microRNA在肿瘤中的作用*
2014-05-06张冰洁金由辛马中良
张冰洁金由辛马中良
①硕士研究生,②教授,③副教授,上海大学生命科学学院,上海 200444
*上海市科委自然科学基金(11ZR141220)和国家自然科学基金(31170750)资助
microRNA在肿瘤中的作用*
张冰洁①金由辛②马中良③
①硕士研究生,②教授,③副教授,上海大学生命科学学院,上海 200444
*上海市科委自然科学基金(11ZR141220)和国家自然科学基金(31170750)资助
miRNA;肿瘤;预测;诊断;治疗;预后
miRNA(microRNAs)是一类由内源基因编码的长度为18~23个碱基的非编码单链RNA分子,它们在转录后水平调节基因的表达。据报道,在多种癌症中发现了miRNA表达量改变,提示其可能与癌症的发病机理、肿瘤生长和转移等相关,起到癌基因或抑癌基因的作用。大量证据表明,miRNA的异常表达会促进肿瘤的发生和发展,因此在包括非小细胞肺癌、乳腺癌和前列腺癌等多种癌症中具有临床价值。笔者主要阐述miRNA的生成过程,以及目前已知的相关miRNA在肺癌、乳腺癌、前列腺癌的预测、诊断、治疗和预后中的临床应用及相关分子机制。
癌症是严重影响人类健康的重大疾病,其中肺癌、乳腺癌、前列腺癌等均为常见的恶性肿瘤。miRNA的发现为癌症的治疗开发了新的途径。miRNA几乎参与了所有的基础信号通路,包括调控与肿瘤发生有关的许多重要基因的表达[1]。笔者将探讨目前已知的相关miRNA在肺癌、乳腺癌、前列腺癌等的预测、诊断、治疗和预后中的临床应用及相关分子机制。
1 miRNA的特性、发现与命名
miRNA是一类由内源基因编码的长度为18~23个碱基的非编码单链RNA分子,它们参与生物体内的多种进程,如器官形态形成与改变、免疫系统发育、造血、新陈代谢、应激反应、过渡转变、双边不对称、细胞分化、增殖和凋亡等。miRNA能在转录后水平调节基因表达,从而对细胞进行基础性调控。
miRNA具有高度保守性、序列同源性、时序性及组织特异性,这些性质与其功能密切相关。生物信息学数据显示,miRNA至少能够调节20%~30%的人类基因。单个miRNA能调节多达100个不同的mRNA,并且数据显示有1 000个以上的mRNA受同一个miRNA的调控。miRNA参与多种细胞进程的调控,如细胞增殖、凋亡、细胞周期和分化等。miRNA异常表达调控与肿瘤、心脑血管疾病等多种重大疾病的发生有关。miRNA既可作为抑癌基因下调原癌基因活性,也可作为癌基因下调抑癌基因活性,还可调节肿瘤相关基因的表达,其自身突变、缺失、易位及相互调控异常等还可导致相关基因异常表达。大量研究表明[2-4],miRNA在人类多种肿瘤中表达的改变,与肿瘤发生、发展、诊断、治疗及预后密切相关。
Victor Ambros 实验室于 1993 年在秀丽隐杆线虫 (Caenorbabditis elegans) 中首次发现了miRNA,并将之命名为lin-4,它是线虫发育时序的一个调控因子[5]。与此同时,Gary Ruvkun的实验室鉴定了首个miRNA的靶标基因[6]。这两个重要的发现共同确认了一种新的转录后基因调控机制。然而,当时科学家们并未察觉到miRNA的重要性。时隔7年,当Reinhart及其同事在线虫中鉴定出第2个miRNA——let-7[7],以及let-7与当时已引起人们兴趣的另一类小RNA——siRNA的关系时,科学家们才意识到miRNA的重要性,由此开启了一个全新的转录后基因调控时代。随后,在包括从线虫、果蝇到人类范围的多个物种中都发现了miRNA[8],表明这些分子代表一个基因家族,由一个古老的祖先小分子RNA基因进化而来。
当实验人员开始对数目日益增多的miRNA进行克隆和测序时,建立一个对已公布miRNA进行命名、注释和分类的专业数据库变得日益迫切,于是miRBase数据库应运而生。miRBase数据库是miRNA序列和注释的主要在线储存库,供人们查询已收录miRNA的序列、基因组定位、前体的基因组织形式等相关信息。要建立数据库首先需对miRNA进行系统命名。miRNA成熟体的命名格式为:XXXX-miR-YYYY。“XXXX”代表由3到4个字母组成的物种代码,“YYYY”是顺序的数字识别号,例如hsa-miR-21。对于高度同源的miRNA,则在其数字识别号后加上英文小写字母加以区分,如hsa-miR-34a、hsa-miR-34b等。由不同转录本加工而成的具有相同成熟序列的miRNA,则在其后加上阿拉伯数字以示区别,如hsa-miR-519a-1、hsa-miR-519a-2。由于miRNA命名原则确定之前,lin-4和let-7已被广泛接受,因此保留原名。
