Structural features of substituted triazole-linked chalcone derivatives as antimalarial activities against D10 strains of Plasmodium falciparum:A QSAR approach
2015-02-01MukeshSharma
Mukesh C. Sharma
Division of Drug Design & Medicinal Chemistry Research Lab, School of Pharmacy, Devi Ahilya University,Takshila Campus, Khandwa Road, Indore (M.P)-452001, India
Structural features of substituted triazole-linked chalcone derivatives as antimalarial activities against D10strains ofPlasmodium falciparum:A QSAR approach
Mukesh C. Sharma
Division of Drug Design & Medicinal Chemistry Research Lab, School of Pharmacy, Devi Ahilya University,Takshila Campus, Khandwa Road, Indore (M.P)-452001, India
A quantitative structure–activity relationship (QSAR) was performed to analyze antimalarial activities against the D10 strains ofPlasmodium falciparumof triazole-linked chalcone and dienone hybrid derivatives using partial least squares regression coupled with stepwise forward–backward variable selection method. QSAR analyses were performed on the available IC50D10strains ofPlasmodium falciparumdata based on theoretical molecular descriptors. The QSAR model developed gave good predictive correlation coefficient (r2) of 0.8994, significant cross validated correlation coefficient (q2) of 0.7689,r2for external test set (2)rpred of 0.8256, coefficient of correlation of predicted data setof 0.3276. The model shows that antimalarial activity is greatly affected by donor and electron-withdrawing substituents. The study implicates that chalcone and dienone rings should have strong donor and electron-withdrawing substituents as they increase the activity of chalcone. Results show that the predictive ability of the model is satisfactory, and it can be used for designing similar group of antimalarial compounds. The findings derived from this analysis along with other molecular modeling studies will be helpful in designing of the new potent antimalarial activity of clinical utility.
quantitative structure–activity relationship (QSAR); chalcone; antimalarial;Plasmodium falciparum; stepwise forward–backward; partial least squares
1 Introduction
Malaria, a devastating infectious disease caused by the protozoaPlasmodium falciparum, affects 200−500 million people worldwide annually.[1] The current global situation with respect to malaria indicates that about two billion people are exposed to the disease and more than one million people die from it every year[2].Malaria is caused by five species of parasites of the genusPlasmodiumthat affect humans:Plasmodium falciparum, Plasmodium vivax, Plasmodium malariae,Plasmodium ovalandPlasmodium knowlesi. The situation is rapidly worsening mainly due to non-availability of effective drugs and development of drug resistance in areas where malaria is frequently transmitted[3−4]. Malaria due toPlasmodium falciparumis the most deadly variety as it is responsible for the majority of malaria deaths. ThePlasmodium falciparumspecies, which is the most virulent and deadly of the malaria parasites, is responsible in more than 90%of the cases. Many commonly available antimalarial drugs and therapies are becoming ineffective because of the emergence of multidrug resistantPlasmodium falciparum, which drives the need for the development of new antimalarial drugs[4]. In spite of the intensive efforts to combat malaria, the incidence of malaria has not decreased, especially in the tropical and subtropical areas[5]. The primary drugs for treatment of malaria have been the quinolones chloroquine (CQ), quinine (QN)and mefloquine; the antifolate combination of pyrimethamine and sulfadoxine; and others[6]. However,during the past three decades,P. falciparumhas developed resistance to every commonly available antimalarial drug[7], including chloroquine-resistant(CQ-R), multi-drug-resistant (MDR) and others[8−10].Ethnic medicine has provided two of the most efficacious drugs, quinine and artemisinin (and its analogs) and the ongoing screening of medicinal plants yields new lead compounds[11]. Quantitative structure activity relationships are the most important applications of chemo metrics, giving information useful for the design of new compounds acting on a specific target.QSAR (quantitative structure-activity relationship)attempts to find a consistent relationship between biological activity and molecular properties[12].Descriptors are generally used to describe different characteristics/attributes of the chemical structure in order to yield information about the activity/property being studied. In the present work, QSAR studies havebeen performed on triazole-linked chalcone and dienone hybrid derivatives as potential antimalarial[13] to explore important molecular properties as well as the interaction patterns between the D10strain ofPlasmodium falciparumpotency and ligands at the molecular level for design of new potent antimalarial activities. We have used this partial least squares analysis in this venture for QSAR modeling and to predict the drug activity of a series of newly synthesized triazole-linked chalcone derivatives.
