Inhibition of human esophageal squamous cell carcinomas by targeted silencing of tumor enhancer genes: an overview
2014-03-29JalilPirayeshIslamianMohsenMohammadiBehzadBaradaran
Jalil Pirayesh Islamian, Mohsen Mohammadi, Behzad Baradaran
1Tabriz University of Medical Sciences, School of Medicine, Tabriz, East Asarbeidjan, Iran;
2Department of Radiation, Medicine-Shahid Beheshti University of Medical Sciences, Tehran, Iran;
3Immunology Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
Inhibition of human esophageal squamous cell carcinomas by targeted silencing of tumor enhancer genes: an overview
Jalil Pirayesh Islamian1, Mohsen Mohammadi2, Behzad Baradaran3
1Tabriz University of Medical Sciences, School of Medicine, Tabriz, East Asarbeidjan, Iran;
2Department of Radiation, Medicine-Shahid Beheshti University of Medical Sciences, Tehran, Iran;
3Immunology Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
Esophageal cancer has been reported as the ninth most common malignancy and ranks as the sixth most frequent cause of death worldwide. Esophageal cancer treatment involves surgery, chemotherapy, radiation therapy, or combination therapy. Novel strategies are needed to boost the oncologic outcome. Recent advances in the molecular biology of esophageal cancer have documented the role of genetic alterations in tumorigenesis. Oncogenes serve a pivotal function in tumorigenesis. Targeted therapies are directed at the unique molecular signature of cancer cells for enhanced e ffi cacy with low toxicity. RNA interference (RNAi) technology is a powerful tool for silencing endogenous or exogenous genes in mammalian cells. Related results have shown that targeting oncogenes with siRNAs, speci fi cally the mRNA, e ff ectively reduces tumor cell proliferation and induces apoptotic cell death.is article will brie fl y review studies on silencing tumor enhancer genes related to the induction of esophageal cancer.
Esophageal carcinoma; ionizing radiation (IR); oncogene; targeted therapy; siRNA
Introduction
Esophageal carcinoma, as a malignancy with poor prognosis, is the sixth leading cause of cancer-related death worldwide1. The incidence rate is close to the prevalence rate, indicating a short overall survival time2. Adenocarcinoma and squamous cell carcinoma (SCC) are the dominant histologies for esophageal carcinoma patients3. Evidence-based studies have suggested that genetic polymorphisms in carcinogen-metabolizing enzymes are important in determining an individual’s susceptibility to cancer4.
Management of esophageal carcinoma is based on the tumor extent according to the TNM classification and is divided into curative and palliative treatments. Patients with loco-regional disease (Stage I,II), in good medical condition, are oen o ff ered curative treatment.
Surgery remains the first choice for patients with early-stage cancers and is the standard against which all other treatment regimens are compared. Commonly used techniques for the resection of localized esophageal carcinoma are the transhiatal and right transthoracic approaches5. Although surgical techniques and postoperative care have improved, the reported mortality rates during operation remain as high as 4% to 10%3. Cisplatin in combination with 5-fluorouracil (5-FU) is considered the standard chemotherapy for esophageal carcinoma.e response rate for cisplatin, as a single agent, is approximately 20%6,7. The combination of cisplatin and continuous-infusion 5-FU has shown a response rate ranging from 35% to 65%8,9. Curatively intended radiation therapy can be performed as a conventional external radiotherapy, as intra-luminal brachytherapy, or in combination. However, the e ffi cacy of conventional radiotherapy is limited by the following factors: (1) the presence of hypoxic, intrinsically radio-resistant, and repair-proficient tumor cells; (2) genetic, metabolic, and microenvironmental heterogeneity of tumors; and (3) undesirable damage to the normal healthy tissues10. Therefore, a significant improvement in therapeutic e ffi cacy can be only achieved by developing e ff ective approachesbased on a comprehensive understanding of the radiobiology of tumor and normal tissues to enhance the radiation damage in tumors selectively while reducing the damage to normal tissues.
Adjuvant modality, a combination of chemotherapy and radiation therapy, is often used for down-staging of the tumor and for improving prognosis after surgery. Despite surgery or chemoradiotherapy, the prognosis of esophageal cancer treatment still remains poor, with a 5-year survival of approximately 10%.e failure of conventional therapy mainly occurs because tumors develop resistance to chemotherapy or radiation, and attempts to overcome resistance with higher doses of radiation and chemotherapeutics inevitably result in an unacceptable degree of toxicity and damage to normal tissues.e major limitation of all these treatments and their combinations is the lack of speci fi city for the tumor cell and toxicity to the patient11.
