciens by TLC-bioautography assayNote:The developing solvent system in TLCwas CH2 Cl2-MeOH(15∶1,v/v).A,TLC results of themyceliamethanol extracts of the fungal isolates Nitaf 05,Nitaf 07,Nitaf 20 and Nitaf22 observed under UV 254 nm;B,antibacterial activity of the same TLC plate.The positive control(CK+)was streptomycin sulfate which was only sampled on the TLC plate and showed antibacterial activity.
The antibacterial activity results of the crude extracts by TLC-bioautography assay were shown in Table 2.The examples of antibacterial component screening of the extractswere shown in Fig.1.The Rf values of the antimicrobial areas usually revealed the relative polarity of the active compounds in the samples,and the diameters can indicate the relative antimicrobial activity of the compounds[13,14].Most of the extracts showed antibacterial activity to a certain extent except for the extracts of the isolates Nitaf06,Nitaf08,Nitaf11,Nitaf15 and Nitaf16.The compounds with antibacterial activity existed in both mycelia and filtrate extracts.The extracts of five fungal isolates(Nitaf05,Nitaf07,Nitaf13,Nitaf20 and Nitaf22)exhibited stronger antibacterial activity than those of the others.

Table 2 Antibacterial activity of the fungal extracts by TLC-bioautography-MTT assay

nd 0.65(++) 0.8(+) nd nd 0.71(+)Nitaf02 M nd nd 0.85(+) nd nd 0.69-0.92(+)F nd 0.77(+) nd nd nd 0.69(+)Nitaf03 M nd nd 0.38-0.65(+) nd nd nd F 0-0.29(+) nd nd 0(++) 0(++) nd Nitaf04 M 0.95(+) nd 0.15(+) nd nd nd F 0-0.58(+) 0(++) 0-0.58(++) 0(++) 0(+) 0(+)Nitaf05 M 0.77(+++) 0.62(+) 0.60(+) 0.30(+) 0.25(+) 0.48(+)F 0.57(++)0.60-0.77(+++)0.60(+) 0.50(+) 0.58(+++) 0.48(+)Nitaf06 M nd nd nd nd nd nd F nd nd nd nd nd nd Nitaf07 M 0.38-0.71(+)0.57-0.77(+) 0.62(+) 0.42(+) 0.25-0.50(+)0.38-0.75(++)F 0.50(+) 0.55(++)0.51-0.77(++) 0.42(+) 0.5(+) 0.38-0.75(++)Nitaf08 M nd nd nd nd nd nd F nd nd nd nd nd nd Nitaf09 M nd nd 0.52(+) nd nd nd F nd nd nd nd nd nd Nitaf10 M nd nd nd nd nd 0.69(+)F nd 0-0.23(+) 0-0.15(+) nd nd 0-0.23(++)Nitaf11 M nd nd nd nd nd nd F nd nd nd nd nd nd Nitaf12 M nd nd nd nd nd nd F nd 0.62(+) nd nd 0.33(+) 0.45(+)Nitaf13 M 0.54(+) 0(++) 0(++) 0(++) 0(++) 0(++)F 0(++) 0.2-0.51(++) 0-0.15(+) 0(++) 0(+++) 0-0.18(++)Nitaf14 M nd nd nd nd nd 0.62(+)F nd 0.71(+) 0.62(+) nd nd 0.65(+)Nitaf15 M nd nd nd nd nd nd F nd nd nd nd nd nd Nitaf16 M nd nd nd nd nd nd F nd nd nd nd nd nd Nitaf17 M nd nd nd nd nd 0.69(+)F nd nd nd nd nd 0.69(+)Nitaf18 M nd nd 0.68(+) 0.58(+) 0.58(+) nd F nd nd 0.68(+) 0-0.67(+) 0.58(+) nd Nitaf19 M nd nd nd nd nd 0.75(+)F nd nd nd nd nd nd Nitaf20 M 0-0.38(++)0.10-0.50(++)0.71-0.85(+) 0(+++) 0(+++) 0(++)F 0.17-0.58(+++)0.10-0.46(++) 0(+) 0-0.42(++)0-0.42(+++)0(++),0.94(+)Nitaf21 M nd nd 0.37(+) nd nd nd F nd nd 0.30(+) nd nd nd Nitaf22 M 0-0.65(+) 0.70(++) 0.74(++) 0.70(++) 0.70(++) 0.63(++)F