2 miRNA的生成与作用机制
在哺乳动物中,miRNA的合成分别在细胞质和细胞核中进行(图1)[9]。在细胞核中,编码miRNA的基因首先在RNA聚合酶II的作用下,产生初始miRNA(pri-miRNA)。然后,在Drosha核酸酶及DGCB8组成的复合体作用下,pri-miRNA被剪切成长度为70~100个核苷酸、具茎环结构的前体miRNA(pre-miRNA)。pre-miRNA被Ran-GTP依赖的转运蛋白Exportin-5特异性识别后从细胞核运送到细胞质内。随后,pre-miRNA在胞质中被Dicer核酸酶切除末端茎环结构,并在双链RNA结合蛋白Loquacious的辅助下,释放出约22 bp miRNA: miRNA*双体。之后,成熟的miRNA进入RNA诱导的基因沉默复合物(RNA-induced silencing complex, RISC)并参与形成非对称RISC复合物(asymmetric RISC assembly)。成熟的miRNA单链与靶mRNA的3'-UTR上的同源序列部分或完全结合,阻碍翻译的继续或引起mRNA降解(少数情况下),从而发挥广泛的生物学作用。双体中只有一条链能结合到RISC上形成成熟的miRNA,另一条链则被降解。两条链形成成熟miRNA的机会是不均等的,这与它们的稳定性不同密切相关。miRNA: miRNA*双螺旋中两条链并非完全配对,每条链的3'端均有2个游离的核苷酸,但miRNA链上靠近5'端有一个不与miRNA*配对的小突起,它明显减弱了miRNA 5'端的稳定性。由于成熟miRNA的产生总是偏向于选择不稳定的5'端,因此miRNA链被选中的概率远高于miRNA*链。

图1 miRNA生成与作用机制
miRNA对靶基因调控主要通过与mRNA的3'-UTR互补配对来实现。若互补程度高,则降解靶mRNA;但在多数情况下,miRNA与靶mRNA为不完全互补配对,抑制其转录后的翻译,而mRNA水平无显著变化。2010年,Khraiwesh等[10]对miRNA的转录后调控机制做了更为深入的阐明。他们在小立碗藓(Physcomitrella patens)中研究发现,Dicer-like1基因编码蛋白中,DICER-LIKE1a蛋白促进miRNA成熟,而DICER-LIKE1b蛋白并不负责miRNA的成熟,控制miRNA对靶位的调控作用。一旦DICER-LIKE1b蛋白突变,将加剧miRNA:靶RNA双链形成,导致编码靶RNA的基因超甲基化,最终导致基因沉默,这些靶序列的转录率急剧降低。另有研究发现,miRNA与靶mRNA识别过程中,miRNA5'端的4~8位碱基在与靶mRNA的结合中比3'端序列更为重要,这段序列被称为种子序列(seed sequence),它在miRNA和靶mRNA配对中起十分关键的作用。由于大多数miRNA抑制基因的功能依赖于部分序列互补,单个miRNA能够靶定多个mRNA,而多个miRNA也能同时作用于一个mRNA,从而在不同的组织和细胞中协同调节基因的表达强度[11]。所以,miRNA可能对蛋白质编码基因具有广泛的微调作用。miRNA的发现改变了后基因组时代对基因调节的认识和理解。
3 miRNA在肿瘤中的作用
全基因组研究显示,miRNA基因通常定位于基因组中出现杂合缺失或扩增的位点、脆性位点、病毒整合位点或其他癌症相关的基因组区域,提示扰乱miRNA的表达和作用将促进癌症的发生与发展[12]。根据早期在肿瘤中的研究,通过对多种miRNA的表达进行分析和研究发现,在多种实体瘤中miRNA的表达呈现大幅度改变现象。与传统mRNA表达分析相比,miRNA表达谱能够更好的分辨不同发育起源的肿瘤[13]。Volinia等[14]在540例肿瘤样本(包括肺癌、乳腺癌、胃癌、前列腺癌、结肠癌和胰腺癌)中进行大规模的miRNA表达分析,发现在实体瘤中大部分miRNA过表达这一特征。
