Phylogenetic Analysis of Epibacterial Communities on the Surfaces of Four Red Macroalgae
2014-04-26WUHongqingLIUMinZHANGWuchangandXIAOTian
WU Hongqing, LIU Min, ZHANG Wuchang and XIAO Tian
1) Key Laboratory of Marine Ecology and Environmental Sciences, Institute of Oceanology, Chinese Academy of Sciences (CAS), Qingdao 266071, P. R. China
2) School of Marine Science and Engineering, Hebei University of Technology, Tianjin 300130, P. R. China
3) Graduate University of Chinese Academy of Sciences, Beijing 100049, P. R. China
4) Key Laboratory of Biology and Genetic Resources of Tropical Crops, Institute of Tropical Bioscience and Biotechnology, Chinese Academy of Tropical Agricultural Science (CATAS), Haikou 571101, P. R. China
Phylogenetic Analysis of Epibacterial Communities on the Surfaces of Four Red Macroalgae
WU Hongqing1),2),3), LIU Min1),4), ZHANG Wuchang1), and XIAO Tian1),*
1) Key Laboratory of Marine Ecology and Environmental Sciences, Institute of Oceanology, Chinese Academy of Sciences (CAS), Qingdao 266071, P. R. China
2) School of Marine Science and Engineering, Hebei University of Technology, Tianjin 300130, P. R. China
3) Graduate University of Chinese Academy of Sciences, Beijing 100049, P. R. China
4) Key Laboratory of Biology and Genetic Resources of Tropical Crops, Institute of Tropical Bioscience and Biotechnology, Chinese Academy of Tropical Agricultural Science (CATAS), Haikou 571101, P. R. China
Macroalgal surfaces are prone to being attached by bacteria. Epibacterial community structures on marine macroalgae are host-specific but temporally and spatially variable. In this study, we investigated the structure of epibacterial communities on the surfaces of four red macroalgae, Gracilaria lemaneiformis, Gloiopeltis furcata, Mazzaella sp. and Porphyra yezoensis, by analyzing the sequences of 16S rRNA gene libraries. Healthy individuals of all macroalgae species were collected in winter from a farm at Dalian, China. The results showed that the epibacterial communities were mainly dominated by α-Proteobacteria, γ-Proteobacteria and Bacteroidetes. Deinococcus-Thermus, Spirochaetes and ε-Proteobacteria were also found. The majority of cloned sequences shared the greatest similarity to those of culturable organisms. A large portion of sequences from the α-Proteobacteria homed in Roseobacter clade, i.e., genera Ahrensia, Roseovarius, Litoreibacter, Octadecabacter, Thaiassobacter and Sulfitobacter, while members of Bacteroidetes mainly belonged to family Flavobacteriaceae. The cloned sequences could be separated into 66 OTUs at 0.01 distance value, and rare common OTUs were found among libraries. At genus level, Pseudoalteromonas dominated Gr. lemaneiformis and Gl. furcata libraries, accounting for 72.2% and 47.3%, respectively. Sulfitobacter dominated P. yezoensis library, accounting for 35.4%. A previously undefined cluster within Deinococcus-Thermus dominated Mazzaella sp. library, accounting for 24.6% of the all. These results indicated that a broad range of bacteria inhabited the surfaces of these macroalgae.
epibacterial community; red alga; 16S rRNA gene
1 Introduction
Macroalgae are important natural resources for a wide variety of products, such as food, alginate, agar, carrageenan, fertilizers and animal feed additives. The vast majority of macroalgae for these products are produced by aquaculture. Macroalgae cultivation industry is developing fast in many countries in the world and algal cultivation in China accounts for a large portion of global annual production (Roesijadi et al., 2010). Macroalgae can only live in euphotic zone and many vital processes take place at algal surface, e.g., the exudation and uptake of nutrients, gases and the absorption of light. They release a large amount of organic carbon into surrounding environment (Sieburth, 1969), thus their surfaces are prone to being attached by bacteria. Furthermore, these bacteria may influence the interaction between macroalgae and environment (Wahl, 2008). Controlling the epibacterialcommunity is substantial for individual algae, and macroalgae have different strategies of modulating the growth of surface bacteria (Keats et al., 1993; Xu et al., 2003; Bhadury and Wright, 2004; Nylund and Pavia, 2005).
Epibacterial communities on marine macroalgae are host-specific, which may vary temporally (Ashen and Goff, 2000; Lachnit et al., 2009, 2011). Recently, many scientists are engaging in the detailed investigation of the epibacterial community of macroalgae. Yang et al. (2008) isolated 63 bacterial strains from Porphyra yezoensis. These isolates had high similarities with the members of 10 genera such as Pseudoalteromonas, Psychrobacter and Bacillus. Lachnit et al. (2011) reported that epibacterial community of Gracilaria vermiculophylla was dominated by α-Proteobacteria and Bacteroidetes in summer. The proportion of α-Proteobacteria increased while the abundance of Bacteroidetes decreased, and particularly the phylum of Deinococcus presented only in winter. Epibacteria are required for some macroalgae as they can provide essential vitamins and growth factors to macroalgae (Tsavkelova et al., 2006; Hodson et al., 2007). Some bacteria can release chemicals, preventing the hostalgae from being biofouled by other organisms (Holmström and Kjelleberg, 1999; Rao et al., 2007). However, some bacteria on algal thalli are known as pathogens. Pseudoalteromonas tetraodonis was assumed to cause‘yellow spot disease’ in Porphyra yezoensis (Wang et al., 2011), while Pseudoalteromonas elyakovii and P. bacteriolytica were believed to associate with ‘red spot diseases’ in Laminaria japonica (Sawabe et al., 1998, 2000). In addition, some isolates of Vibrio sp. were identified as agar-digesting bacteria, which associated with ‘rotten thallus syndrome’ of Gracilaria sp. (Celia, 1992).