2 Materials and method
QSAR studies were performed using the Molecular Design Suite VLife MDS software package, version 3.5 from VLife Sciences, Pune, India[14]. All computational work was performed on a HP Compaq PC running on Intel Pentium-D processor.
2.1 Biological activity dataset for analysis
The QSAR studies were performed on a series of triazole-linked chalcone and dienone hybrid analogs derivatives as antimalarial activity, to which the in vitro antimalarial activities against the D10strain ofPlasmodium falciparumpotency values (measured by IC50) were collected from Ref.[13]. The D10(IC50μM)values were expressed in negative logarithmic units,pIC50(lgIC50) and used as dependent variables in QSAR analysis. The chemical structures and corresponding pIC50are listed in Fig. 1 and Table 1.

Fig. 1 Structure of triazole-linked chacone and dienone hybrid analogs derivative
2.2 Training and test set
Sphere exclusion method[15] was adopted for division of training and test set. Sphere exclusion method is used for creating training and test sets from the data.This is a rational selection method which takes into consideration both biological and chemical spaces for division of dataset. Dissimilarity value provides handle to vary train/test set size. It needs to be adjusted by trial and error until a desired division of train and test set is achieved. As a rule, increase in dissimilarity value will lead to increase in number of molecules in the test set.The compounds of both training and test sets were randomly selected subject to the constraint to ensure complete and representative coverage across the entire range of pIC50values. The models were externally validated using a test set with 10 compounds (Table 1)and were not included in the QSAR models development process.
2.3 Two-dimensional QSAR studies

Table 1 Structures and in vitro antimalarial activities against D10 strains of P. falciparum
All the molecules were constructed using the standard geometry with 2D molecular module of molecular design suite. Three-dimensional structures were drawn for each molecule and the molecular geometries optimized using Monte Carlo conformational search[16]. All the compounds were batch optimized for the minimization of energies and geometry optimization using Merck molecular force field followed by considering distance-dependent dielectric constant of 1.0,convergence criterion or root-mean-square (RMS)gradient at 0.01 kcal/(mol·Å) and the iteration limit to 10000[17]. The purpose of molecular descriptor is to calculate the properties of molecules that serve as numerical characterizations of molecules in other calculations, such as QSAR, diversity analysis.
2.4 Calculation of 2D descriptors
The energy-minimized geometry was used for the calculation of the various 2D descriptors (Individual, Chi,ChiV, Kappa, element count, estate number, estate contribution, Polar surface area and Alignment independent) and was considered independent variables in the present work. The electrostatic descriptors constitute charged polarization, polarity parameter, local dipole index, maximum positive charge, maximum negative charge, total absolute atomic charge, total negative charge, total positive charge. The preprocessing of the independent variables (i.e. descriptors) was done by removing invariable (constant column), which resulted in a total of 250 descriptors to be used for QSAR analysis. The various alignment-independent (AI)descriptors[18] were also calculated. For calculation of AI descriptors every atom in the molecule was assigned at least one and at most three attributes. After all atoms have been assigned their respective attributes, selective distance count statistics for all combinations of different attributes are computed. To calculate AI descriptors, we have used following attributes, 2 (double bonded atom),3 (triple bonded atom), C, N, O, S, H, F, Cl, Br and I and the distance range of 0−7.
2.5 Model validation
Internal validation was carried out using leave-oneout (q2, LOO) method[19]. To calculateq2, each molecule in the training set was sequentially removed,the model refit using same descriptors, and the biological activity of the removed molecule predicted using the refit model. Theq2can be calculated using Eq. (1).

whereyiandyˆiare the actual and predicted activity of thei-th molecule in the training set, respectively;ymeanis the average activity of all molecules in the training set.For external validation, activity of each molecule in the test set was predicted using the model generated from the training set. The2rpredvalue is calculated as

whereyiandyˆiare the actual and predicted activity of thei-th molecule in the test set, respectively;ymeanis the average activity of all molecules in the training set. Both summations are over all molecules in the test set.