肿瘤相关的miRNA被称为肿瘤microRNA组学(onco-miR)。癌基因性质的miRNA过表达会促进肿瘤发生,它们通过作用于信号通路促进细胞增殖、逃避细胞凋亡、激发无限复制潜能、促进血管生成、侵袭和转移等。同样地,抑癌基因性质的miRNA低表达也能产生相似的影响[15]。以miR-17~92家族为例,它们为癌基因性质的miRNA,在肺癌、B细胞淋巴瘤、乳腺癌和胰腺癌等几种癌症中,此家族的表达上调[14,16]。miR-17~92家族的靶基因包括E2F1、PTEN 和p21[17]。miR-92在肺癌,尤其是非小细胞肺癌(non-small-cell lung cancer,NSCLC)中高表达,它能在体外促进细胞增殖[18]。在小鼠动物实验中,miR-92表达缺失的转基因组小鼠与正常小鼠相比表现出致死性,这些小鼠体型明显偏小且肺脏严重发育不完全。除此之外,突变型小鼠的心脏在发育过程中也有缺陷[19]。Lu等[20]发现,miR-17~92在肺上皮组织中特定过表达导致胚胎致死性表型。其机制可能为miR-92通过调节Rb、Rbl1、Rbl2、PTEN基因从而起作用。
下面将分别介绍miRNA与肺癌、乳腺癌及前列腺癌的发病机理。
3.1 肺癌相关miRNA
目前,肺癌死亡率一直高居癌症死亡率榜首[21],患者5年生存率不到15%。约85%的肺癌为非小细胞肺癌,与小细胞癌相比其癌细胞生长分裂较慢,扩散转移相对较晚。早期诊断发现可能发病的患者是NSCLC治疗的最大挑战之一。如果在原发位阶段将其诊出,则5年生存率将提升至近50%;若在发生淋巴结转移和远处转移时才发现,那么其5年生存率将大大降低。寻找分子生物学标记并将其应用于诊断、治疗和预后将极大地提高患者的生存率。
随着基因组学和蛋白质组学的发展,许多新的候选生物标记物被发现,但仍然缺乏可靠的标记物,对非小细胞肺癌的诊断依旧十分困难[13]。miRNA的发现与鉴定为肺癌的诊断提供了新的思路。
miR-21[22-29]、miR-34a[30-34]、miR-126[35-41]、miR-155[42-43]、miR-130[34]等是肺癌相关miRNA (表1)。miR-21在癌症中一般为高表达。miR-21的表达上调通常与NSCLC病例的恶性结果相关[22-24]。miR-21可作为NSCLC复发与低生存率的预测因子[35-36]。miR-34a在肺癌中低表达,表明其在肺癌中起抑癌基因作用(笔者所在实验室的研究也证明了这一点)。通过多变量分析发现,miR-34a有望成为NSCLC复发的独立预测因子,并且可作为肺癌治疗靶点。miR-126为抑癌基因,在肺癌中表达下调[35-41]。miR-126与NSCLC差预后性相关,证明其可作为NSCLC的特异性独立预后因子。miR-155在NSCLC细胞中一般为高表达[42],在NSCLC病例血清中低表达[43]。miR-155在临床诊断及治疗方面具有较高价值,深入研究其在肺癌中的作用机理、分子机制有助于明确其在肺癌中的作用及其在临床方面的应用。

表1 肺癌相关miRNA
3.2 乳腺癌相关miRNA
乳腺癌是女性最常见的恶性肿瘤,在女性中的死亡率仅次于肺癌[21,45]。2013年,美国新增乳腺癌病例达232 340例,占女性新发恶性肿瘤的29%,排名女性恶性肿瘤发病率首位[21]。在中国,乳腺癌的发病率和死亡率也居高不下,且发病率呈逐年升高趋势。导致乳腺癌患者死亡的主要原因是癌症转移引起的并发症[21]。乳腺癌的转移是一个由多种基因调控、多步骤逐渐发展的复杂过程,涉及肿瘤细胞生长、迁移和侵袭等一系列过程。miR-21[46-57]、miR-34a[58-62]、miR-126[63-66]、miR-155[67-69]、miR-200c[70-74]、miR-221/222[75-78]等多种miRNA参与了乳腺癌转移的基因表达调控和信号通路调控等[63,70,79],在肿瘤转移过程中发挥重要作用(表2)。
miR-21在乳腺癌等多种癌症中普遍高表达,已明确其为原癌基因作用的miRNA。miR-34a在多种癌症中普遍低表达,是较为明确的抑癌基因。miR-34a表达激活是侵袭性乳腺癌表型的标记之一,同时,还可作为乳腺癌低复发风险和低死亡率的独立预测因子。miR-126在乳腺癌中低表达,且与乳腺癌的转移密切相关。miR-126有可能是潜在的乳腺癌转移生物标记物。miR-155在乳腺癌中显著高表达,是明确的原癌基因。Mattiske等[69]对miR-155进行了深入全面的调查,总结了其在乳腺癌中的作用、分子机制和靶基因等,miR-155的靶基因有147个,如RhoA[67]、FOXO3A[68]、SOCS1[69]等。miR-200c在乳腺癌中低表达,提示其为抑癌基因作用的miRNA。miR-200c能够抑制乳腺癌EMT进程[70,80]。 miR-221/222在乳腺癌中普遍高表达。多项研究表明[75-76],miR-221/222与乳腺癌的药物抗性密切相关。