To avoid the diseases caused by epibacteria, it is necessary to investigate the distribution of epibacteria on algal surface. Red macroalgae, especially some species of genus Eucheuma, Gloiopeltis, Gracilaria, Grateloupia, Mazzaella and Porphyra, are widely cultured in China, whereas studies dealing with the epibacterial communities on the surfaces of these algae are scarce. In this study, the composition of epibacterial communities on the surfaces of four red algae, Gracilaria lemaneiformis, Gloiopeltis furcata, Mazzaella sp. and Porphyra yezoensis, was unraveled by constrcting and sequencing 16S rRNA gene libraries.
2 Materials and Methods
2.1 Sample Collection
Four species of marine red algae, Gracilaria lemaneiformis, Gloiopeltis furcata, Mazzaella sp. and Porphyra yezoensis, were collected on February 22nd, 2008 from a farm at Dalian, China. The samples were stored in separate sterile plastic bags and placed on ice for transportation to laboratory within 12 h after sampling.
2.2 Detachment of Epibacteria
Bacteria were detached from samples with a procedure described by Matsuo et al. (2003) with some modifications. In detail, 10 g of thalli cut from different individuals (>10) were rinsed gently with sterile seawater three times to remove debris and loosely attached bacteria. Rinsed thalli were then transferred to a sterile 500 mL Erlenmeyer flask containing 200 mL seawater and 20 g glass beads (4–5 mm in diameter). The epibacterial components were detached by shaking (150 r min-1) for 10 min and the suspension was collected. The remainder of the algae was treated with the same procedure four times. Totally, about 1 L suspension containing the detached bacteria was collected and then filtered through a 0.22 μm filter (Millipore). The filters attached with bacteria collected from macroalgae were stored at -20℃ for DNA extraction.
2.3 DNA Extraction, PCR and Construction of Libraries
Filters of the same algae were cut into pieces and mixed under sterile conditions. Total DNA was extracted following Fuhrman et al. (1988). Bacterial 16S rRNA gene was amplified by PCR with universal bacterial primers 27F (5’-AGAGTTTGATCCTGGCTCAG-3’) and 1492R (5’-GGTTACCTTGTTACGACTT-3’) (Weisburg et al., 1991). The amplification condition was as follows: an initial denaturation at 94℃ for 10 min, followed by 30 cycles of denaturation at 94℃ for 1 min, annealing at 50℃ for 1 min and extension at 72℃ for 1.5 min, and an extra extension at 72℃ for 10 min. PCR products were electrophorezed in 0.8% low-melting agarose gel and purified with UNIQ-10 DNA Gel Extraction Kit (Sangon, China). Purified PCR product was ligated with pMD18-T vector (TaKaRa, China) and transferred into E. coli Top10 competent cells. The ampicillin-resistant transformants were picked out according to colonial color randomly.
2.4 Phylogenetic Analyses
The 16S rRNA gene inserts were sequenced using primer 27F in SinoGenoMax Co., Ltd. (Beijing, China). The resulting sequences were screened by Mallard (Ashelford et al., 2006) and Bellerophon (Huber et al., 2004). Chimeras and other anomalies were excluded in further analysis. Valid sequences were compared to GenBank entries using basic local alignments tool (BLAST) to determine their approximate phylogenetic affiliation and 16S rRNA gene sequence similarities. DOTUR software (Schloss and Handelsman, 2005) was used to assign sequences to operational taxonomic units (OTUs). Duplicate sequences were grouped into an OTU at phylogenetic distance value of 0.01. Representative sequences were aligned by Bioedit program. Phylogenetic trees were constructed with MEGA software version 5.0 (Tamura et al., 2011) using Neighbor-Joining method with Tamura-Nei model. The robustness of tree topologies was tested by 1000 bootstrap analyses.
2.5 Estimation of Bacterial Diversity
Coverage was calculated as:

where n1was the number of OTUs represented by one clone in a library and N was library size (Good, 1953). The Shannon-Wiener diversity index (H') was calculated according to equation

where piis the proportion of total clones belonging to ith OTU. The nonparametric SChao1estimator of species richness was calculated using the equation:

where Sobsis the total number of OTUs, F1is the number of OTUs observed only once, and F2is the number of OTUs observed twice (Kemp and Aller, 2004).
2.6 Nucleotide Sequence Accession Numbers
All partial 16S rRNA gene sequences generated in this study were submitted to GenBank database under the accession numbers JX437116 to JX437131 and JX49-4993 to JX495040.