3 Results and discussion
A QSAR analysis has been performed to study the quantitative effects of the molecular structure of the substituted triazole-linked chalcone and dienone hybrid on their antimalarial evaluation. Stepwise forward–backward based feature selection coupled with partial least squares was used as a chemometric tool for QSAR modeling. The below models are validated by predicting the biological activities of the training set and test molecules, as indicated in Table 2. Several 2D QSAR models were constructed, and the best three-regression equation obtained is represented as
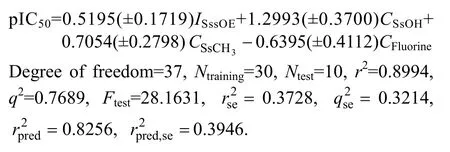
The developed QSAR models are evaluated using the following statistical measures:n(the number of compounds in regression); optimum component (number of optimum PLS components in the model);r2(the squared correlation coefficient),Ftest(the Fischer value for statistical significance),q2(cross-validated correlation coefficient);(r2for external test set).The regression coefficientr2is a relative measure of fit by the regression equation.
Model 1 generated using the SW-PLS method with 0.8994 as the coefficient of determination (r2) was considered the best model using the same molecules in the test and training sets as in QSAR (Table 3). The model can explain 90% of the variance in the observed activity values. The model shows an internal predictive power (q2=0.7689) of 76% and a predictivity for the external test setof about 82%. The activity contribution chart for 2D-QSAR model is shown in Fig. 2. Figure 3 shows the fit plot of experimental vs predicted pIC50values for the training as well as the test sets by the best QSAR Model 1.
The descriptorISssOEwhich is electrotopological state indices for number of oxygen atom connected with two single bonds showed positive contribution with
contribution of ~30%. Such positive effect indicated that the antimalarial activity increased with the presence of methoxy groups such as compounds 1−4; 7−9; 13−20 and 29−31; 35−37. It emphasizes that increase in methoxy of compound will favor the biological activity.The good activity of molecules of 1−4, 7−9, 13−20 and 29−31, 35−37 over other molecules justifies this finding.The next descriptorCSsOH(signifies