Preparation of full-length deubiquitinating complex Ubp3/Bre5 and characterization of interaction with Cdc48
2015-03-06SuWenchengLyuCaoShiLiliJingXiaofeiGaiYuanmingZhangJieTanHuanboWangPengjuXiaLixinZouPeijianandQinGang
Su Wencheng, Lyu Cao, Shi Lili, Jing Xiaofei,Gai Yuanming, Zhang Jie, Tan Huanbo, Wang Pengju,Xia Lixin, Zou Peijian, and Qin Gang†
1) College of Bioengineering, Tianjin University of Science and Technology, Tianjin 300457, P.R.China 2) National Engineering Laboratory for Industrial Enzymes, Tianjin Institute of Industrial Biotechnology,Chinese Academy of Sciences, Tianjin 300308, P.R.China 3) Tianjin Key Laboratory of Molecular Design and Drug Discovery, Tianjin Institute of Pharmaceutical Research,Tianjin 300193, P.R.China 4) Health Science Center, Shenzhen University, State Key Laboratory of Respiratory Disease for Allergy at Shenzhen University,Shenzhen 518060, P.R.China

【生物工程 / Bioengineering】
Preparation of full-length deubiquitinating complex Ubp3/Bre5 and characterization of interaction with Cdc48
Su Wencheng1,2, Lyu Cao1,2, Shi Lili3, Jing Xiaofei1,2,Gai Yuanming2, Zhang Jie2, Tan Huanbo2, Wang Pengju2,Xia Lixin4, Zou Peijian2, and Qin Gang2†
1) College of Bioengineering, Tianjin University of Science and Technology, Tianjin 300457, P.R.China 2) National Engineering Laboratory for Industrial Enzymes, Tianjin Institute of Industrial Biotechnology,Chinese Academy of Sciences, Tianjin 300308, P.R.China 3) Tianjin Key Laboratory of Molecular Design and Drug Discovery, Tianjin Institute of Pharmaceutical Research,Tianjin 300193, P.R.China 4) Health Science Center, Shenzhen University, State Key Laboratory of Respiratory Disease for Allergy at Shenzhen University,Shenzhen 518060, P.R.China
Ubiquitination modification is a dynamic process essential for eukaryotic cell physiology. Ubp3, theSaccharomycescerevisiaehomologueofhumandeubiquitinaseUSP10,togetherwithitscofactorBre5,playsanactiveroleinnumerouscellularprocesses.AlthoughBre5isessentialforUbp3functioninvivo,unfortunately,duetodifficultyinpreparingcriticalquantitiesofintactfunctionalUbp3andUbp3/Bre5reconstitute,systemiccharacterizationonthiscomplexislacking.Hence,howexactlyBre5regulatesUbp3activitystillremainselusive.Tofillthisgap,wereportthesuccessfulexpressionandpurificationofrecombinantUbp3andBre5inEscherichiacoliinmonomericandcomplexform.Toourknowledge,thisisthefirstreportthesuccessfulpreparationoffull-lengthUbp3/Bre5proteincomplexinlargescale,whichallowsustoobtainfurtherunderstandingofmolecularbases.ThestoichiometricinteractionbetweenpurifiedUbp3andBre5confirmedproperfoldingoftheseproteins.ToassesstheproposeddirectinteractionsbetweenUbp3andBre5withtheubiquitinselectiveATPaseassociatedwithavarietyofcellularactivities(AAAATPase)Cdc48,seriesofpull-downassaysareperformed;resultsrevealthat,neitherUbp3norBre5aloneisabletobindCdc48.However,theUbp3/Bre5complexcouldbindCdc48efficiently,whichprovidsnovelinsightonUbp3/Bre5-Cdc48interactionmode.Insummary,ourresultslaythefoundationforfuturemechanisticevaluationbybothbiochemicalandstructuralmeans.
protein binding; deubiquitinase Ubp3; cofactor Bre5; ATPase Cdc48; deubiquitinating complex; GST-pulldown; direct interaction
The dynamic balance between ubiquitination and deubiquitination, the two reversal post-translational regulation processes, plays a vital role in eukaryotic cell physiology facilitated by highly specific catalytic machinery[1]. Deubiquitination is catalyzed by deubiquitinating proteases (DUBs), which are mainly categorized into five groups based on structural homology: ubiquitin-specific processing proteases (USPs/UBPs), ubiquitin C-terminal hydrolases (UCHs), ovarian tumor domain-containing proteases (OTUs), Ataxin-3-like proteases and Jab1/Mov34/Mpr1 Pad1 N-terminal+ (MPN+)(JAmmol/L) proteases[2-3]. Among DUBs, UBPs represent the most abundant type. In theSaccharomycescerevisiaegenome,atleast16UBPencodingsequenceshavebeendiscovered.Noneofthemisessentialforcellviability,butcertainindividualmutantsexhibitpleiotropicabnormalities,implicatingimportantandwidespreadrolesincellularfunctions[4-5].
Ubp3,theSaccharomycescerevisiaehomologueofhumanUSP10,hasbeenshowntobeinvolvedinregulatingmultiplecellularprocesses,includingDNArepair[6-7],transcriptionregulation[8-9],signaltransduction[10-11],anterograde/retrogradetransport[12]andribophagy[13].OnepositiveregulatornamedBre5directlyinteractswithUbp3andisindispensableforUbp3function[12].InitialstructuralcharacterizationhasrevealedthatUbp3andBre5formasymmetricheterotetramerinwhichtheBre5NTF2-likedomaindimerinteractswithtwoN-terminalmotifsofUbp3withapparent1∶1stoichiometry[14-15].TheeffectofcofactorBre5onUbp3istoeitherfacilitatesubstratetargeting,ortomodulateitscatalyticactivityortoachieveboth;unfortunately,directprooffrombiochemicalandstructuralaspectsisstilldeficientduetodifficultyinpreparingthelargeUbp3andfull-lengthfunctionalUbp3/Bre5reconstitute.Recently,oneintriguingconnectionoftheubiquitin-selectivechaperonCdc48,anditscofactorUfd3toUbp3/Bre5mediatedribophagywasproposed.Ubp3andBre5wereshowntointeractwithCdc48andUfd3directly[16].However,themolecularbasisontheseinteractionsandfunctionalmechanismunderlyingthemremaintobepreciselyevaluated.