3 Results
3.1 Clone Libraries and Statistical Analysis
Four 16S rRNA gene libraries (Gracilaria lemaneiformis, Gloiopeltis furcata, Mazzaella sp. and Porphyra yezoensis) were constructed, resulting in a total of 256 reliable sequences. The total number of positive clones from each sample varied between 55 and 79 (Table 1), generating 11–26 OTUs (defined by 0.01 distance value). Good’s coverage values ranged from 73.68% to 93.67%. The Shannon diversity indices for Mazzaella sp. and P. yezoensis libraries were higher than those of Gr. lemaneiformis and Gl. Furcata (Table 1). Rarefaction analysis indicated that bacterial phylotypes were not saturated in all four clone libraries and consequently the full diversity of the bacterial communities was far from being covered (Fig.1).

Table 1 Diversity indices (calculated at 0.01 distance value) of 16S rRNA gene libraries
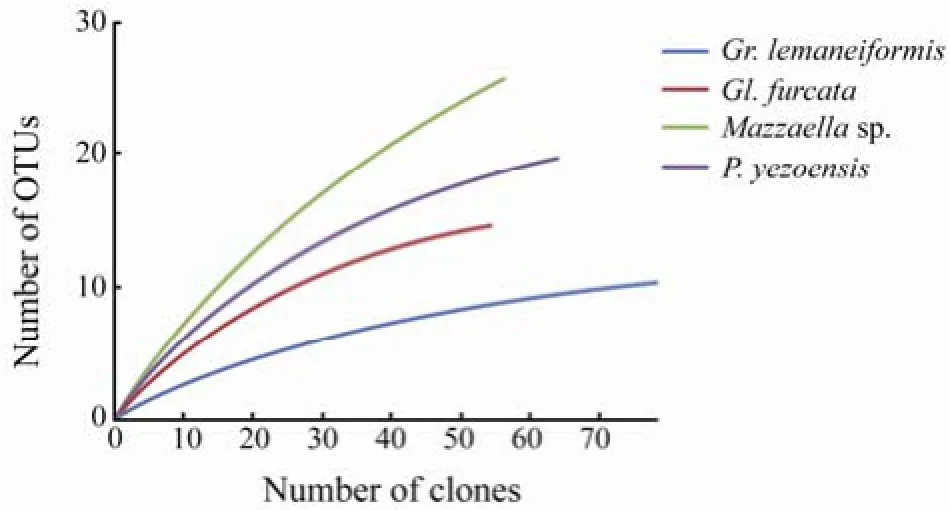
Fig.1 Rarefaction curves of the four 16S rRNA gene libraries at 0.01 distance value.
3.2 Phylogenetic Analysis of Sequences
All the 256 sequences were assigned to 66 OTUs. Only one each OTU was used for tree calculations. Phylogenetic analysis of the partial 16S rRNA gene revealed that libraries were mainly dominated by α-, γ-Proteobacteria, and Bacteroidetes (Fig.2). In addition, a few sequences were affiliated to ε-Proteobacteria, Spirochaetales and Deinococcus-Thermus. The most obvious difference in the composition of libraries was the relative abundance of each group, as shown in Fig.2. Overall, 3–5 groups of bacteria were found in Gl. furcata, Mazzaella sp. and P. yezoensis libraries whereas all sequences from Gr. lemaneiformis fell into the γ-Proteobacteria. The Gr. lemaneiformis library showed low diversity at the bacterial phylum level.
A total of 24 OTUs were grouped within α-Proteobacteria accounting for 32.7%, 21.1% and 26.2% of the sequences from Gl. furcata, Mazzaella sp. and P. yezoensis libraries, respectively. Most sequences were found to cluster in proximity to seven genera: Robiginitomaculum, Ahrensia, Roseovarius, Litoreibacter, Octadecabacter, Thalassobacter, and Sulfitobacter (Fig.3a). Three closely related OTUs (JX494967, JX495005 and JX495011) from Mazzaella sp. and Gl. furcata libraries were tentatively identified as α-Proteobacteria, but could not be affiliated to any known group. Their closest matches in GenBank (more than 99% similarity) were unculturable α-Proteobacteria. Sequences affiliated to Robiginitomaculum, Litoreibacter, Thalassobacter were only found in Mazzaella sp. library whereas no special genus within α-Proteobacteria was found in Gl. furcata and P. yezoensis libraries. Sequences related to Sulfitobacter were observed in all the three libraries.
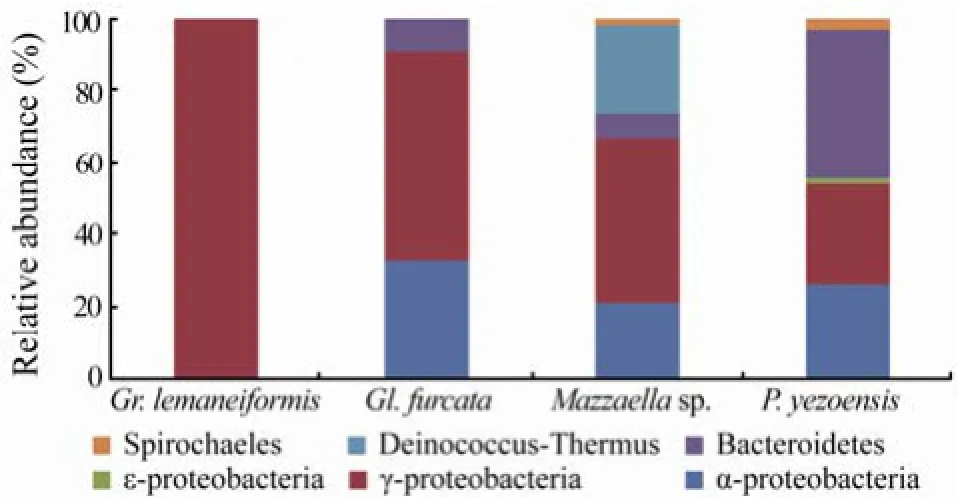
Fig.2 Phylogenetic distribution of the four 16S rRNA gene libraries.