Neoadjuvant chemoradiotherapy for esophageal squamous cell carcinoma (ESCC) is bene fi cial in the seing of a complete pathological response. A gene expression study has shown that Rad51 expression is a useful predictive factor for the e ffi cacy of neoadjuvant chemoradiotherapy in ESCC. Rad51 expression a ff ects both chemo- and radio-sensitivity in various cancers12.
Targeted therapy
Overall survival rates for ESCC patients remain substantially unchanged over the past several decades despite aggressive multimodality intervention13. However, recent insights into the epigenetic mechanisms associated with multi-step carcinogenesis14and delineation of signal transduction pathways conferring chemo/radiation resistance in cancer cells15-17provide new opportunities for the development of potentially effective targeted molecular therapies for ESCC and Barres esophagus. Modern cancer therapies have evolved from non-specific cytotoxic agents that affect both normal and cancer cells to targeted therapies and personalized medicine.
Targeted therapies are directed at the unique molecular signatures of cancer cells to achieve signi fi cant e ffi cacy with low toxicity18. Molecular studies of human esophageal tumors have revealed frequent genetic abnormalities (Table 1)4. Regardless of patient origin and suspected etiological factors, genetic changes that are consistently observed in ESCC are as follows: (1) alterations in tumor suppressor genes, specifically p53, resulting in altered DNA replication and repair, cell proliferation, and apoptosis; (2) disruption of the G1/S cell cycle checkpoint and loss of cell cycle control; and (3) alterations in oncogene function resulting in deregulation of cell signaling cascades19,20. In a recent comprehensive genomic analysis of 158 ESCC cases, as part of the International Cancer Genome Consortium research project, whole-genome sequencing was applied to 17 ESCC cases and whole-exome sequencing to 71 cases, of which 53 cases plus an additional 70 ESCC cases were unused in whole-genome and whole-exome sequencing, were subjected to array comparative genomic hybridization analysis. Eight significantly mutated genes were identified, among of which six genes are well-known tumor-associated genes (TP53, RB1, CDKN2A, PIK3CA, NOTCH1, and NFE2L2), whereas two have not previously been described in ESCC (ADAM29 and FAM135B)21. In particular, FAM135B is identified as a novel cancer-implicated gene that promotes the malignancy of ESCC cells. In addition, a correlation between CD133 expression and the immunolocalization of several markers, such as p53, p16, p27, murine double minute 2 (MDM2), Ki-67, and epidermal growth factor receptor (EGFR), was observed.ese indicators are known as prognostic markers or tumor proliferation factors in ESCC22. In a study by Nam et al.23, positive expressions of p53, Rb tumor suppressor protein (pRb), hMLH1, and MDM2 were observed in 40%, 46.7%, 40%, and 66.7% of the tissue specimens, respectively.
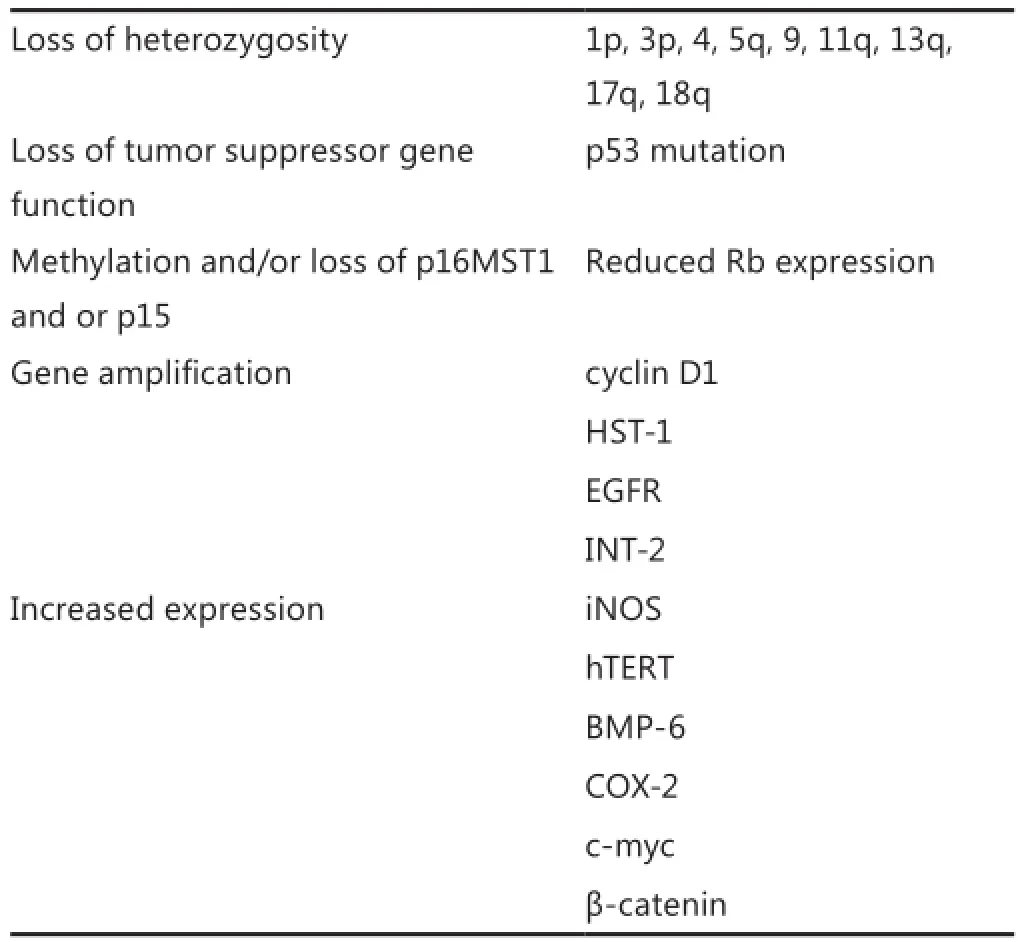
Table 1Molecular alterations in human esophageal squamous cell carcinoma4