total number of hydroxy group connected with one single bond) reveals that hydroxy group should be directly attached with chalcone ring for the maximal antimalarial activity and suggests that the increased number of hydroxy atoms will augment the potency of the compounds. The next descriptor isdefining the total number of —CH3group connected with single bond. The positive coefficient of this descriptor signifies the importance of methyl group for antimalarial activities. The next descriptor(~20%) directly proportional to the activity shows the role of the total number of fluorine atom in a molecule. It reveals that presence of electron withdrawing groups over the chalcone and dienone is favorable for the activity (like compounds 6, 11, 12, 22,27, 28, 33, 34, 39 and 40).Table 3 Correlation matrix indicating inter-correlation between descriptors used in model 1
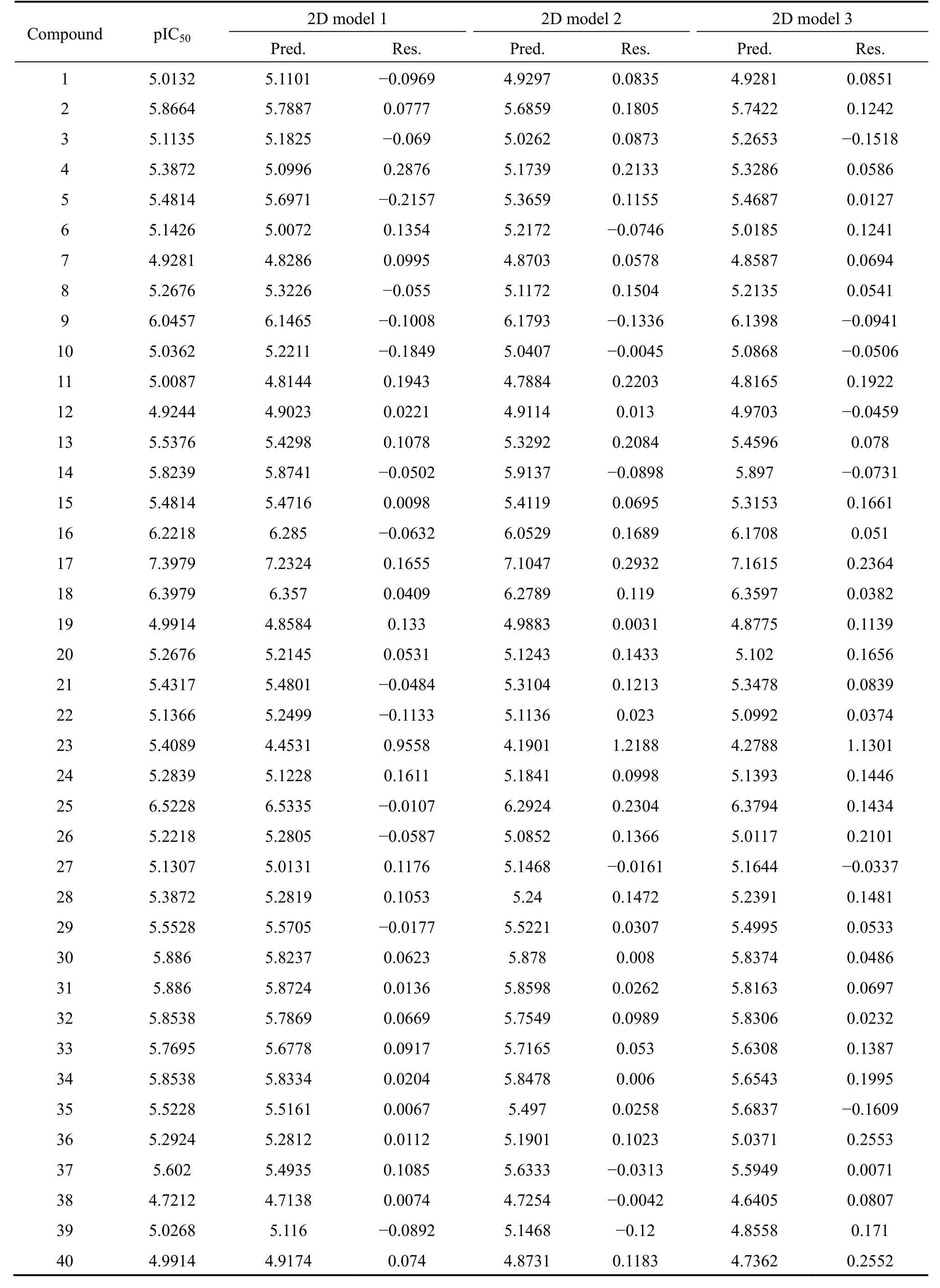
Table 2 Comparative observed and predicted activities of triazole-linked chalcone and dienone derivatives as D10 strains of P.falciparum
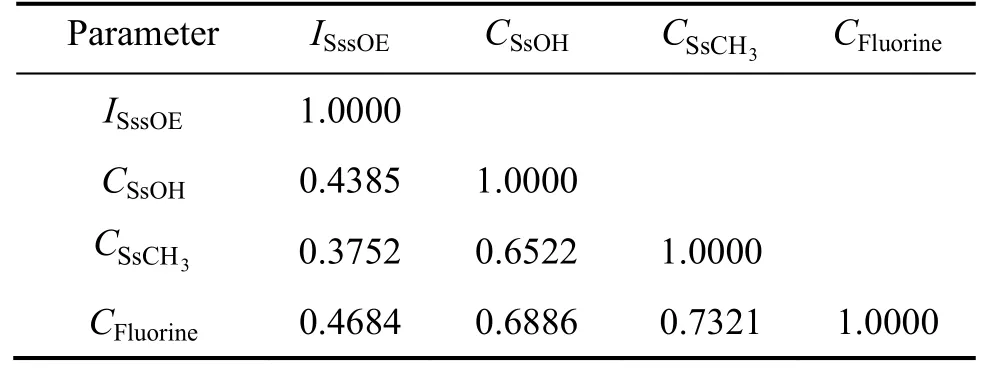
Parameter ISssOE CSsOH 3 C CFluorine SsCH ISssOE 1.0000 CSsOH 0.4385 1.0000 C 0.3752 0.6522 1.0000 CFluorine 0.4684 0.6886 0.7321 1.0000 SsCH3

Fig. 2 Contribution charts of descriptors for 2D model 1

Fig. 3 Graph of observed vs predicted activities of QSAR model 1

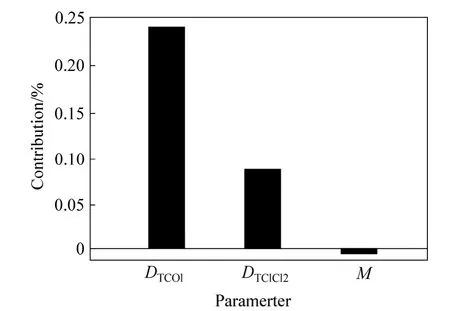
Fig. 4 Contribution charts of descriptors for 2D model 2

Fig. 5 Graph of observed vs predicted activity of QSAR model 2
The developed SW-PLS model reveals that the descriptorDTCO1carbon atoms (single double or triple bonded) separated from any oxygen atom (single or double bonded) by bond distance in a molecule plays most important role (~31%) in determining activity.The alignment-independent descriptorDTClCl2showed positive contribution approx ~25%, which reveals the count of number of chlorine atoms separated from any other chlorine atom by 2 bonds in a molecule results increases in activity as augment by the molecules 5, 10,21, 26, 32 and 38.Msignifies relative molecular mass of a compound. This descriptor is inversely proportional to the activity (~15%) and indices the presence heavy or bulky group which decreases the activity.