γ-Proteobacteria was frequently encountered in all four libraries. Thirty five OTUs covered 100%, 58.2%, 45.6%, and 27.7% of the sequences from Gr. lemaneiformis, Gl. furcata, Mazzaella sp., and P. yezoensis libraries, respectively (Fig.2). Most sequences were related to established phylogenetic groups containing culturable representatives, i.e., Colwellia, Alteromonas, Pseudoalteromonas, Psychromonas, Vibrio, Aliivibrio, Photobacterium, Moritella, Oceanisphaera, Oleispira, Marinomonas, Psychrobacter, Cocleimonas and Leucothrix (Fig.3b). Four OTUs (JX49-5029, JX494971, JX494988 and JX495019) from P. yezoensis and Mazzaella sp. libraries, containing 10 sequences, were grouped with the clones obtained with culture-independent methods, suggesting that a few previously unrecognized γ-Proteobacteria groups exist on the surfaces of these algae. Sequences affiliated to Vibrio, Photobacterium and Oceanisphaera were only found in Gr. lemaneiformis library, while Alteromonas and Marinomonas were only found in Gl. furcata library, Aliivibrio and Oleispira were only included in Mazzaella sp. library, and Moritella and Cocleimonas belonged to P. yezoensis library. No genus was found in all four libraries.A single sequence related to Arcobacter of ε-Proteobac- teria was found in P. yezoensis library.
Nine OTUs were assigned to the phylum Bacteroidetes. Bacteroidetes was the dominant group of P. yezoensis library (41.5% of the clones) and also abundant in Gl. furcata and Mazzaella sp. libraries (9.1% and 7.0% respectively). Sequences related to Winogradskyella and Olleya were only found in P. yezoensis library while Polaribacter and Lacinutrix belonged to Mazzaella sp. and Gl. furcata libraries, respectively. The remaining se-quences could not be affiliated to any known genus.
Two OTUs found in libraries of Mazzaella sp. and P. yezoensis (1 and 2 sequences, respectively) were tentatively identified as Spirochaetes, and they were distinct from nearest relatives in GenBank (lower than 97% similarity).
One OTU covered 24.6% of the whole sequence of Mazzaella sp. library was affiliated to Deinococcus-Thermus. The closest match in GenBank (99% similarity) was unculturable clone GU451443 from macroalgal surface (Lachnit et al., 2011).
4 Discussion
Phylogenetic analysis revealed that α-, γ-Proteobacteria and Bacteroidetes were the predominant groups on the surfaces of the examined macroalgae. Generally, the three phylogenetic groups found in the present study take part in organic material degradation (Cottrell and Kirchman, 2000). The bacteria of these groups have already been described in other investigations dealing with the epibacterial community of algae (Sapp et al., 2007).
Clones affiliated with α-Proteobacteria occurred more frequently in the libraries of Gl. furcata (32.7%), Mazzaella sp. (21.1%) and P. yezoensis (26.2%), but were not found in Gr. lemaneiformis library. Roseobacter clade (Rhodobacteraceae) within α-Proteobacteria represents one of the most abundant, metabolically versatile and ecologically important bacteria groups commonly found in marine habitats, no matter whether cultivation-dependent or independent assays were applied (González and Moran, 1997). They occur in a broad variety of marine environment (including plankton, sediments, sea ice, macroalgal surface, etc.) and constitute about 25% of marine communities, especially in coastal and Polar Regions (Buchan et al., 2005; Piekarski et al., 2009). In the present study, the majority of sequences falling within α-Proteobacteria were related to Roseobacter clade, i.e., Ahrensia, Roseovarius, Litoreibacter, Octadecabacter, Thaiassobacter and Sulfitobacter (Fig.3a), which was in agreement with the previous observations on bacterial communities from some Ulvacean alga (Tujula et al., 2010). Tujula et al. (2010) speculated that high proportion of Alphaproteobacteria may be linked to DMSP utilization. Many species of Roseobacter clade had the property of DMSP degradation and assimilation with high efficiency. Besides, bacteria within this clade were also known to be ubiquitous and rapid colonizers of surfaces in coastal environments (Dang and Lovell, 2000). In Fig.3a, the presence of sulfite bacteria in the microflora of the investigated red algae was remarkable, which was consistent with the finding of Beleneva and Zhukova (2006). Clones affiliated to Sulfitobacter dominated the α-Proteobacteria group of Gl. furcata, P. yezoensis libraries, and also were found in Mazzaella sp. library. These gram-negative aerobic heterotrophs played an important role in cycling organic sulfur and were originally isolated from seawater (Sorokin, 1995). They were also isolated from starfish, seagrass (Ivanova et al., 2004) and found on the surface of macroalgae (Allgaier et al., 2003; Beleneva and Zhukova, 2006). Seven clones of Gl. furcata library were closely related to Sulfitobacter sp. LM-16 (AJ534237) which was isolated from surface of brown algae Laminaria sp. (Allgaier et al., 2003). Two clones in Mazzaella sp. library were affiliated to Sulfitobacter and closely related to an uncultured Ulva prolifera associated clone (HM437338) (Liu et al., 2011). Detailed analysis of sequences (grouped at 0.01 distance level, Figs.3a–c) revealed that rare OTUs were common among libraries. These results showed that significant difference existed in epibacterial community structure on the four algae. Many studies had demonstrated that the structure of epibacterial communities was host-specific but temporally variable (Lachnit et al., 2011).