Other genetic alterations that are commonly associated with clinical tumors include p53 mutations24,25; loss of p16MST1 and/ or p1526, and/or RARβ expression27; ampli fi cation of cyclin D1, HST-1, EGFR and INT-228-31; and elevations in iNOS, hTERT, BMP-6, COX-2 and c-Myc expression32-36; as well as cytoplasmicβ-catenin levels37. One or several of these alterations contribute to the growth and metastatic potential of these tumors.
Two other putative tumor suppressor genes, namely, FEZ-1 on chromosome 8q22 and DLC1 on 3p21, have been identi fi ed as novel candidates that may serve a function in esophageal carcinogenesis, given that their expressions are absent in some sporadic tumors38,39. In contrast to the extensive literature on genetic alterations in frank tumors, limited information is available on genetic alterations in precancerous lesions of the esophagus.
The p53 gene results in cell cycle arrest through p21WAF1 induction, which sequesters CDKs by down-regulating bcl-2 (known as a key molecule in the regulation of apoptosis or programmed cell death), while up-regulating bax40.is process induces apoptosis18. MDM2, also known as HDM2 in humans, is a negative regulator of the tumor suppressor p5318. MDM2 belongs to a large family of ring- fi nger-containing proteins and functions mainly, if not exclusively, as an E3 ligase41,42. MDM2 targets p53 for mono- and/or poly-ubiquitylation, thereby controlling its localization and/or levels through proteasomedependent degradation. MDM2-mediated mono-ubiquitylation of p53 results in cytoplasmic sequestration, whereas polyubiquitylation triggers p53 degradation. MDM2 also suppresses p53 function by binding to p53, thereby hindering its capability to interact with the basal transcriptional machinery and transcriptional co-activators, such as p30042,43. In response to DNA damage, phosphorylation of p53 on Ser20 and of MDM2 on Ser395, as mediated by kinases such as ATM, interrupts the p53-MDM2 interaction, thus resulting in the nuclear accumulation of p53 and the activation of its transcriptional program44. MDM2 is overexpressed in a various human cancers, including melanoma, non-small cell lung cancer, breast cancer, esophageal cancer, leukemia, non-Hodgkin’s lymphoma, and sarcoma18. The tumor suppressor p53 is a powerful anti-tumor molecule that is frequently inactivated by mutations or deletions in cancer. However, half of the human tumors express wild type (wt) p53, and its activation by antagonizing its negative regulator MDM2 might o ff er a new therapeutic strategy45. Proofof-concept experiments have demonstrated the feasibility of this approach in vitro, but the development of pharmacological inhibitors remains a challenge. Potent and selective smallmolecule MDM2 inhibitors have recently been identified46,47. Studies of these compounds have strengthened the concept that selective and non-genotoxic p53 activation is a viable alternative to current cytotoxic chemotherapy. However, clinical validation remains pending. As a single agent, Nutlin-3a only increases apoptosis and decreases survival preferentially in wt p53-expressing cells. Although the present data support the notion that initiating the growth suppressive and pro-apoptotic activity of p53 by MDM2 antagonists is a potentially valuable strategy for treating OS with wt p53, further studies are needed to reveal the true therapeutic potential of this approach48.