表2 乳腺癌相关miRNAs
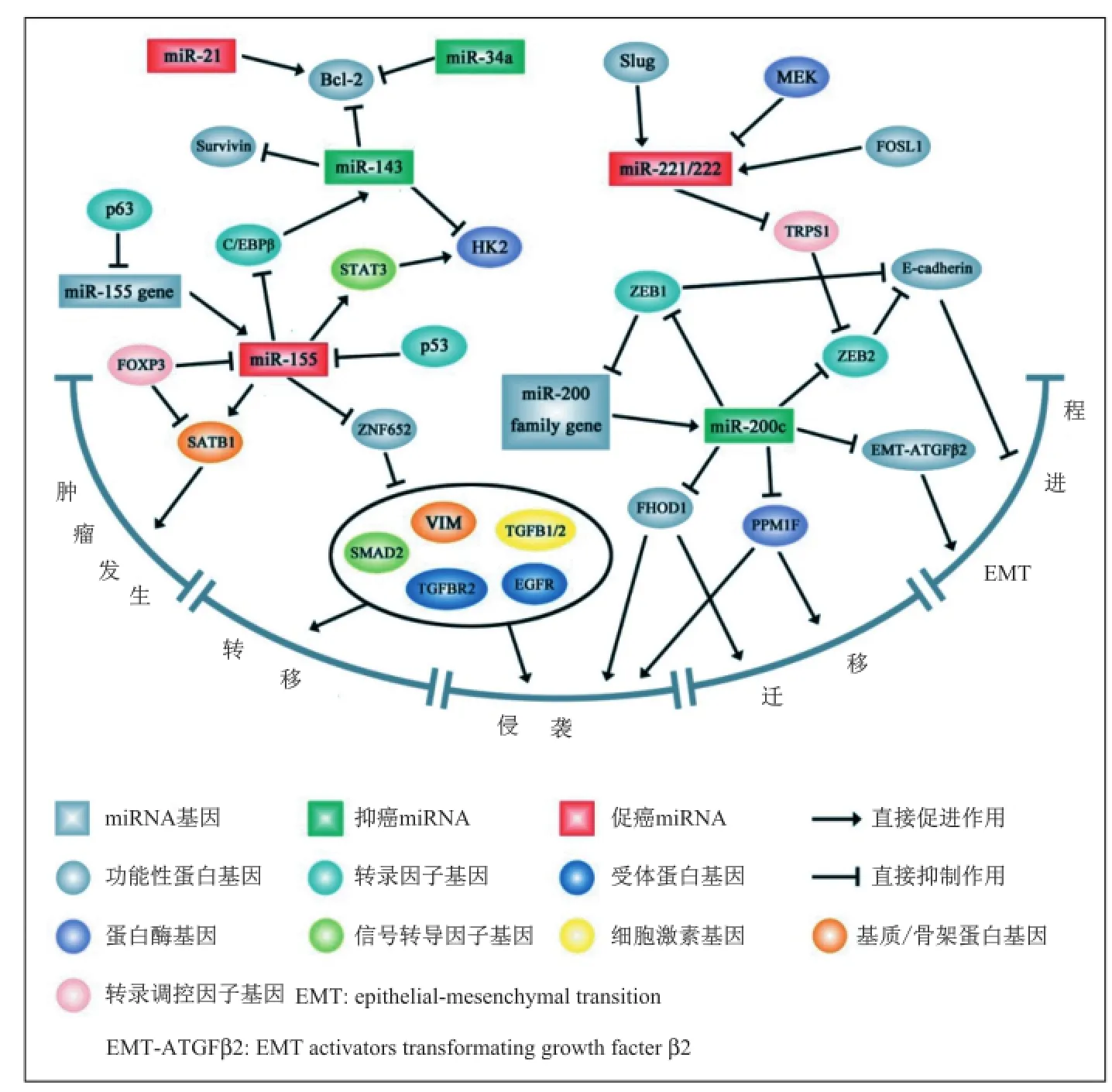
图2 部分相关miRNA在乳腺癌中的调控分子机理
3.3 前列腺癌相关miRNA前列腺癌是男性最常见的恶性肿瘤,是男性因癌症死亡的第2大原因,仅次于肺癌[21,45]。2013年,美国新增前列腺癌病例238 590例,占男性新发恶性肿瘤的28%[21]。经证实[81-83],miRNA参与了前列腺癌发生、发展和转移等一系列过程。miR-21[84-90]、miR-34a[81,91-94]、miR-221/222[95-97]、miR-205[46-48,95,98-100]等miRNA在前列腺癌中的作用详见表3。

表3 前列腺癌相关miRNAs
4 展望
miRNA是一类非编码单链RNA分子,它们通过调控基因表达和细胞内信号通路参与肿瘤的发生和发展等一系列过程。miRNA的独特生物学特性使其成为最为重要的肿瘤预测、诊断和预后的生物标记物及治疗靶点。然而,miRNA在肿瘤中的作用机制十分复杂,其所参与的调控网络十分广泛,同一miRNA在不同癌症中的表达水平和作用可能不同,甚至同一miRNA在某种癌症的不同分期/分型中的表达水平和作用也可能不同。深入透彻地研究特定miRNA在特定癌症中的作用、分子机制和调控网络有助于更好地了解癌症的发病机理,并能更好地找出相应的治疗方法。
(2013年11月5日收稿)■
[1] AMBROS V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing [J]. Cell, 2003, 113: 673-676.
[2] LU J, GETZ G, MISKA E A, et al. MicroRNA expression profiles classify human cancers [J]. Nature, 2005, 435: 834-838.
[3] VOORTMAN J, GOTO A, MENDIBOURE J, et al. MicroRNA expression and clinical outcomes in patients treated with adjuvant chemotherapy after complete resection of non-small cell lung carcinoma [J]. Cancer Res, 2010, 70: 8288-8298.
[4] LIU J, ZHENG M, TANG Y L, et al. MicroRNAs, an active and versatile group in cancers [J]. Int J Oral Sci, 2011, 3: 165-175.
[5] LEE R C, FEINBAUM R L, AMBROS V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14 [J]. Cell, 1993, 75: 843-854.
[6] WIGHTMAN B, HA I, RUVKUN G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans [J]. Cell, 1993, 75: 855-862.
[7] REINHART B J, SLACK F J, BASSON M, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans [J]. Nature, 2000, 403: 901-906.
[8] LAGOS-QUINTANA M, RAUHUT R, MEYER J, et al. New microRNAs from mouse and human [J]. RNA, 2003, 9: 175-179.
[9] AMBROS V. microRNAs: tiny regulators with great potential [J]. Cell, 2001, 107: 823-826.
[10] KHRAIWESH B, ARIF M A, SEUMEL G I, et al. Transcriptional control of gene expression by microRNAs [J]. Cell, 2010, 140: 111-122.