The most abundant group of bacteria detected on the surface of four red algae was γ-Proteobacteria. They dominated three libraries, Gr. lemaneiformis (100%), Gl. furcata (58.2%) and Mazzaella sp. (45.6%), and were the second dominant group in P. yezoensis library (27.7%). The clones related to this group were mainly affiliated to Colwellia, Pseudoalteromonas, Psychromonas, Moritella and Leucothrix, etc. Pseudoalteromonas is a newly established genus (Gauthier et al., 1995) and members of Pseudoalteromonas have been found on living surfaces especially of macroalgae (Skovhus et al., 2004, 2007; Vynne et al., 2011). Previous studies have reported that many species of Pseudoalteromonas can produce antifouling agents against settlement of bacteria, fungi, algal spores and invertebrate larvae (Holmström and Kjelleberg, 1999; Egan et al., 2001; Holmström et al., 2002). Other reports stated that lipases and proteinases secreted by Pseudoalteromonas could cause disease by degrading the cell wall of macroalgae (Sawabe et al., 1998; Ivanova et al., 2002; Alekseeva et al., 2004). In our investigation, clones belonging to Pseudoalteromonas dominated Gr. lemaneiformis library obviously (57 clones, 72.2% of the total). One OTU covering 55 clones shared 99% similarity with P. haloplanktis and P. elyakovii (Fig.3a). The former is a psychrophilic bacterium isolated from Antarctica, which lives on organic remains of algae and can convert the cellulose into an immediate nutritive form (Violot et al., 2005). P. elyakovii had been isolated from spot-wound fronds of L. japonica (Sawabe et al., 2000). Furthermore, clones assigned to Pseudoalteromonas also dominated Gl. furcata library (47.3% of total clones). Their close relative was P. nigrifaceus (99% similarity), which lacked the ability to hydrolyze most of the algal polysaccharides (Ivanova et al., 2003). In a study of Smolina et al. (2005), P. nigrifaciens KMM 156 lipopolysaccharide and its fragments were able to inhibit prokaryote and eukaryote cells adhesion. It was difficult to relate the dominance observed in this study to particular functional phenotypes, because the Pseudoalteromonas are metabolically extremely diverse. As for the other two clone libraries, Pseudoalteromonas-related clones were rarely found. In a previous study, Skovhus et al. (2007) suggested that samples with the highest degree of fouling displayed the highest Pseudoalteromonas diversity. The relative abun-dance of Pseudoalteromonas on the four red algae maybe indicated different degree of fouling.
Bacteria belonging to Bacteroidetes group are highly diverse and are important members of the marine bacterioplankton. They are found in a wide range of habitats and are involved in organic material degradation (Fuhrman and Hagström, 2008). Members of Flavobacteriaceae family within Bacteroidetes are commonly found on surfaces of marine macroalgae (Hengst et al., 2010; Tujula et al., 2010; Lachnit et al., 2011). Many literatures have documented that some species of this group are important for morphogenesis of macroalgae (Nakanishi et al., 1996; Matsuo et al., 2003; Marshall et al., 2006). In this study, Bacteroidetes was the most abundant group within clone library of P. yezoensis and also found in the libraries of Gl. furcata and Mazzaella sp. (Fig.2). Clones of this group were mainly assigned to Polaribacter, Winogradskyella, Olleya and Lacinutrix. Twenty three clones related to Olleya prevailed in the clone library of P. yezoensis (35.4% of the total library). Their close relative in Gen-Bank was Olleya sp. WT-MY15 (JQ327134), which was isolated from wood falls in the South Sea in Korea (Lee et al., 2013).
Fourteen clones (24.6% of the total) of Mazzaella sp. library were assigned to Deinococcus-thermus phylum. They couldn’t be related to any known isolates or clones in the GenBank. Their presence is especially interesting as Deinococcus species have been isolated from extreme environments such as thermal springs and radioactive residues (Makarova et al., 2001).
In summary, this study unraveled the composition of epibacterial communities on the surfaces of four red algae sampled from a farm at Dalian, China. However, the epibacterial communities on macroalgae are quite complex and dynamic. To better understand the subtle interaction between epibacteria and host species, more detailed studies are needed.
Acknowledgements
This work was supported by the Foundation for Innovative Research Groups of the National Natural Science Foundation of China (No. 41121064) and the National High Technology Research and Development Program of China (863 Program) (No. 2007AA09Z434).
Alekseeva, S. A., Bakunina, I. Y., Nedashkovskaya, O. I., Isakov, V. V., Mikhailov, V. V., and Zvyagintseva, T. N., 2004. Intracellular alginolytic enzymes of the marine bacterium Pseudoalteromonas citrea KMM 3297. Biochemistry (Moscow), 69 (3): 262-269.