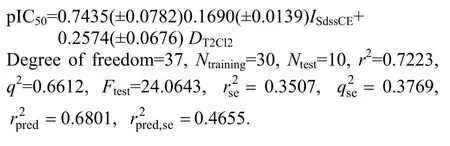
In 2D QSAR model,r2>0.5 suggests significant percentage of the total variance in biological activity is accounted by the model. The stability of model judged by leave-one-out procedure is fairly good (q2>0.6),suggesting that the model can be utilized for predictions.Model 3 was generated using the partial least squares regression method with 0.7223 as the coefficient of determination (r2) was considered using the same molecules in the test and training sets. The model can explain 72 % of the variance in the observed activity values. The model shows an internal predictive power(q2=0.6612) of 66% and predictivity for the external test setof about 68%. The descriptorwhich signifies estate contributions defining electro topologic state indices for the number of =CH2groups attached to two single bonds, also showed a positive contribution. The nextISdssCE, an electrotopological parameter, which defines the total number of carbon atoms connected with one double and two single bonds. The descriptor shows the highest negative correlation among the parameters selected for the derived QSAR model. The negative coefficient suggests that inclusion of such carbon atoms in the molecules lead to decreased antimalarial activity shown by substituted chalcone derivatives.DT2Cl2alignment-independent descriptor which means the count of number of single-bonded atoms separated from chlorine atom by two bonds in a molecule to be detrimental for the activity was exhibited by the compounds. It emphasizes that increase inDT2Cl2of compound will favor the biological activity. The activity contribution chart for QSAR model is shown in Fig. 6 and plots of observed vs predicted values of pIC50are shown in Fig. 7.

Fig. 6 Contribution charts of descriptors for 2D model 3

Fig. 7 Graph of observed vs. predicted activity of QSAR Model 3
4 Conclusions
A quantitative structure–activity relationship(QSAR) study is applied to diverse set of potentially active compounds against the D10, strains ofPlasmodium falciparumstrains of malaria. In pursuit of better antimalarial drugs, a quantitative structure-activity relationship analysis using a novel set of 2D descriptors electrostatic, topological, constitutional, geometrical, and physicochemical descriptors is performed on a series of antimalarial activity triazole-linked chalcone and dienone hybrid. The QSAR models discussed above explains how electron withdrawing, and H-donor properties should be modified to achieve better antimalarial activity. The present work reveals that presence of methoxy, hydroxy groups and less bulky groups at R position of chalcone scaffold increase the antimalarial activity. It shows the requirement of electropositive groups such as methyl,ethyl, propyl, and butyl or less electronegative groups such as cyanide, hydroxyl, amino, nitro, etc. For each set,statistically significant models were obtained using the stepwise forward–backward variable method encoded in software. These models may be considered as mathematical equations for the prediction of antimalarial activities of the compounds structurally similar to those used. We have reported herein the QSAR models for antimalarial activity to update the design process to develop some novel and potent antimalarial agents.
The author is thankful to Vlife Science Technologies Pvt. Ltd. (Pune India) for providing the facility.
[1]World Health Organization:World Malaria Report 2010[R]. Geneva,Switzerland, 2010.
[2]TRIGG P I, KONDRACHINE A V. Malaria parasite biology,pathogenesis and protection:The current global malaria situation[M].Washington, DC:ASM, 1998:11−22.
[3]WHITE N J. Drug resistance in malaria[J]. British Medical Bulletin,1998, 54:703−715.