[11] TRANG P, WIGGINS J F, DAIGE C L, et al. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice [J]. Mol Ther, 2011, 19: 1116-1122.
[12] CALIN G A, CROCE C M. MicroRNA signatures in human cancers [J]. Nat Rev Cancer, 2006, 6: 857-866.
[13] LUDWIG J A, WEINSTEIN J N. Biomarkers in cancer staging, prognosis and treatment selection [J]. Nat Rev Cancer, 2005, 5: 845-856.
[14] VOLINIA S, CALIN G A, LIU C G, et al. A microRNA expression signature of human solid tumors defines cancer gene targets [J]. Proc Natl Acad Sci USA, 2006, 103: 2257-2261.
[15] HANAHAN D, WEINBERG R A. The hallmarks of cancer [J]. Cell,2000, 100: 57-70.
[16] OTA A, TAGAWA H, KARNAN S, et al. Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma [J]. Cancer Res, 2004, 64: 3087-3095.
[17] MENDELL J T. miRiad roles for the miR-17-92 cluster in development and disease [J]. Cell, 2008, 133: 217-222.
[18] NANA-SINKAM S P, KARSIES T, RISCILI B, et al. Lung microRNA: from development to disease [J]. Expert Rev Respir Med, 2009, 3: 373-385.
[19] VENTURA A, YOUNG A G, WINSLOW M M, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters [J]. Cell, 2008, 132: 875-886.
[20] LU Y, THOMSON J M, WONG H Y, et al. Transgenic over-expression of the microRNA miR-17-92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells [J]. Dev Biol, 2007, 310: 442-453.
[21] SIEGEL R, NAISHADHAM D, JEMAL A. Cancer statistics, 2013 [J]. CA Cancer J Clin, 2013, 63: 11-30.
[22] FREZZETTI D, DE MENNA M, ZOPPOLI P, et al. Upregulation of miR-21 by Ras in vivo and its role in tumor growth [J]. Oncogene, 2011, 30: 275-286.
[23] HATLEY M E, PATRICK D M, GARCIA M R, et al. Modulation of K-Ras-dependent lung tumorigenesis by microRNA-21 [J]. Cancer Cell, 2010, 18: 282-293.
[24] LIU Z L, WANG H, LIU J, et al. MicroRNA-21 (miR-21) expression promotes growth, metastasis, and chemo- or radioresistance in nonsmall cell lung cancer cells by targeting PTEN [J]. Mol Cell Biochem, 2013, 372: 35-45.
[25] ZHONG Z, DONG Z, YANG L, et al. miR-21 induces cell cycle at S phase and modulates cell proliferation by down-regulating hMSH2 in lung cancer [J]. J Cancer Res ClinOncol, 2012, 138: 1781-1788.
[26] SAITO M, SCHETTER A J, MOLLERUP S, et al. The association of microRNA expression with prognosis and progression in early-stage, non-small cell lung adenocarcinoma: a retrospective analysis of three cohorts [J]. Clin Cancer Res, 2011, 17: 1875-1882.
[27] MA X L, LIU L, LIU X X, et al. Prognostic role of microRNA-21 in non-small cell lung cancer: a meta-analysis [J]. Asian Pac J Cancer Prev, 2012, 13: 2329-2334.
[28] LIU X G, ZHU W Y, HUANG Y Y, et al. High expression of serum miR-21 and tumor miR-200c associated with poor prognosis in patients with lung cancer [J]. Med Oncol, 2012, 29: 618-626.
[29] YANG M, SHEN H, QIU C, et al. High expression of miR-21 and miR-155 predicts recurrence and unfavourable survival in non-small cell lung cancer [J]. Eur J Cancer, 2013, 49: 604-615.
[30] GALLARDO E, NAVARRO A, VINOLAS N, et al. miR-34a as a prognostic marker of relapse in surgically resected non-small-cell lung cancer [J]. Carcinogenesis, 2009, 30: 1903-1909.
[31] MUDDULURU G, CEPPI P, KUMARSWAMY R, et al. Regulation of Axl receptor tyrosine kinase expression by miR-34a and miR-199a/b in solid cancer [J]. Oncogene, 2011, 30: 2888-2899.
[32] BANDI N, VASSELLA E. miR-34a and miR-15a/16 are co-regulated in non-small cell lung cancer and control cell cycle progression in a synergistic and Rb-dependent manner [J]. Mol Cancer, 2011, 10: 55.
[33] JI X, WANG Z, GEAMANU A, et al. Delta-tocotrienol suppresses Notch-1 pathway by upregulating miR-34a in nonsmall cell lung cancer cells [J]. Int J Cancer, 2012, 131: 2668-2677.
[34] AHN Y H, GIBBONS D L, CHAKRAVATI D, et al. ZEBI drives prometastatic actin cytoskeletal remodeling by downregulating miR-34a expression [J]. The Journal of Clinical Investigation, 2012, 122: 3170-3183.
[35] ACUNZO M, VISONE R, ROMANO G, et al. miR-130a targets MET and induces TRAIL-sensitivity in NSCLC by downregulating miR-221 and 222 [J]. Oncogene, 2012, 31: 634-642.
[36] DONNEM T, LONVIK K, EKLO K, et al. Independent and tissuespecific prognostic impact of miR-126 in nonsmall cell lung cancer: coexpression with vascular endothelial growth factor-A predicts poor survival [J]. Cancer, 2011, 117: 3193-3200.