Allgaier, M., Uphoff, H., Felske, A., and Wagner-Döbler, I., 2003. Aerobic anoxygenic photosynthesis in Roseobacter clade bacteria from diverse marine habitats. Applied and Environmental Microbiology, 69 (9): 5051-5059.
Ashelford, K. E., Chuzhanova, N. A., Fry, J. C., Jones, A. J., and Weightman, A. J., 2006. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Applied and Environmental Microbiology, 72 (9): 5734-5741.
Ashen, J. B., and Goff, L. J., 2000. Molecular and ecological evidence for species specificity and coevolution in a group of marine algal-bacterial symbioses. Applied and Environmental Microbiology, 66 (7): 3024-3030.
Beleneva, I. A., and Zhukova, N. V., 2006. Bacterial communities of brown and red algae from Peter the Great Bay, the Sea of Japan. Microbiology, 75 (3): 410-419.
Bhadury, P., and Wright, P. C., 2004. Exploitation of marine algae: Biogenic compounds for potential antifouling applications. Planta, 219 (4): 561-578.
Buchan, A., González, J. M., and Moran, M. A., 2005. Overview of the marine Roseobacter lineage. Applied and Environmental Microbiology, 71 (10): 5665-5677.
Celia, R. L., 1992. Agar-digesting bacteria associated with‘rotten thallus syndrome’ of Gracilaria sp. Aquaculture, 102 (1-2): 1-7.
Cottrell, M. T., and Kirchman, D. L., 2000. Natural assemblages of marine Proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecularweight dissolved organic matter. Applied and Environmental Microbiology, 66 (4): 1692-1697.
Dang, H. Y., and Lovell, C. R., 2000. Bacterial primary colonization and early succession on surfaces in marine waters as determined by amplified rRNA gene restriction analysis and sequence analysis of 16S rRNA genes. Applied and Environmental Microbiology, 66 (2): 467-475.
Egan, S., James, S., Holmström, C., and Kjelleberg, S., 2001. Inhibition of algal spore germination by the marine bacterium Pseudoalteromonas tunicata. FEMS Microbiology Ecology, 35 (1): 67-73.
Fuhrman, J. A., and Hagström, Å., 2008. Bacterial and archaeal community structure and its patterns. In: Microbial Ecology of the Oceans. Kirchman, D. L., ed., John Wiley & Sons, Inc., New Jersey, 45-90.
Fuhrman, J. A., Comeau, D. E., Hagström, Å., and Chan, A. M., 1988. Extraction from natural panktonic microorganisms of DNA suitable for molecular biological studies. Applied and Environmental Microbiology, 54 (6): 1426-1429.
Gauthier, G., Gauthier, M., and Christen, R., 1995. Phylogenetic analysis of the genera Alteromonas, Shewanella, and Moritella using genes coding for small-subunit rRNA sequences and division of the genus Alteromonas into two genera, Alteromonas (emended) and Pseudoalteromonas gen. nov., and proposal of twelve new species combinations. International Journal of Systematic Bacteriology, 45 (4): 755-761.
González, J. M., and Moran, M. A., 1997. Numerical dominance of a group of marine bacteria in the alpha-subclass of the class Proteobacteria in coastal seawater. Applied and Environmental Microbiology, 63 (11): 4237-4242.
Good, I. J., 1953. The population frequencies of species and the estimation of population parameters. Biometrika, 40 (3-4): 237-264.
Hengst, M. B., Andrade, S., González, B., and Correa, J. A., 2010. Changes in epiphytic bacterial communities of intertidal seaweeds modulated by host, temporality, and copper enrichment. Microbial Ecology, 60 (2): 282-290.
Hodson, S., Croft, M., Deery, E., Smith, A., and Warren, M., 2007. Algae acquire Vitamin B12through a symbiotic relationship with bacteria. Comparative Biochemistry and Physiology-Part A: Molecular & Integrative Physiology, 146 (4):S222.
Holmström, C., and Kjelleberg, S., 1999. Marine Pseudoalteromonas species are associated with higher organisms and produce biologically active extracellular agents. FEMS Microbiology Ecology, 30 (4): 285-293.
Holmström, C., Egan, S., Franks, A., McCloy, S., and Kjelleberg, S., 2002. Antifouling activities expressed by marine surface associated Pseudoalteromonas species. FEMS Microbiology Ecology, 41 (1): 47-58.
Huber, T., Faulkner, G., and Hugenholtz, P., 2004. Bellerophon: A program to detect chimeric sequences in multiple sequence alignments. Bioinformatics, 20 (14): 2317-2319.
Ivanova, E. P., Bakunina, I. Y., Nedashkovskaya, O. I., Gorshkova, N. M., Alexeeva, Y. V., Zelepuga, E. A., Zvaygintseva, T. N., Nicolau, D. V., and Mikhailov, V. V., 2003. Ecophysiological variabilities in ectohydrolytic enzyme activities of some Pseudoalteromonas species, P. citrea, P. issachenkonii, and P. nigrifaciens. Current Microbiology, 46 (1): 6-10.