[4]LI Jia-zhong, LI Shu-yan, BAI Chong-liang, LIU Huan-xiang,PAOLA GRAMATICA. Structural requirements of 3-carboxyl-4(1H)-quinolones as potential antimalarials from 2D and 3D QSAR analysis[J]. Journal of Molecular Graphics and Modelling,2013, 44:266−277.
[5]BEALES P F, BRABIN B, DORMAN E, GILLES H M, LOUTAIN L, MARSH K, MOLYNEUX M E, OLLIARO P, SCHAPIRA A,TOUZE J E, HIEN T T, WARRELL D A, WHITE N. Severe falciparum malaria[J]. Transactions of the Royal Society of Tropical Medicine and Hygiene, 2000, 94:1−90.
[6]WINTER R W, KELLY J X, SMILKSTEIN M J, DODEAN R,HINRICHS D, RISCOE M K. Antimalarial quinolones:Synthesis,potency, and mechanistic studies[J]. Experimental Parasitology,2008, 118:487−497.
[7]CROSS R M, MONASTYRSKYI A, MUTKA T S, BURROWS J N,KYLE D E, MANETSCH R. Endochin optimization:Structure–activity, structure–property relationship studies of 3-substituted 2-methyl-4(1H)-quinolones with antimalarial activity[J]. Journal of Medicinal Chemistry, 2010, 53:7076−7094.
[8]WELLEMS T E, PLOWE C V. Chloroquine-resistant malaria[J].Journal of Infectious Diseases, 2001, 184:770−776.
[9]SIDHU A B S, VERDIER-PINARD D, FIDOCK D A. Chloroquine resistance inPlasmodium falciparummalaria parasites conferred by pfcrt mutations[J]. Science, 2002, 298:210−213.
[10] HYDE J E. Drug-resistant malaria[J]. Trends in Parasitology, 2005,21:494−498.
[11] WILLCOX M L, BODEKER G. Traditional herbal medicines for malaria[J]. BMJ, 2004, 329:1156.
[12] KARELSON M. Molecular Descriptors in QSAR/QSPR[M]. New York:Wiley-Interscience, 2000.
[13] GUANTAI E M, NCOKAZI K, EGAN T J, GUT J, ROSENTHAL P J, SMITH P J, CHIBALE K. Design, synthesis and in vitro antimalarial evaluation of triazole-linked chalcone and dienone hybrid compounds[J]. Bioorganic and Medicinal Chemistry, 2010,18:8243−8256.
[14] VLIFE MDS 3.5:Molecular design suite[M]. Pune, India:Vlife Sciences Technologies Pvt Ltd, 2008.
[15] GOLBRAIKH A, TROPSHA A. Predictive QSAR modeling based on diversity sampling of experimental datasets for the training and test set selection[J]. Journal of Computer-Aided Molecular Design,2002, 16:357−369.
[16] METROPOLIS N, ROSENBLUTH A W, ROSENBLUTH M N,TELLER A H, TELLER E. Equation of state calculations by fast computing machines[J]. Journal of Chemical Physics, 1953, 21:1087−1092.
[17] HALGREN T A. Merck molecular force field II. MMFF94 vander Waals and electrostatic parameters for intermolecular interactions[J].Journal of Computational Chemistry, 1996, 17:520−552
[18] BAUMANN K. An alignment-independent versatile structure descriptor for QSAR and QSPR based on the distribution of molecular features[J]. Journal of Chemical Information and Modeling, 2002, 42:26−35
[19] CRAMER R D, PATTERSON D E, BUNCE J D. Comparative molecular field analysis (CoMFA) 1. Effect of shape on binding of steroids to carrier proteins[J]. Journal of the American Chemical Society, 1998, 110:5959−5967.
© Central South University Press and Springer-Verlag Berlin Heidelberg 2015
10.1007/s11771-015-2917-8
date:2014−12−12; Accepted date:2015−02−22
Mukesh C. Sharma, PhD; Tel:+96−731−2100605; E-mail:mukeshcsharma@yahoo.com
(Edited by DENG Lü-xiang)
杂志排行
Journal of Central South University的其它文章
- Effect of precipitation condition on photosynthesis and biomass accumulation and referring to splash erosion status in five typical evergreen tree species in humid monsoon climatic region of subtropical hill-land
- Effect of wall temperature and random distribution of micro organic dust particles on their combustion parameters