Priorattemptstopurifyrecombinantfull-lengthUbp3werenotverysuccessful[15].WefindthatthiswasmostlyduetotheintrinsicinstabilityofN-terminalregionofUbp3 (ourunpublisheddata).ToinitiatesystemiccharacterizationofthedeubiquitinatingcomplexUbp3/Bre5,wereportsuccessfulexpressionandpurificationofrecombinantUbp3andBre5inamonomericandcomplexform,throughtheEscherichiacoli(E.coli)expressionsystem.Utilizingthesepurifiedsamples,wecarefullyassesstheinteractionsbetweenCdc48andUbp3/Bre5invitro.Asfarasweknow,thisisthefirstreportonthesuccessfulpreparationofafull-lengthUbp3/Bre5complexinlargescale,whichallowsustoobtainfurtherunderstandingofmolecularbasisoftheUbp3/Bre5viabiochemicalandstructuralbiologymeans.
1 Materials and methods
1.1Materials
E.coliDH5α,E.coliBL21 (DE3),E.colitrx(DE3)andT4DNALigasewerepurchasedfromBeijingTransGenBiotech(Beijing,China).pGEX-4T-1,pET-28aandpET-32-Bre5 were obtained from our laboratory. Restriction enzymes were purchased from Fermentas Life Sciences (Vilnius, Lithuania). Es Taq DNA Polymerases were purchased from Beijing CoWin Bioscience Co., Ltd (Beijing, China). Extraction Kit, Plasmid Mini Kit and Cycle-pure Kit were purchased from OMEGA Bio-Tek (Norcross,GA). Primers were ordered from Shanghai Sangon Biotechnology (Shanghai, China). IPTG was purchased from Sigma (CA, USA). Recombinant glutathione S-transferase (GST), GST-Cdc48 and 6×His-Cdc48 were previously prepared as reported[17].
1.2 Construction of the expression plasmids
TheUbp3encodingsequencewasamplifiedfromSaccharomycescerevisiaegenome,withprimersdesignedwithrestrictionendonucleasecloningsitesEcoRIandXhoI(Table1).ThePCRreactionswerecarriedoutas:Step1 94 ℃,2min;Step2 94 ℃,30s, 55 ℃,30s, 72 ℃,2min;Step3 72 ℃, 10min,with25cyclesofstep2.TherecoveredPCRproductandvectorpGEX-4T-1weredigestedwithEcoRIandXhoIrestrictionenzymesandwereligatedviaT4ligase,thentransformedintoE.coliDH5αcells.PositiveclonesdesignatedaspGEX-4T-1-Ubp3wereselectedbycolonyPCRandwereverifiedbyDNAsequencing.
DNAfragmentsencodingBre5genewasamplifiedbyPCRfromapreviouslyconstructedpET-32-Bre5 plasmid with primers designed with restriction endonuclease cloning sitesNcoIandXhoI(Table1).PCRreactionswerecarriedoutas:Step1 95 ℃, 2min;Step2 94 ℃, 30s, 55 ℃, 30s, 72 ℃, 1min30s;Step3 72 ℃, 10min,with25cyclesofstep2.TheamplifiedBre5fragmentwasclonedintoapET-28avectorbythesimilarprocessdescribedabove.Positivecloneswereselectedviadoubledigestion,andthesequencingverifiedplasmidwasdesignatedaspET-28a-Bre5.
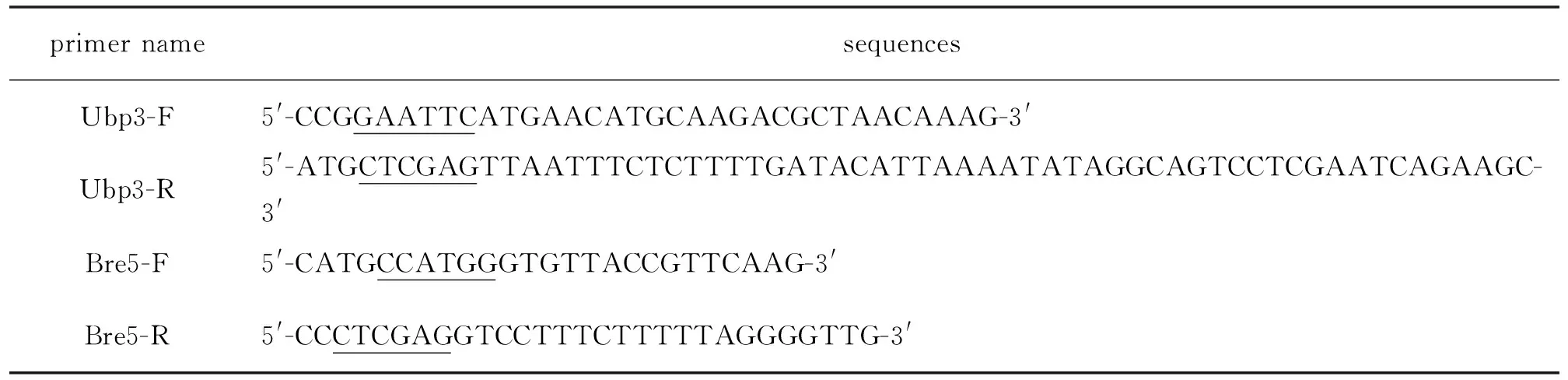
Table 1 PCR primer sequences1)图1 PCR扩增使用引物
1)Restriction sites are underlined.
1.3 Expression trials of recombinant Ubp3 and Bre5
pGEX-4T-1-Ubp3 and pET-28a-Bre5 plasmids were transformed intoE.colitrx(DE3)andE.coliBL21(DE3)cellsrespectively.AsinglecolonywasinoculatedintoaLBmediumsupplementedwithproperantibiotics(100μg/mLampicillinor50μg/mLkanamycin)andcultivatedovernightat37 ℃withvigorousshaking.Theovernightculturesweredilutedinfreshpre-warmedmedium(includingproperantibiotics)andgrownat37 ℃withvigorousshaking,untiltheOD600reached0.5-0.7.ProteinexpressionwasinducedbyaddingIPTG(finalconcentrationat0.2mmol/LforGST-Ubp3 induction and 0.1 mmol/L for 6×His-Bre5 induction), and cultures were collected after overnight growth at either 25 ℃ or 16 ℃. Samples were sonicated and fractionated, and whole cell lysate, supernatant, and pellet fractions were analyzed by means of sodium dodecyl sulfate polyacrylamide gel electropheresis (SDS-PAGE) and Coomassie staining.