[37] ZHU X, LI H, LONG L, et al. miR-126 enhances the sensitivity of non-small cell lung cancer cells to anticancer agents by targeting vascular endothelial growth factor A [J]. ActaBiochimBiophys Sin (Shanghai), 2012, 44: 519-526.
[38] JUSUFOVIC E, RIJAVEC M, KESER D, et al. let-7b and miR-126 are down-regulated in tumor tissue and correlate with microvessel density and survival outcomes in non-small-cell lung cancer [J]. PloS One, 2012, 7: e45577.
[39] DONNEM T, FENTON C G, LONVIK K, et al. MicroRNA signatures in tumor tissue related to angiogenesis in non-small cell lung cancer [J]. PloS One, 2012, 7: e29671.
[40] SUN Y, BAI Y, ZHANG F, et al. miR-126 inhibits non-small cell lung cancer cells proliferation by targeting EGFL7 [J]. Biochem Biophys Res Commun, 2010, 391: 1483-1489.
[41] ZHONG M, MA X, SUN C, et al. MicroRNAs reduce tumor growth and contribute to enhance cytotoxicity induced by gefitinib in nonsmall cell lung cancermiR-126 inhibits non-small cell lung cancer cells proliferation by targeting EGFL7 [J]. Chemico-Biological Interactions, 2010, 184: 431-438.
[42] YANG J, LAN H, HUANG X, et al. MicroRNA-126 inhibits tumor cell growth and its expression level correlates with poor survival in non-small cell lung cancer patients [J]. PloS One, 2012, 7: e42978.
[43] DONNEM T, EKLO K, BERG T, et al. Prognostic impact of MiR-155 in non-small cell lung cancer evaluated by in situ hybridization [J]. Journal of Translational Medicine, 2011, 9: 6.
[44] HEEGAARD N H, SCHETTER A J, WELSH J A, et al. Circulating micro-RNA expression profiles in early stage nonsmall cell lung cancer [J]. Int J Cancer, 2012, 130: 1378-1386.
[45] SIEGEL R, WARD E, BRAWLEY O, et al. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths [J]. CA Cancer J Clin, 2011, 61: 212-236.
[46] MAJID S, DAR A A, SAINI S, et al. MicroRNA-205-directed transcriptional activation of tumor suppressor genes in prostate cancer [J]. Cancer, 2010, 116: 5637-5649.
[47] GANDELLINI P, PROFUMO V, CASAMICHELE A, et al. miR-205 regulates basement membrane deposition in human prostate: implications for cancer development [J]. Cell Death Differ, 2012, 19: 1750-1760.
[48] TUCCI P, AGOSTINI M, GRESPI F, et al. Loss of p63 and its microRNA-205 target results in enhanced cell migration and metastasis in prostate cancer [J]. Proc Natl Acad Sci USA, 2012, 109: 15312-15317. [49] LEE J H, VOORTMAN J, DINGEMANS A M, et al. MicroRNA expression and clinical outcome of small cell lung cancer [J]. PloS One, 2011, 6: e21300.
[50] QI L, BART J, TAN L P, et al. Expression of miR-21 and its targets (PTEN, PDCD4, TM1) in flat epithelial atypia of the breast in relation to ductal carcinoma in situ and invasive carcinoma [J]. BMC Cancer, 2009, 9: 163.
[51] YANG Y, CHAERKADY R, BEER M A, et al. Identification of miR-21 targets in breast cancer cells using a quantitative proteomic approach [J]. Proteomics, 2009, 9: 1374-1384.
[52] YAN L X, WU Q N, ZHANG Y, et al. Knockdown of miR-21 in human breast cancer cell lines inhibits proliferation, in vitro migration and in vivo tumor growth [J]. Breast Cancer Res, 2011, 13: R2.
[53] WICKRAMASINGHE N S, MANAVALAN T T, DOUGHERTY S M, et al. Estradiol downregulates miR-21 expression and increases miR-21 target gene expression in MCF-7 breast cancer cells [J]. Nucleic Acids Res, 2009, 37: 2584-2595.
[54] SELCUKLU S D, DONOGHUE M T, KERIN M J, et al. Regulatory interplay between miR-21, JAG1 and 17beta-estradiol (E2) in breast cancer cells [J]. Biochem Biophys Res Commun, 2012, 423: 234-239.
[55] WALTER B A, GOMEZ-MACIAS G, VALERA V A, et al. miR-21Expression in pregnancy-associated breast cancer: a possible marker of poor prognosis [J]. J Cancer, 2011, 2: 67-75.
[56] RASK L, BALSLEV E, JORGENSEN S, et al. High expression of miR-21 in tumor stroma correlates with increased cancer cell proliferation in human breast cancer [J]. APMIS, 2011, 119: 663-673.
[57] MAR-AGUILAR F, LUNA-AGUIRRE C M, MORENO-ROCHA J C, et al. Differential expression of miR-21, miR-125b and miR-191 in breast cancer tissue [J]. Asia Pac J Clin Oncol, 2013, 9: 53-59.
[58] LI X J, JI M H, ZHONG S L, et al. Microrna-34a modulates chemosensitivity of breast cancer cells to adriamycin by targeting notch1 [J]. Arch Med Res, 2012, 43: 514-521.