Note:A.t.=Agrobacterium tumefaciens;B.s.=Bacillus subtilis;P.s.=Pseudomonas lachrymans;R.s.=Ralstonia solanacearum;S.h.=Staphylecoccus hacmolyticus;X.v.=Xanthomonas vesicatoria;M,mycelia methanol extract;F,filtrate ethyl acetate extract;Developing solvent system in TLC was CH2 Cl2-MeOH(15∶1,v/v);nd,no antibacterial activity was detected;+ ,the diameter of the antimicrobial activity area was0-5 mm;++ ,the diameter of the antimicrobial activity area was 5-10 mm;+++,the diameter of the antimicrobial activity area wasmore than 10 mm.
Conclusion
In this study,we reported the endophytic fungi isolated from the healthy roots and stems of N.tabacum and their antibacterial activity.23 representative fungal isolateswere identified based on theirmorphological features and ITS-rDNA analysis.18 generawere obtained,among which,Acremonium (isolates Nitaf01 and Nitaf02),Fusarium(isolates Nitaf07,Nitaf08,and Nitaf09),Penicillium(isolates Nitaf18 and Nitaf20)and Plectosphaerella(isolates Nitaf11 and Nitaf12)were dominant genera.13 genera,namely Acremonium,Cladosporium,Clonostachys,Ilyonectria,Mortierella,Myriodontium,Petriella,Plectosphaerella,Podospora,Purpureocillium,Rhizopycnis,Stephanonectria and Thielavia were isolated from N.tabacum for the first time.The endophytic fungi obtained in this study were different from the previous reports[7,8].The reasons for this might be resulted from the cultivar of tobacco aswell as sampling time and place[19].The crude extracts of five fungal isolates(Colletotrichum sp.Nitaf05,Fusarium sp.Nitaf07,Purpureocillium sp.Nitaf13,Penicillium sp.Nitaf20 and Rhizopycnis sp.Nitaf22)showed strong antibacterial activity against six bacteria.The antibacterial compounds of the endophytic fungi existed in both extracts ofmycelia and filtrate.The results suggest that there is a diversity of the endophytic fungi in N.tabacum.Furthermore,some fungi(i.e.,isolates Nitaf05,Nitaf07,Nitaf13,Nitaf20 and Nitaf22)have the potential to produce natural antimicrobial compounds.Future study will focus on identification of the antibacterial compounds from these fungiaswell as their application as the biocontrol agents[20].
1 Saikonen K,Wali P,Helander M,et al.Evolution of endophyte-plant-symbioses.Trends Plant Sci,2004,9:275-280.
2 Porras-Alfaro A,Bayman P.Hidden fungi,emergent properties:endophytes and microbiomes.Annu Rev Phytopathol 2011,49:291-315.
3 Zhao J,Shan T,Mou Y,et al.Plant-derived bioactive compounds produced by endophytic fungi.Min-Rev Med Chem,2011,11:159-168.
4 Gutierrez RMP,Gonzalez AMN,Ramirez AM.Compounds derived from endophytes:a review of phytochemistry and pharmacology.Curr Med Chem,2012,19:2992-3030.
5 Debbab A,Aly A,Proksch P.Mangrove derived fungal endophytes– a chemical and biological perception.Fungal Divers,2013,61:1-27.
6 Wang X,Bennetzen JL.Current status and prospects for the study of Nicotiana genomics,genetics,and nicotine biosynthesis genes.Mol Genet Genomics,2015,290:11-21.
7 Pei Z,Zhang M.Distribution characteristics of endophyic fungi in tobacco.Henan Agric Sci,2009,6:97-99.
8 LiW,Qian Z,Jin R,et al.Diversity and distribution characteristics of endophytic fungi in Nicotiana tabacum in Dali District,Yunnan Province.Microbiol China,2013,40:783-791.
9 Ainsworth GC,Sparrow FK,Sussman AS.The Fungi-an Advanced Treatise.Volume IV A,a Taxonomic Review with Keys:Ascomycetes and Fungi Imperfecti.New York:Academic Press,1973.
10 PhotitaW,Taylor PWJ,Ford R,et al.Morphological and molecular characterization of Colletotrichum species from herbaceous plants in Thailand.Fungal Divers,2005,18:117-133.
11 Li J,Zhao J,Xu L,et al.Endophytic fungi from rhizomes of Paris polyphylla var.yunnanensis.World JMicrobiol Biotechnol,2008,24:733-737.
12 Xu L,Zhou L,Zhao J,et al.Fungal endophytes from Dioscorea zingiberensis rhizomes and their antibacterial activity.Lett Appl Microbiol,2008,46:68-72.
13 Zhong L,Zhou Y,Gao S,etal.Endophytic fungi from the hybrid‘Neva’of Populus deltoides Marsh x Populus nigra L.and their antimicrobial activity.Afr JMicrobiol Res,2011,5:3924-3929.
14 Lou J,Fu L,Luo R,et al.Endophytic fungi from medicinal herb Salvia miltiorrhiza Bunge and their antimicrobial activity.Afr JMicrobiol Res,2013,7:5343-5349.
15 Wang H,Qi M,Cutler AJ.A simplemethod of preparing plant samples for PCR.Nucleic Acids Res,1993,21:4153-4154.
16 Jasalavich CA,Ostrofsky A,Jellison J.Detection and identification of decay fungi in spruce wood by restriction fragment length polymorphism analysis of amplified genes encoding rRNA.Appl Environ Microbiol,2000,66:4725-4734.
17 Zhao J,Xu L,Huang Y,etal.Detection of antimicrobial compounds from extracts of the endophytic fungi associated with Paris polyphylla var.yunnanensis using TLC-bioauthography-MTT assay.Nat Prod Res Dev(天然产物研究与开发),2008,20:28-32.
18 Bernas T,Dobrucki JW.The role of plasma membrane in bioreduction of two tetrazolium salts,MTT,and CTC.Arch Biochem Biophys,2000,380:108-116.
19 Rodriguez RJ,White Jr.JF,Arnold AE,et al.Fungal endophytes:diversity and functional roles.New Phytol,2009,182:314-330.
20 Nisa H,Kamili AN,Nauchoo IA,et al.Fungal endophytes as prolific source of phytochemicals and other bioactive natural products:a review.Microb Pathogenesis,2015,82:50-59.