The co-expression experiment was essentially described above, except that both pGEX-4T-1-Ubp3 and pET-28a-Bre5 were co-transformed intoE.coliBL21(DE3)inthepresenceofantibiotics(100μg/mLampicillinplus50μg/mLkanamycin).
1.4PurificationofUbp3andBre5
TopurifyrecombinantGST-Ubp3inlargescale,cultivatedcellswithoptimalinductionwereharvestedbycentrifugation(4 500r/min, 20min, 4 ℃)andre-suspendedinicecoldbuffer(PBS,pH=7.4, 0.4mmol/LofPMSF, 1×proteaseinhibitor, 3mmol/LDTT)andlysedviaFrenchpress.Thelysatewasthencentrifugedat15 000r/minfor30minat4 ℃,withthesupernatantappliedtoGlutathioneSepharoseresin(GEHealthcarecat. #17-5132-03)pre-equilibratedwithPBSandincubatedwithrotatingfor4hat4 ℃.Thecolumnwaswashedwithequilibrationbuffer(PBScontaining3mmol/LDTT, 0.1%TritonX-100),andtheproteinwaselutedwithelutionbuffer(50mmol/LTris-HCl,pH=8.0, 200mmol/LNaCl, 10mmol/Lglutathione).TheelutionfractionswereanalyzedviaSDS-PAGEandCoomassiestaining.Selectedelutionfractionswerecombinedanddialyzedagainstice-colddialysisbuffer(50mmol/LTris-HCl,pH=7.5, 50mmol/LNaCl, 5%glycerol),thenappliedtotheSPSepharoseFF(GEHealthcarecat. #17-0929-01)pre-equilibratedwithanice-colddialysisbuffer.Afterbeingwashedwith10bedvolumesofthesamebuffer,thecolumnwaselutedwithanelutionbuffercontainingastepwiseincreaseinsaltconcentration(0.2, 0.3and0.4mol/LNaCl,respectively).TheelutionfractionswereanalyzedviaSDS-PAGEandCoomassiestaining.TheconcentrationofpurifiedrecombinantGST-Ubp3 protein was determined by using bovine serum albumin as a standard.
To purify recombinant 6×His-Bre5 on a in large scale, cells were harvested by centrifugation (4 500 r/min, 20 min, 4 ℃) and pellets were re-suspended in 100 mL of lysis buffer (50 mmol/L Tris-HCl, pH=7.5, 150 mmol/L NaCl and 20 mmol/L imidazole, 0.4 mmol/L of PMSF, 5 mmol/L β-mercaptoethanol). Cells were lysed via French Press. Lysates were clarified (15 000 r/min, 30 min, 4 ℃), and the supernatants were transferred to Ni Sepharose FF (GE Healthcare cat. #17-5318-03) pre-equilibrated with lysis buffer and rotated for 2 h at 4 ℃. The column was sequentially washed with a wash buffer (50 mmol/L Tris, pH=7.5, 150 mmol/L NaCl, 0.1% Triton X-100, 5 mmol/L β-mercaptoethanol) containing a stepwise increase of imidazole concentrations (20, 50 and 100 mmol/L). Then the 6×His-Bre5 protein was eluted with elution buffer (50 mmol/L Tris, pH=7.5, 150 mmol/L NaCl, 250 mmol/L imidazole, 5 mmol/L β-mercaptoethanol), and elution fractions were analyzed via SDS-PAGE and Coomassie staining. The concentration of purified recombinant 6×His-Bre5 protein was determined by using bovine serum albumin as a standard.
1.5 Preparation of Ubp3 /Bre5 complex
For the large-scale Ubp3/Bre5 complex purification, GST-Ubp3 was purified as described above, except that after GST-Ubp3 binding, sufficient amount of purified 6×His-Bre5 was applied to glutathione column and incubated for 1 h at 4 ℃.
1.6 LC-MS/MS analysis of recombinant Ubp3 and Bre5
For protein identification LC-MS/MS analysis was conducted using LTQ XL from Thermo Fisher (ESI-MS/MS). The instrument was operated with a spray voltage of 3.5 kV and an ion transfer tube temperature of 25 ℃. The information-dependent acquisition (IDA) mode of operation was employed in which a survey scan fromm/z400to1 800wasacquiredfollowedbycollision-induceddissociation(CID),andforMS/MS,usinganormalizedcollisionenergyof35%withanactivationqof0.25for30ms.IonselectionthresholdsforMSandMS/MSwere1 000and500counts,respectively.
TandemmassspectrawereextractedbytheXcaliburversion1.0.0.2.AllMS/MSsampleswereanalyzedusingSequest.IodoacetamidederivativeofCys,de-amidationofAsnandGln,oxidationofMetwerespecifiedinSequestasvariablemodifications.ProteomeDiscoverer1.2wasusedtovalidateMS/MSbasedpeptideandproteinidentifications.Peptideidentificationswereacceptediftheycouldbeestablishedatprobabilitygreaterthan95.0%asspecifiedbytheresultfilter,whichisXcorr> 1.9 if the charge is 1,Xcorr> 2.2 if the charge is 2,Xcorr> 3.75 if the charge is 3. Protein identifications were accepted if they were established at probability greater than 99.0% and contained at least 2 identified unique peptides.
1.7 GST-pulldown experiments
For pull-down assays, GST or GST fusion proteins were first incubated with 50 μL of pre-equilibrated glutathione-Sepharose beads in buffer A (50 mmol/L Tris, 100 mmol/L NaCl, 1 mmol/L DTT, 0.1% triton X-100, pH=7.5) for 1 h at 4 ℃. The beads were washed once with 500 μL of buffer A to remove unbound material and then incubated with prey proteins for 1 h at 4 ℃. Beads were washed three times with 1 mL of buffer A, followed by three times of wash with buffer B (50 mmol/L Tris, 100 mmol/L NaCl, 1 mmol/L DTT, pH=7.5), then mixed with an SDS-PAGE loading buffer and analyzed on SDS-PAGE.