Ivanova, E. P., Gorshkova, N. M., Sawabe, T., Zhukova, N. V., Hayashi, K., Kurilenko, V. V., Alexeeva, Y., Buljan, V., Nicolau, D. V., Mikhailov, V. V., and Christen, R., 2004. Sulfitobacter delicatus sp. nov. and Sulfitobacter dubius sp. nov., respectively from a starfish (Stellaster equestris) and sea grass (Zostera marina). International Journal of Systematic and Evolutionary Microbiology, 54 (2): 475-480.
Ivanova, E. P., Sawabe, T., Alexeeva, Y. V., Lysenko, A. M., Gorshkova, N. M., Hayashi, K., Zukova, N. V., Christen, R., and Mikhailov, V. V., 2002. Pseudoalteromonas issachenkonii sp. nov., a bacterium that degrades the thallus of the brown alga Fucus evanescens. International Journal of Systematic and Evolutionary Microbiology, 52 (1): 229-234.
Keats, D. W., Groener, A., and Chamberlain, Y. M., 1993. Cell sloughing in the littoral zone coralline alga, Spongites yendoi (Foslie) Chamberlain (Corallinales, Rhodophyta). Phycologia, 32 (2): 143-150.
Kemp, P., and Aller, J., 2004. Estimating prokaryotic diversity: When are 16S rDNA libraries large enough? Limnology and Oceanography: Methods, 2: 114-125.
Lachnit, T., Blümel, M., Imhoff, J. F., and Wahl, M., 2009. Specific epibacterial communities on macroalgae: Phylogeny matters more than habitat. Aquatic Biology, 5 (2): 181-186.
Lachnit, T., Meske, D., Wahl, M., Harder, T., and Schmitz, R., 2011. Epibacterial community patterns on marine macroalgae are host-specific but temporally variable. Environmental Microbiology, 13 (3): 655-665.
Lee, M. H., Jung, Y. T., Park, S., and Yoon, J. H., 2013. Olleya namhaensis sp. nov., isolated from wood falls, and emended description of the genus Olleya Mancuso Nichols et al. 2005 emend. Lee et al. 2010. International Journal of Systematic and Evolutionary Microbiology, 63 (Pt 5): 1610-1615.
Liu, M., Dong, Y., Zhao, Y., Zhang, G. T., Zhang, W. C., and Xiao, T., 2011. Structures of bacterial communities on the surface of Ulva prolifera and in seawaters in an Ulva blooming region in Jiaozhou Bay, China. World Journal of Microbiology and Biotechnology, 27 (7): 1703-1712.
Makarova, K. S., Aravind, L., Wolf, Y. I., Tatusov, R. L., Minton, K. W., Koonin, E. V., and Daly, M. J., 2001. Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiology and Molecular Biology Reviews: MMBR, 65 (1): 44-79.
Marshall, K., Joint, I., Callow, M. E., and Callow, J. A., 2006. Effect of marine bacterial isolates on the growth and morphology of axenic plantlets of the green alga Ulva linza. Microbial Ecology, 52 (2): 302-310.
Matsuo, Y., Suzuki, M., Kasai, H., Shizuri, Y., and Harayama, S., 2003. Isolation and phylogenetic characterization of bacteria capable of inducing differentiation in the green alga Monostroma oxyspermum. Environmental Microbiology, 5 (1): 25-35.
Nakanishi, K., Nishijima, M., Masamichi, N., Kazuyoshi, K., and Naotsune, S., 1996. Bacteria that induce morphogenesis in Ulva Pertusa (Chlorophyta) grown under axenic conditions. Journal of Phycology, 32 (3): 479-482.
Nylund, G. M., and Pavia, H., 2005. Chemical versus mechanical inhibition of fouling in the red alga Dilsea carnosa. Marine Ecology Progress Series, 299: 111-121.
Piekarski, T., Buchholz, I., Drepper, T., Schobert, M., Wagner-Doebler, I., Tielen, P., and Jahn, D., 2009. Genetic tools for the investigation of Roseobacter clade bacteria. BMC Microbiology, 9: 265.
Rao, D., Webb, J. S., Holmström, C., Case, R., Low, A., Steinberg, P., and Kjelleberg, S., 2007. Low densities of epiphytic bacteria from the marine alga Ulva australis inhibit settlement of fouling organisms. Applied and Environmental Microbiology, 73 (24): 7844-7852.
Roesijadi, G., Jones, S. B., Snowden-Swan, L. J., and Zhu, Y., 2010. Macroalgae as a Biomass Feedstock: A Preliminary Analysis. Prepared for the U.S. Department of Energy under Contract DE-AC05-76RL01830 by Pacific Northwest National Laboratory.
Sapp, M., Schwaderer, A. S., Wiltshire, K. H., Hoppe, H. G., Gerdts, G., and Wichels, A., 2007. Species-specific bacterial communities in the phycosphere of microalgae? Microbial Ecology, 53 (4): 683-699.
Sawabe, T., Makino, H., Tatsumi, M., Nakano, K., Tajima, K., Iqbal, M. M., Yumoto, I., Ezura, Y., and Christen, R., 1998. Pseudoalteromonas bacteriolytica sp. nov., a marine bacterium that is the causative agent of red spot disease of Laminaria japonica. International Journal of Systematic Bacteriology, 48 (3): 769-774.