[59] LI L, YUAN L, LUO J, et al. MiR-34a inhibits proliferation and migration of breast cancer through down-regulation of Bcl-2 and SIRT1 [J]. Clin Exp Med, 2013, 13: 109-117.
[60] KASTL L, BROWN I, SCHOFIELD A C. miRNA-34a is associated with docetaxel resistance in human breast cancer cells [J]. Breast Cancer Res Treat, 2012, 131: 445-454.
[61] PEURALA H, GRECO D, HEIKKINEN T, et al. MiR-34a expression has an effect for lower risk of metastasis and associates with expression patterns predicting clinical outcome in breast cancer [J]. PloS one, 2011, 6: e26122.
[62] YANG S, LI Y, GAO J, et al. MicroRNA-34 suppresses breast cancer invasion and metastasis by directly targeting Fra-1 [J]. Oncogene, 2013, 32: 4294-4303.
[63] PNG K J, HALBERG N, YOSHIDA M, et al. A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells [J]. Nature, 2012, 481: 190-194.
[64] WANG F, ZHENG Z, GUO J, et al. Correlation and quantitation of microRNA aberrant expression in tissues and sera from patients with breast tumor [J]. Gynecol Oncol, 2010, 119: 586-593.
[65] ZHANG J, DU Y Y, LIN Y F, et al. The cell growth suppressor, mir-126, targets IRS-1 [J]. Biochem Biophys Res Commun, 2008, 377: 136-140.
[66] ZHU N, ZHANG D, XIE H, et al. Endothelial-specific intron-derived miR-126 is down-regulated in human breast cancer and targets both VEGFA and PIK3R2 [J]. Mol Cell Biol, 2011, 351: 157-164.
[67] KONG W, YANG H, HE L, et al. MicroRNA-155 is regulated by the transforming growth factor beta/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA [J]. Mol Cell Biol, 2008, 28: 6773-6784.
[68] ZHU J, HU XQ, GUO GL, et al. [Expression and its clinical significance of miR-155 in human primary breast cancer] [J]. Zhonghua wai ke za zhi [Chinese Journal of Surgery], 2010, 48: 205-208.
[69] MATTISKE S, SUETANI R J, NEILSEN P M, et al. The oncogenic role of miR-155 in breast cancer [J]. Cancer Epidemiol Biomarkers Prev, 2012, 21: 1236-1243.
[70] HOWE E N, COCHRANE D R, RICHER J K. Targets of miR-200c mediate suppression of cell motility and anoikis resistance [J]. Breast Cancer Res, 2011, 13: R45.
[71] JURMEISTER S, BAUMANN M, BALWIERZ A, et al. MicroRNA-200c represses migration and invasion of breast cancer cells by targeting actin-regulatory proteins FHOD1 and PPM1F [J]. Mol Cell Biol, 2012, 32: 633-6
[72] LIN J, LIU C, GAO F, et al. MiR-200c enhances radiosensitivity of human breast cancer cells [J]. J Cell Biochem, 2013, 114: 606-615.
[73] HOWE E N, COCHRANE D R, CITTELLY D M, et al. miR-200c targets a NF-kappaB up-regulated TrkB/NTF3 autocrine signaling loop to enhance anoikis sensitivity in triple negative breast cancer [J]. PloS One, 2012, 7: e49987.
[74] KOPP F, OAK P S, WAGNER E, et al. miR-200c sensitizes breast cancer cells to doxorubicin treatment by decreasing TrkB and Bmi1 expression [J]. PloS One, 2012, 7: e50469.
[75] LU Y, ROY S, NUOVO G, et al. Anti-microRNA-222 (anti-miR-222) and -181B suppress growth of tamoxifen-resistant xenografts in mouse by targeting TIMP3 protein and modulating mitogenic signal [J]. J Biol Chem, 2011, 286: 42292-42302.
[76] RAO X, DI LEVA G, LI M, et al. MicroRNA-221/222 confers breast cancer fulvestrant resistance by regulating multiple signaling pathways [J]. Oncogene, 2011, 30: 1082-1097.
[77] ZHAO J J, LIN J, YANG H, et al. MicroRNA-221/222 negatively regulates estrogen receptor alpha and is associated with tamoxifen resistance in breast cancer [J]. J Biol Chem, 2008, 283: 31079-31086.
[78] STINSON S, LACKNER M R, ADAI A T, et al. miR-221/222 targeting of trichorhinophalangeal 1 (TRPS1) promotes epithelial-tomesenchymal transition in breast cancer [J]. Sci Signal, 2011, 4: pt5.
[79] TAVAZOIE S F, ALARCON C, OSKARSSON T, et al. Endogenous human microRNAs that suppress breast cancer metastasis [J]. Nature, 2008, 451: 147-152.
[80] RADISKY D C. miR-200c at the nexus of epithelial-mesenchymal transition, resistance to apoptosis, and the breast cancer stem cell phenotype [J]. Breast Cancer Res, 2011, 13: 110.
[81] FUJITA Y, KOJIMA K, HAMADA N, et al. Effects of miR-34a on cell growth and chemoresistance in prostate cancer PC3 cells [J]. Biochem Biophys Res Commun, 2008, 377: 114-119.
[82] GANDELLINI P, FOLINI M, LONGONI N, et al. miR-205 exerts tumor-suppressive functions in human prostate through downregulation of protein kinase Cepsilon [J]. Cancer Res, 2009, 69: 2287-2295.