2 Results
2.1 Construction of the expression plasmids
Due to the exceptional ability of GST tag to greatly enhance the solubility and stability of fused proteins, GST tag has been widely used for facilitating recombinant protein preparation; therefore we introduce a GST domain fused at the N-term ofUbp3. TheUpb3 gene was amplified usingSaccharomycescerevisiaegenomeDNAasatemplate,asinglebandatabout2.8kbwasobtained,inaccordancewiththesizeofUpb3 coding region (Fig.1(a)); the encoding fragment was inserted into bacterial expression vector pGEX-4T-1, and a positive plasmid designated as pGEX-4T-1-Ubp3 was selected via colony PCR (Fig.1(b)) and verified via DNA sequencing. TheBre5genewasamplifiedsimilarly,withafragmentofabout1.5kbobtained(Fig.1(c)),theencodingfragmentwasinsertedintopET-28atointroducea6×HistagatN-termofBre5.Thepositiveplasmid,designatedpET-28a-Bre5, was selected by double restriction enzyme digestion (Fig.1(d)) and confirmed via DNA sequencing.
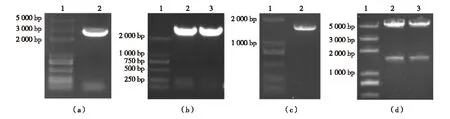
(a) PCR amplification of Ubp3 coding region from Saccharomyces cerevisiae genome. Lane 1, DNA marker; Lane 2, PCR product. (b) Verification of expression plasmids pGEX-4T-1-Ubp3 by colony PCR. Lane 1, DNA marker; Lane 2-3, PCR amplified fragments verifying two positive clones. (c) PCR amplification of Bre5 coding region from a previously constructed plasmid pET-32-Bre5. Lane 1, DNA marker; Lane 2, PCR product. (d) Verification of expression plasmids pET-28a-Bre5 by restriction enzyme digestion. Lane 1, DNA marker; Lane 2-3, two positive clones digested with Nco I and Xho I.Fig.1 Ubp3 and Bre5 coding fragments amplified by PCR and verification of recombinant expression plasmids图1 PCR扩增Ubp3和Bre5编码片段及质粒构建验证
2.2 Expression and purification of Ubp3
Small scale expression trials of Ubp3 inE.colitrx(DE3)wereperformedatvariousinductionconditions.Comparedtonon-inducedcondition,onebandmigratingatabout130kDabecameapparentafterIsopropylβ-D-1-thiogalactopyranoside(IPTG)inductionwith0.2mmol/LIPTGat16 ℃,whichcorrespondstotheGST-Ubp3fusion,thusresultedinarelativelyhighersolubilityoftheinducedprotein(Fig.2(a));therefore,wechosethesameinductionconditionforlargescalepreparation.AsshowninFig.2(b),afterasingle-stepglutathionecolumnpurification,GST-Ubp3wasenrichedinelutionfractions.However,thesamefractionsalsocontainedseveralcontaminatingcomponentsofdiversemolecularweight.Thesizeofaprominentcontaminantwasabout26kDa(Fig.2(b),Lane6),similartoanintactGSTdomain,whichisnotsurprisingsinceGSTtruncatesarefrequentlyco-purifiedwithGSTfusionproteins,especiallywhenthefusedpartnercontainsdegradation-proneareas.OthermajorcontaminatingproteinsappearedtobethedegradationintermediatesofUbp3,sincethesebandsremainedratherunstable,almostdisappearedduringdialysis(comparedFig.2(b),Lane6withFig.2(c),Lane1).TofurtherimprovethepurityofcombinedGST-Ubp3poolandespeciallytoremoveGSTtruncates,wecontinuedwithion-exchangechromatography.BasedontheestimatedpIsforUbp3andGST(7.9forUbp3versus4.5forGST),SPsepharosewasselected,andtheefficacyofcontaminantremovalwasshowninFig.2(c).GSTtruncatesindialysisbufferremainedpoorlyboundtoSPresinclearlyhencelargelyexistedinflowthrough(Fig.2(c),Lane2);incontrast,mostGST-Ubp3adsorbedtoSPresinatthesamecondition,andwasabletobeefficientlyelutedwhenNaClconcentrationwasincreasedto0.2-0.3mol/L(Fig.2(c),Lane6-9).Recoveredfull-lengthGST-Ubp3exhibitedsignificantimprovementonpurity(>85%),itsidentitywasuniquelyverifiedbyLC-MS/MS(Fig.3).TotalyieldsofrecombinantGST-Ubp3arelistedinTable2.
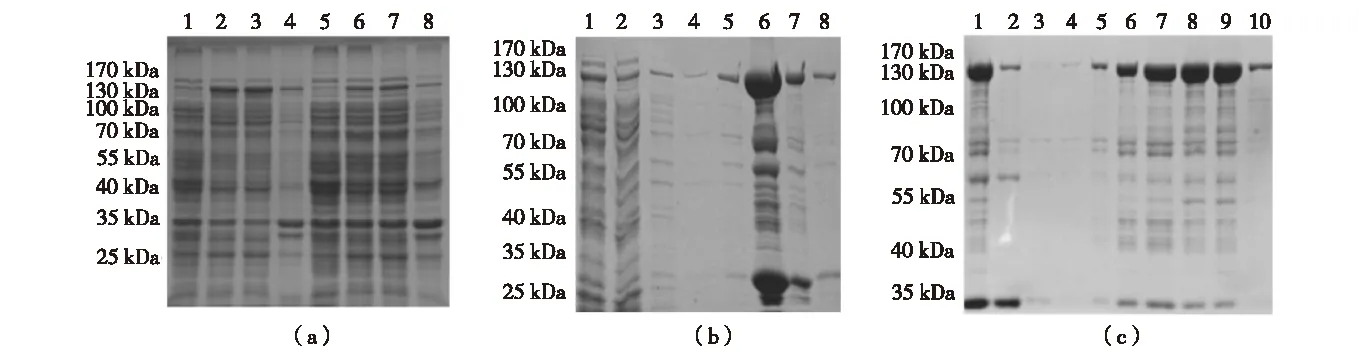