Sawabe, T., Tanaka, R., Iqbal, M. M., Tajima, K., Ezura, Y., Ivanova, E. P., and Christen, R., 2000. Assignment of Alteromonas elyakovii KMM 162T and five strains isolated from spot-wounded fronds of Laminaria japonica to Pseudoalteromonas elyakovii comb. nov. and the extended description of the species. International Journal of Systematic and Evolutionary Microbiology, 50 (1): 265-271.
Schloss, P. D., and Handelsman, J., 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Applied and Environmental Microbiology, 71 (3): 1501-1506.
Sieburth, J. M., 1969. Studies on algal substances in the sea. III. The production of extracellular organic matter by littoral marine algae. Journal of Experimental Marine Biology and Ecology, 3 (3): 290-309.
Skovhus, T. L., Holmström, C., Kjelleberg, S., and Dahllöf, I., 2007. Molecular investigation of the distribution, abundance and diversity of the genus Pseudoalteromonas in marine samples. FEMS Microbiology Ecology, 61 (2): 348-361.
Skovhus, T. L., Ramsing, N. B., Holmström, C., Kjelleberg, S., and Dahllöf, I., 2004. Real-time quantitative PCR for assessment of abundance of Pseudoalteromonas species in marine samples. Applied and Environmental Microbiology, 70 (4): 2373-2382.
Smolina, T. P., Gorshkova, R. P., Nazarenko, E. L., and Besednova, N. N., 2005. Inhibition of prokaryote and eukaryote cells adhesion by sea Proteobacteria Pseudoaltero-monas nigrifaciens KMM 156 lipopolysaccharide and its fragments. Antibiotiki i Khimioterapiya, 50 (5-6): 4-6.
Sorokin, D. Y., 1995. Sulfitobacter pontiacus gen. nov., sp. nov.–A new heterotrophic bacterium from the Black Sea, specialized on sulfite oxidation. Microbiology, 64: 295-305.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S., 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, 28 (10): 2731-2739.
Tsavkelova, E. A., Klimova, S., Cherdyntseva, T. A., and Netrusov, A. I., 2006. Microbial producers of plant growth stimulators and their practical use: A review. Prikladnaia Biokhimiia i Mikrobiologiia, 42 (2): 133-143.
Tujula, N. A., Crocetti, G. R., Burke, C., Thomas, T., Holmström, C., and Kjelleberg, S., 2010. Variability and abundance of the epiphytic bacterial community associated with a green marine Ulvacean alga. The ISME Journal, 4 (2): 301-311.
Violot, S., Aghajari, N., Czjzek, M., Feller, G., Sonan, G. K., Gouet, P., Gerday, C., Haser, R., and Receveur-Bréchot, V., 2005. Structure of a full length psychrophilic cellulase from Pseudoalteromonas haloplanktis revealed by X-ray diffraction and small angle X-ray scattering. Journal of Molecular Biology, 348 (5): 1211-1224.
Vynne, N. G., Månsson, M., Nielsen, K. F., and Gram, L., 2011. Bioactivity, chemical profiling, and 16S rRNA-based phylogeny of Pseudoalteromonas strains collected on a global research cruise. Marine Biotechnology, 13 (6): 1062-1073.
Wahl, M., 2008. Ecological lever and interface ecology: Epibiosis modulates the interactions between host and environment. Biofouling, 24 (6): 427-438.
Wang, H. B., Li, X. S., Xia, Y. M., and Yan, B. L., 2011. Isolation, identification and biological pathogen of yellow spot disease in conchocelis of Porphyra yezoensis. Marine Environmental Science, 30 (3): 361-364.
Weisburg, W. G., Barns, S. M., Pelletier, D. A., and Lane, D. J., 1991. 16S ribosomal DNA amplification for phylogenetic study. Journal of Bacteriology, 173 (2): 697-703.
Xu, N., Fan, X., Yan, X., Li, X., Niu, R., and Tseng, C. K., 2003. Antibacterial bromophenols from the marine red alga Rhodomela confervoides. Phytochemistry, 62 (8): 1221-1224.
Yang, R., Fang, W. Y., Shan, Y. Y., Chen, H. M., Sun, X., and Ye, Y. F., 2008. Genetic diversity of epiphytic bacteria in Porphyra yezoensis. Acta Oceanologica Sinica, 30 (4): 161-168.
(Edited by Qiu Yantao)
(Received March 13, 2013; revised May 2, 2013; accepted July 9, 2014)
© Ocean University of China, Science Press and Spring-Verlag Berlin Heidelberg 2014
* Corresponding author. E-mail: txiao@ms.qdio.ac.cn
杂志排行
Journal of Ocean University of China的其它文章
- The Formation of Wind Curl in the Marine Atmosphere Boundary Layer over the East China Sea Kuroshio in Spring
- Wind Wave Characteristics and Engineering Environment of the South China Sea
- Prediction of the Mooring Force of a 2-D Floating Oil Storage Tank
- An Experimental Study on the Wave-Induced Pore Water Pressure Change and Relative Influencing Factors in the Silty Seabed
- Fe-Si-Mn-Oxyhydroxide Encrustations on Basalts at East Pacific Rise near 13˚N: An SEM – EDS Study
- Seasonal Changes in Phytoplankton Biomass and Dominant Species in the Changjiang River Estuary and Adjacent Seas: General Trends Based on Field Survey Data 1959 - 2009