[83] SHEN J, HRUBY G W, MCKIERNAN J M, et al. Dysregulation of circulating microRNAs and prediction of aggressive prostate cancer [J]. The Prostate, 2012, 72: 1469-1477.
[84] LU Z, LIU M, STRIBINSKIS V, et al. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene [J]. Oncogene, 2008, 27: 4373-4379.
[85] FINLAYSON A E, FREEMAN K W. A cell motility screen reveals role for MARCKS-related protein in adherens junction formation and tumorigenesis [J]. PloS One, 2009, 4: e7833.
[86] RIBAS J, NI X, HAFFNER M, et al. miR-21: an androgen receptorregulated microRNA that promotes hormone-dependent and hormoneindependent prostate cancer growth [J]. Cancer Res, 2009, 69: 7165-7169.
[87] SHI G H, YE D W, YAO X D, et al. Involvement of microRNA-21 in mediating chemo-resistance to docetaxel in androgen-independent prostate cancer PC3 cells [J]. Acta Pharmacologica Sinica, 2010, 31: 867-873.
[88] DARIMIPOURAIN M, WANG S, ITTMANN M, et al. Transcriptional and post-transcriptional regulation of Sprouty1, a receptor tyrosine kinase inhibitor in prostate cancer [J]. Prostate Cancer Prostatic Dis, 2011, 14: 279-285.
[89] REIS S T, PONTES-JUNIOR J, ANTUNES A A, et al. miR-21 may acts as an oncomir by targeting RECK, a matrix metalloproteinase regulator, in prostate cancer [J]. BMC Urology, 2012, 12: 14.
[90] LI J, WANG Y, LUO J, et al. miR-134 inhibits epithelial to mesenchymal transition by targeting FOXM1 in non-small cell lung cancer cells [J]. FEBS letters, 2012, 586: 3761-3765.
[91] KOJIMA K, FUJITA Y, NOZAWA Y, et al. MiR-34a attenuates paclitaxel-resistance of hormone-refractory prostate cancer PC3 cells through direct and indirect mechanisms [J]. Prostate, 2010, 70: 1501-1512.
[92] LIU C, KELNAR K, LIU B, et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44 [J]. Nat Med, 2011, 17: 211-215.
[93] KASHAT M, AZZOUZ L, SARKAR SH, et al. Inactivation of AR and Notch-1 signaling by miR-34a attenuates prostate cancer aggressiveness [J]. Am J Transl Res, 2012, 4: 432-442.
[94] YAMAMURA S, SAINI S, MAJID S, et al. MicroRNA-34a modulates c-Myc transcriptional complexes to suppress malignancy in human prostate cancer cells [J]. PloS One, 2012, 7: e29722.
[95] SPAHN M, KNEITZ S, SCHOLZ C J, et al. Expression of microRNA-221 is progressively reduced in aggressive prostate cancer and metastasis and predicts clinical recurrence [J]. Int J Cancer, 2010, 127: 394-403.
[96] ZHENG C, YINGHAO S, LI J. MiR-221 expression affects invasion potential of human prostate carcinoma cell lines by targeting DVL2 [J]. Med Oncol, 2012, 29: 815-822.
[97] CHEN Y, ZAMAN M S, DENG G, et al. MicroRNAs 221/222 and genistein-mediated regulation of ARHI tumor suppressor gene in prostate cancer [J]. Cancer Prevention Research, 2011, 4: 76-86.
[98] BHATNAGAR N, LI X, PADI S K, et al.Downregulation of miR-205 and miR-31 confers resistance to chemotherapy-induced apoptosis in prostate cancer cells [J]. Cell Death Dis, 2010, 1: e105.
[99] HULF T, SIBBRITT T, WIKLUND E D, et al. Epigenetic-induced repression of microRNA-205 is associated with MED1 activation and a poorer prognosis in localized prostate cancer [J]. Oncogene, 2013, 32: 2891-2899.
[100] SZCZYRBA J, NOLTE E, HART M, et al. Identification of ZNF217, hnRNP-K, VEGF-A and IPO7 as targets for microRNAs that are downregulated in prostate carcinoma [J]. Int J Cancer, 2013, 132: 775-784.
The roles of microRNAs in cancer
ZHANG Bing-jie①, JIN You-xin②, MA Zhong-liang③
①Master Candidate, ②Professor, ③Associate Professor, School of Life Sciences, Shanghai University, Shanghai 200444, China
miRNAs, 18~23 nucleotide are a class of single stranded small non-coding RNA encoded by endogenous genes, which regulate gene expression at the post-transcriptional level. Abberant expressions of miRNAs reported in many kinds of human cancers may associate with cancer pathogenesis, tumor growth and metastasis, thus they can function as either oncogenes or antioncogenes. Numerous evidences indicated that the disregulation of miRNAs contribute to tumor initiation and progression. So these miRNAs can be used as clinical diagnosis biomarkers in many cancers, including non-small-cell lung cancer, breast cancer and prostate cancer. We introduce the clinical applications of the current known of miRNAs in early diagnosis, therapy and prognosis of non-small-cell lung cancer, breast cancer and prostate cancer, as well as their molecular mechanisms.
miRNA, cancer, predict, diagnosis, therapy, prognosis
(编辑:段艳芳)
10.3969/j.issn.0253-9608.2014.05.008