(a)Small scale expression trials of GST-Ubp3 induced at 16 ℃ (Lane 1-4) and 25 ℃ (Lane 5-8) respectively. Lane 1 and 5, total proteins of uninduced cells; Lane 2 and 6, total proteins of induced cells; Lane 3 and 7, soluble fraction of induced cells; Lane 4 and 8, insoluble fraction of induced cells. (b) GST-Ubp3 purification through glutathione column chromatography. Lane 1, soluble fraction of induced cells; Lane 2, flowthrough; Lane 3-4, wash; Lane 5-8, elution fraction. (c) Further purification of GST-Ubp3 through SP cation-exchange chromatography. Lane 1, dialyzed GST-Ubp3 pool from glutathione column chromatography; Lane 2, flowthrough; Lane 3, wash; Lane 4-6, 0.2 mol/L NaCl elution fraction; Lane 7-9, 0.3 mol/L NaCl elution fraction; Lane 10, 0.4 mol/L NaCl elution fraction.Fig.2 SDS-PAGE analysis on expression and purification of Ubp3 in E.coli图2 蛋白电泳分析 Ubp3在大肠杆菌内的表达及纯化
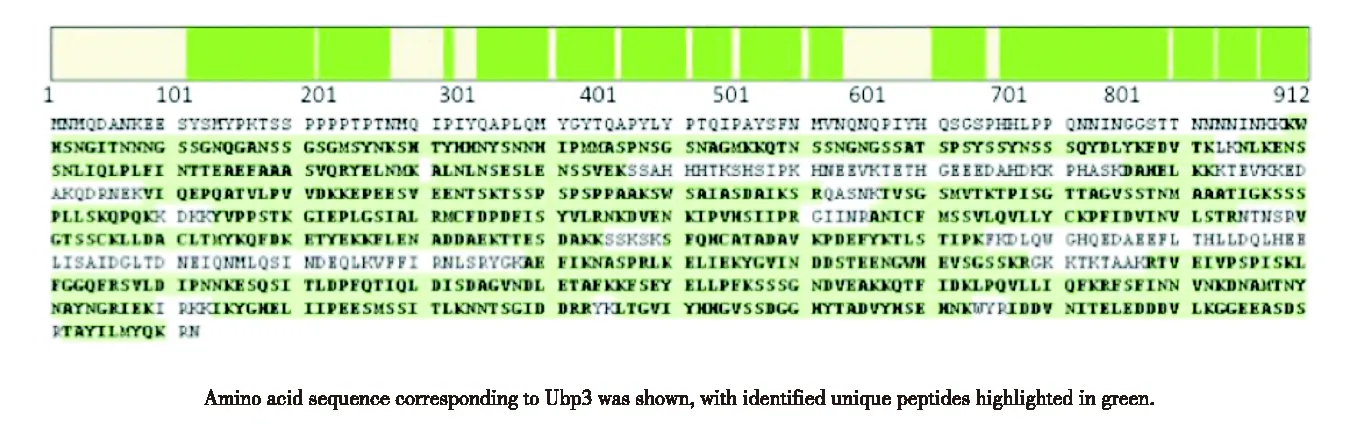
Amino acid sequence corresponding to Ubp3 was shown, with identified unique peptides highlighted in gray.Fig.3 (Color online) Identity verification of purified Ubp3 via tandem MS/MS图3 MS/MS鉴定纯化的Ubp3蛋白

proteinnamepurificationstagesV/mLm(targetprotein)/mgyield/%celllysate10060100glutathioneaffini-tychromatography161627GST-Ubp3Spcation-ex-changechromatography2010176×His-Bre5celllysate100150100nickleaffinitychromatography246040
2.3 Expression and purification of Bre5
In experiments parallel to Ubp3, expression trials of Bre5 inE.coliBL21(DE3)werealsoperformed.Uponinduction,oneproteinwithamolecularweightofabout70kDaappears(Fig.4(a)),whichislargerthantheexpectedsizeof6×His-Bre5 (about58kDa);thisismostlikelyduetounusualmobilityofBre5inSDS-PAGE,sincetheidentityofpurifiedproteinwasconfidentlyverifiedasBre5byLC-MS/MS(Fig.5).Targetproteininducedwith0.1mmol/LIPTGat16 ℃exhibitedrelativelybettersolubility(Fig.4(a)),sameinductionconditionwasalsoappliedtolargescalepurification. 6×His-Bre5waspreparedaccordingtostandardone-stepnickelaffinitychromatographyprocedure,asshowninFig.4(b).Theimidazoleelutionfractionswerepooledanddialyzed,totalyieldsofrecombinant6×His-Bre5aresummarizedinTable2.
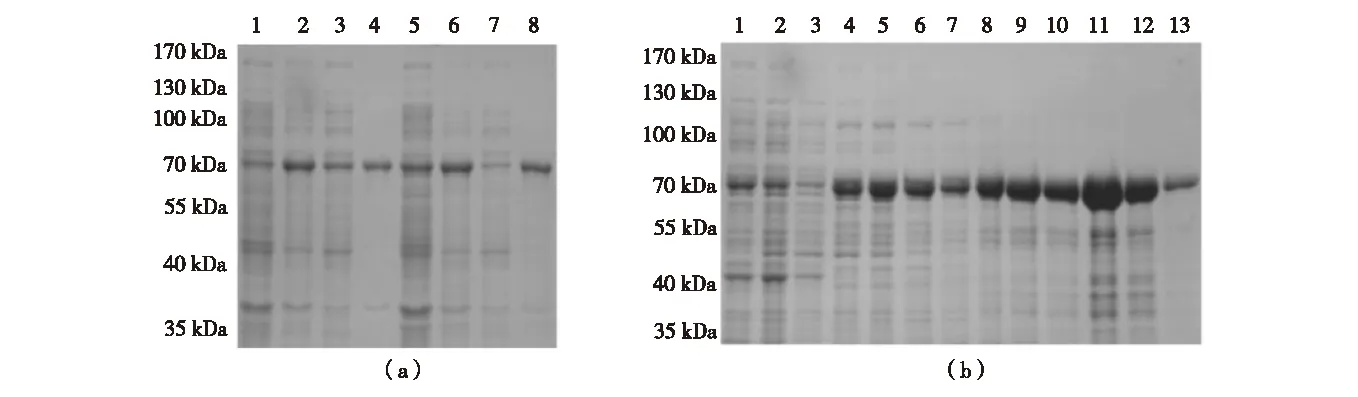
(a) Small scale expression trials of 6×His-Bre5 induced at 16 ℃ (Lane 1-4) and 25 ℃ (Lane 5-8) respectively. Lane 1 and 5, total proteins of uninduced cells; Lane 2 and 6, total proteins of induced cells; Lane 3 and 7, soluble fraction of induced cells; Lane 4 and 8, insoluble fraction of induced cells. (b) 6×His-Bre5 purification through nickel affinity chromatography. Lane 1, soluble fraction of induced cells; Lane 2, flowthrough; Lane 3, wash; Lane 4, 50 mmol/L imidazole elution fraction; Lane 5-6, 80 mmol/L imidazole elution fraction; Lane 7-9, 100 mmol/L imidazole elution fraction; Lane 10-12, 250 mmol/L imidazole elution fraction; Lane 13, 500 mmol/L imidazole elution fraction.Fig.4 SDS-PAGE analysis on expression and purification of Bre5 in E.coli图4 蛋白电泳分析Bre5蛋白在大肠杆菌内的表达及纯化

Amino acid sequence corresponding to Bre5 was shown, with identified unique peptides highlighted in gray.Fig.5 (Color online) Identity verification of purified Bre5 via tandem MS/MS图5 MS/MS鉴定纯化的Bre5蛋白
2.4FunctionaltestofrecombinantUbp3andBre5
HavingsuccessfullyobtainedsolubleUbp3andBre5inhighpurity,wesoughttoconfirmwhethertheyareproperlyfoldedornot.Previously,ithasbeenwellestablishedthatUbp3andBre5physicallyinteractwitheachotherinvivoandinvitro[12, 14-15];thus,weperformedaGST-pulldowntodirectlyexaminetheinteraction.AsshowninFig.6,incontrasttoGST(Lane4),GST-Ubp3displaysastoichiometricinteractionwithBre5 (Lane5),consistentwithpreviousreports[14-15].Basedonthesedata,weconcludthatourpreparedrecombinantUbp3andBre5arefunctional.
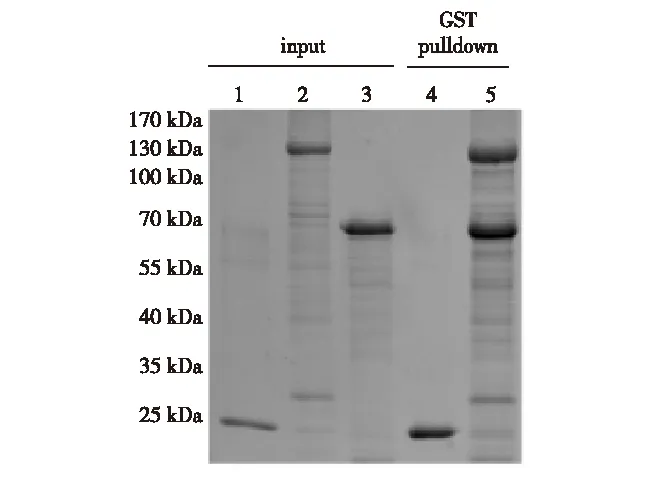
Lane 1, GST; Lane 2, GST-Ubp3; Lane 3, 6×His-Bre5; Lane 4, pulldown sample using GST as bait and 6×His-Bre5 as prey; Lane 5, pulldown sample using GST-Ubp3 as bait and 6×His-Bre5 as prey.Fig.6 GST-pulldown assay between recombinant Ubp3 and Bre5图6 GST-pulldown 检测Ubp3与Bre5结合
2.5Large-scalepreparationofUbp3/Bre5complex
ThesuccessfulpurificationoffunctionalUbp3andBre5individuallypromptedustotrypreparingUbp3/Bre5complexdirectly,whichisessentialforfurtherfunctionalandstructuralcharacterization.Initially,wetookoureffortonco-expressingpGEX-4T-1-Ubp3 and pET-28a-Bre5 inE.coli.UnfortunatelytheinductionofUbp3andBre5isnotatcomparablelevel(Fig.7(a)),GST-Ubp3inductionisnearlyundetectable),preventingproductivecomplexassemblyinvivo.Tosolvethisproblem,wedevelopeda‘hybrid’procedureasshowninFig.7(b),GST-Ubp3waspurifiedaccordingtotheestablishedtwo-stepproceduredescribedabove,exceptthatafterGST-Ubp3bindingtoglutathionecolumn,sufficientamountofrecombinant6×His-Bre5wasaddedtotriggertheon-columncomplexassembly.Byfollowingthisstrategy,theglutathioneelutionfractionsdisplayednearlystoichiometricdistributions
ofUbp3andBre5,indicatingsuccessfulcomplexformationonglutathionecolumn(Fig.7(c)).Moreover,successiveSPcation-exchangechromatographysignificantlyenhancedthepurityofUbp3/Bre5complexbyefficientlyremovingGSTtruncatesandothercontaminants,aprocedurecomparabletoindividualUbp3purification(Fig.7(d));interestingly,thepreformedUbp3/Bre5complexwelltoleratedhighsaltelutioncondition(0.4-0.5mol/LNaCl),presentingexceptionalstability.

(a) Small-scale co-expression trials of GST-Ubp3 and 6×His-Bre5 induced at 16 ℃ (Lane 1-4) and 25 ℃ (Lane 5-8) respectively. Lane 1 and 5, total proteins of un-induced cells; Lane 2 and 6, total proteins of induced cells; Lane 3 and 7, soluble fraction of induced cells; Lane 4 and 8, insoluble fraction of induced cells. (b) Procedure for two-round GST-Ubp3/6×His-Bre5 complex preparation. (c) The first round of GST-Ubp3/6×His-Bre5 complex preparation through glutathione column chromatography. Lane 1, soluble fraction of induced cells; Lane 2, flowthrough after 6×His-Bre5 incubation with glutathione column ; Lane 3-7, wash; Lane 8-11, elution fraction. (d) The second round of GST-Ubp3/6×His-Bre5 complex purification through SP cation-exchange chromatography. Lane 1, dialyzed GST-Ubp3/6×His-Bre5 pool from glutathione column chromatography; Lane 2, flowthrough; Lane 3-4, wash; Lane 5, 0.2 mol/L NaCl elution fraction; Lane 6-7, 0.3 mol/L NaCl elution fraction; Lane 8-10, 0.4 mol/L NaCl elution fraction.Fig.7 Preparation of recombinant Ubp3/Bre5 complex图7 重组Ubp3/Bre5复合体的制备
2.6 Assessment of interactions between Ubp3 and Bre5 with Cdc48
Recently, the Ubp3/Bre5 complex has been linked to the ATPase associated with a variety of cellular activities (AAA ATPase) Cdc48. Ossareh-Nazari et al[16]proposed a direct interaction between Ubp3 and Bre5 with Cdc48 respectively, hence providing further evidence that Cdc48 has close crosstalk with deubiquitinating pathways. Taking advantages of Ubp3, Bre5 and Ubp3/Bre5 preparations, we directly examined the proposed interactions by performing a series of GST-pulldown experiments. To our surprise, we could hardly observe any interaction between GST-Ubp3 and Cdc48 (Fig.8(a), Lane 5), consistently. GST-Cdc48 also failed to pulldown Bre5 (Fig.8(b), Lane 3); these results are obviously contradictory to the former finding from Ossareh-Nazari et al[16]. Intriguingly, however, when the preformed GST-Ubp3/Bre5 complex was used as pulldown bait, a significant binding of Cdc48 was observed (Fig.8(c), Lane 3), indicating the assembled Ubp3/Bre5 complex is indeed able to physically interact with Cdc48. At this moment, we could not explain the discrepancy, but our results strongly suggest that more careful experiments need to be performed to uncover the real Ubp3/Bre5-Cdc48 interaction mode.
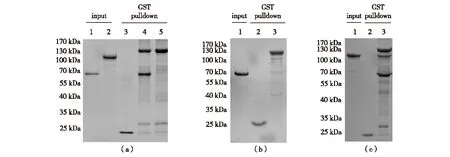
(a) GST-Ubp3 fails to interact with 6×His-Cdc48. Lane 1, 6×His-Bre5; Lane 2, 6×His-Cdc48; Lane 3, pulldown sample using GST as bait and 6×His-Cdc48 as prey; Lane 4, pulldown sample using GST-Ubp3 as bait and 6×His-Bre5 as prey; Lane 5, pulldown sample using GST-Ubp3 as bait and 6×His-Cdc48 as prey. (b) GST-Cdc48 fails to interact with 6×His-Bre5. Lane 1, 6×His-Bre5; Lane 2, pulldown sample using GST as bait and 6×His-Bre5 as prey; Lane 3, pulldown sample using GST-Cdc48 as bait and 6×His-Bre5 as prey. (c) GST-Ubp3 /6×His-Bre5 complex interacts with 6×His-Cdc48. Lane 1, 6×His-Cdc48; Lane 2, pulldown sample using GST as bait and 6×His-Cdc48 as prey; Lane 3, pulldown sample using GST-Ubp3/6×His-Bre5 complex as bait and 6×His-Cdc48 as prey.Fig.8 GST-pulldown assays between Ubp3 and Bre5 with Cdc48图8 GST-pulldown检测Ubp3 /Bre5 与Cdc48的结合
3 Discussions and conclusions
The pleiotropic defects ofubp3mutantindicateitswidespreadcellularfunctions.DespitetheinvolvementofdeubiquitinatingactivityofUbp3/Bre5inmultiplecellularprocessesandpartialresolutionofUbp3/Bre5interactionmode,themolecularbasisonhowBre5regulatesUbp3activityisstilllargelyunknown.TopreciselydissectthepotentialroleofBre5instepofsubstraterecognition,catalyticactivationorcrosstalkwithotherinteractingpartners,asophisticatedinvitroreconstitutionsystemishighlydemanding.ObviouslythesuccessfulpreparationoffunctionalUbp3/Bre5complexinhighhomogeneityisaprerequisite.Inthiswork,wehaveestablishedanexpressionandpurificationsystemtofulfillthisrequirement.Withthedevelopmentofasimpleandefficientpurificationstrategywecouldobtainrecombinantfull-lengthUbp3,Bre5andalsoUbp3/Bre5complexinlargescale,whichtoourknowledgehasnotbeenreportedbefore.
Cdc48,ahighlyconservedcomponentinAAAATPasefamily,wasnoticedandstudiedrecently,becauseofitsdistinctubiquitin-selectiveproperty:thehomohexamerofCdc48canactasageneralplatformformulti-purposedecisionmakingofmolecularevents,dependingonitsabilitytointeractwithplentyofcofactors,amongwhichbothubiquitinligasesandDUBsareincluded[18-19].ThediscoveryofUbp3-Cdc48interactionfurtherenrichesthetoolboxofCdc48andextendsitsactioninribophagypathway.Toconfirmthisimportantnotionandobtaindeeperinsight,wesetupaseriesofinteractionassaystoassesstheproposedone-to-oneinteractionsbetweenUbp3,Bre5andCdc48.Importantly,inoursystemwecouldnotreproducethediscoveredphysicalinteractionbetweenUbp3andBre5withCdc48,however,wecoulddetectastableinteractionbetweenUbp3/Bre5complexandCdc48,implicatingsynergisticactionofUbp3andBre5uponCdc48binding.ThisperspectivecouldpotentiallyexplaintheobligatoryroleofBre5inUbp3functioning.Undoubtedly,revisionofthecurrentworkingmodelawaitsmorecarefulexperiments.
WebelievethatthehighpurityreconstitutesofrecombinantUbp3/Bre5willcontinuouslybringdeeperinsightonmolecularpropertiesofthiscomplexinfuture.Importantly,inanestablishedinvitroenzymaticactivityassay,theUbp3/Bre5complexcanexhibittypicaldeubiquitinatingenzymeactivity(manuscriptinpreparation),whichopensupapathforsystemiccatalyticmechanismcharacterizationofUbp3.
[1] Wilkinson K D. Ubiquitination and deubiquitination: targeting of proteins for degradation by the proteasome[J]. Seminars in Cell & Developmental Biology, 2000, 11(3):141-148.
[2] Nijman S M,Luna-Vargas M P,Velds A,et al.A genomic and functional inventory of deubiquitinating enzymes[J]. Cell, 2005, 123(5):773-786.
[3] Reyes-Turcu F E, Ventii K H, Wilkinson K D. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes[J]. Annual Review of Biochemistry, 2009,78:363-397.
[4] Amerik A Y, Li S J, Hochstrasser M. Analysis of the deubiquitinating enzymes of the yeast Saccharomyces cerevisiae[J]. The Journal of Biological Chemistry, 2000, 381(9/10): 981 -992.
[5] Poulsen J W, Madsen C T, Young C, et al. Comprehensive profiling of proteome changes upon sequential deletion of deubiquitylating enzymes[J]. Journal of Proteomics, 2012, 75(13):3886-3897.
[6] Bilsland E, Hult M, Bell S D, et al. The Bre5/Ubp3 ubiquitin protease complex from budding yeast contributes to the cellular response to DNA damage[J]. DNA Repair (Amst), 2007, 6(10):1471-1484.
[7] Mao P, Smerdon M J. Yeast deubiquitinase Ubp3 interacts with the 26 S proteasome to facilitate Rad4 degradation[J]. Journal of Biological Chemistry, 2010, 285(48): 37542-37550.
[8] Chew B S, Siew W L, Xiao B, et al. Transcriptional activation requires protection of the TATA-binding protein Tbp1 by the ubiquitin-specific protease Ubp3[J]. Biochemical Journal, 2010, 431(3):391-399.
[9] Kvint K, Uhler J P, Taschner M J, et al. Reversal of RNA polymerase II ubiquitylation by the ubiquitin protease Ubp3[J]. Molecular Cell, 2008, 30(4):498-506.
[10] Li Y, Wang Y. Ras protein/cAMP-dependent protein kinase signaling is negatively regulated by a deubiquitinating enzyme, Ubp3, in yeast[J]. Journal of Biological Chemistry, 2013, 288(16):11358-11365.
[11] Wang Y, Zhu M, Ayalew M, et al. Down-regulation of Pkc1-mediated signaling by the deubiquitinating enzyme Ubp3[J]. Journal of Biological Chemistry, 2008, 283(4):1954-1961.
[12] Cohen M, Stutz F, Belgareh N, et al. Ubp3 requires a cofactor, Bre5, to specifically de-ubiquitinate the COPII protein, Sec23[J]. Nature Cell Biology, 2003, 5(7):661-667.
[13] Kraft C, Deplazes A, Sohrmann M, et al. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease[J]. Nature Cell Biology, 2008, 10(5):602-610.
[14] Li K, Ossareh-Nazari B, Liu X, et al. Molecular basis for bre5 cofactor recognition by the ubp3 deubiquitylating enzyme[J]. Journal of Molecular Biology, 2007,372(1):194-204.
[15] Li K, Zhao K, Ossareh-Nazari B, et al. Structural basis for interaction between the Ubp3 deubiquitinating enzyme and its Bre5 cofactor[J]. Journal of Biological Chemistry, 2005, 280(32):29176-29185.
[16] Ossareh-Nazari B, Bonizec M, Cohen M, et al. Cdc48 and Ufd3, new partners of the ubiquitin protease Ubp3, are required for ribophagy[J]. EMBO reports, 2010, 11(7): 548-554.
[17] Rumpf S, Jentsch S. Functional division of substrate processing cofactors of the ubiquitin-selective Cdc48 chaperone[J]. Molecular Cell, 2006, 21(2):261-269.
[18] Hanzelmann P, Buchberger A, Schindelin H. Hierarchical binding of cofactors to the AAA ATPase p97[J]. Structure, 2011, 19(6):833-843.
[19] Stolz A, Hilt W, Buchberger A, et al. Cdc48: a power machine in protein degradation[J]. Trends in Biochemical Sciences, 2011, 36(10):515-523.
【中文责编:晨 兮;英文责编:艾 琳】
去泛素酶复合体Ubp3/Bre5的制备及与Cdc48作用
苏文成1, 2,吕 操1,2,时丽丽3,景晓飞1,2,盖园明2,张 洁2,谭焕波2,王鹏举2, 夏立新4,邹培建2,秦 刚2
1)天津工业技术大学生物技术学院, 天津 300457; 2)中国科学院天津工业生物技术研究所,国家工业酶重点实验室,天津300308; 3)天津药物研究院天津分子设计与药物发现重点实验室, 天津300193;4)深圳大学医学部,呼吸疾病国家重点实验室深圳大学变态反应分室, 深圳 518060
泛素化是一种存在于真核细胞中与生理功能密切相关的蛋白修饰,泛素化与去泛素化处于动态调节过程中. Ubp3是与人USP10同源的酵母去泛素化酶,结合辅引子Bre5在细胞内发挥广泛作用.为研究该复合体的工作机制,制备重组蛋白复合体,在大肠杆菌中成功表达并纯化重组Ubp3与Bre5单体及Ubp3/Bre5复合体,首次成功大规模制备重组Ubp3/Bre5复合体.通过一系列pulldown实验,检验Ubp3/Bre5与AAA家族中泛素选择性ATP酶Cdc48的相互作用模式,结果发现,Ubp3及Bre5无法单独与Cdc48结合,但Ubp3/Bre5复合体可以有效与Cdc48相互作用.提出了Ubp3/Bre5-Cdc48相互作用的新模式,制备了高质量重组Ubp3/Bre5复合体.该研究为通过生化及结构生物学进行分子机制探索奠定了基础.
结合蛋白质;Ubp3去泛素化酶;结合辅因子Bre5;ATP酶Cdc48;去泛素化复合体;与谷光苷肽巯基转移酶沉淀试验;直接相互作用
天津市科技支撑计划资助项目(11ZCZDSY08100); 中国科学院百人计划资助项目(KSCW2-YW-BR-4) ;国家自然科学基金资助项目(81273275)
苏文成(1987—),女(汉族),内蒙古自治区呼和浩特市人,天津工业技术大学硕士,E-mail:woshisuwenc@yahoo.com
/References:
:Su Wencheng, Lyu Cao, Shi Lili, et al.Preparation of full-length deubiquitinating complex Ubp3/Bre5 and characterization of interaction with Cdc48[J]. Journal of Shenzhen University Science and Engineering, 2015, 32(1): 58-67.
Q 513 Document code:A
10.3724/SP.J.1249.2015.01058
Received:2014-01-17;Revised:2014-12-24;Accepted:2014-12-26
Foundation:The Program of Tianjin Municipal Science & Technology Project (11ZCZDSY08100); The Program of “One Hundred Talented People” of the Chinese Academy of Sciences (KSCW2-YW-BR-4); National Natural Science Foundation of China (81273275)
† Corresponding author:Associate professor Qin Gang, E-mail: qing@genequantum.com
引 文:苏文成,吕 操,时丽丽,等. 去泛素酶复合体Ubp3/Bre5的制备及与Cdc48作用[J]. 深圳大学学报理工版,2015,32(1):58-67.(英文版)