RNA interference (RNAi) is the process of sequence-speci fi c, post-transcriptional gene silencing directed by short interfering 21-23 nucleotides (nt) double-stranded RNA (siRNA). A number of studies have demonstrated that the introduction of siRNAs into mammalian and human cells causes specific and effective suppression of the corresponding mRNA molecules. siRNAs can inhibit the in vivo expression of endogenous genes, providing further support to the notion that oncogene-specific siRNAs may be new alternatives for gene-specific therapeutics of human cancers. For example, MDM2 targets p53 protein for degradation in the ubiquitin pathway, resulting in the abrogation of its antiproliferative and apoptosis-promoting effects. In addition, studies have shown that p14 induces the degradation of the proto-oncogene MDM2, which destabilizes p53. Furthermore, cell-cycle arrest mediated by p14 can be terminated in cells lacking functional p53, indicating that p14 may act upstream of p53. Signi fi cant alterations in the chromatin structure occur during multistep esophageal carcinogenesis49. Global DNA demethylation results in de-repression and activation of an operator gene through the deactivation of a repressor gene of imprinted alleles, such as H19 and IGF250,51, as well as the up-regulation of germ cell-restricted genes, many of which are linked to the X chromosome and encode proteins that are recognized by tumor reactive lymphocytes52-54. Paradoxically, site-specific DNA methylation silences a variety of tumor suppressor genes, including p16, RASSF1A, FHIT, E-cadherin, and RARb in esophageal cancers55-58. Hasan et al.59also showed that TC21 knockdown sensitizes ESCC to cisplatin. Several of these tumor suppressor genes are methylated in Barres epithelium, indicating that epigenetic events that perturb cell cycle regulation occur extremely early during esophageal adenocarcinogenesis55,60. By contrast, the effects of CTA expression regarding the malignant phenotype of esophageal cancers have not been clearly established61,62.e implications of the methylation-mediated inactivation of tumor suppressor genes are evident. For example, restoration of p16 or FHIT expression by gene transfer techniques mediates growth arrest and apoptosis in cultured esophageal cancer cells63,64. Furthermore, several studies suggest that the aberrant methylation of tumor suppressor genes coincides with adverse response to therapy in esophageal cancer patients56,65,66. Recent studies suggest that the aberrant activity of DNA methyltransferases and histone deacetylases (HDACs) may contribute to the inactivation of tumor suppressor gene expression and perturbed cell cycle regulationin aerodigestive tract malignancies67,68. In contrast to genes that have been mutated or deleted, the expression of epigenetically silenced tumor suppressors can be restored in cancer cells by pharmacologic compounds, including DNA demethylating agents and HDAC inhibitors69. Although HDAC inhibitors, such as FK228, modulate the chromatin structure through the acetylation of the core histone proteins, these agents also disrupt oncoprotein signaling by mechanisms that are remarkably analogous to those mediated by geldanamycin derivatives70,71. Indeed, FK228-mediated growth arrest in cancer cells coincides with signaling inhibition through the EGFr-ras-raf-Erk and P13K/AKT pathways70. Furthermore, FK228 and other HDAC inhibitors markedly enhance p21expression63,70. Abrogation of p21 expression by flavopiridol enhances depsipeptidemediated apoptosis in malignant pleural mesothelioma cells, thus contributing to cell cycle arrest, but also inhibiting apoptosis of cancer cells following exposure to a variety of conventional chemotherapeutic agents and novel antitumor compounds72.
Discussion
Surgery, radiation therapy, and chemotherapy have been the main modes of treatment for human malignancies for more than 40 years. The use of a combination of radiation and chemotherapy is often called chemoradiation in the medical literature73.e rate of tumor growth and expansion is controlled by the balance between cell proliferation/survival and apoptosis, which are strictly regulated processes in normal cells. However, apoptosis is disrupted in cancer cells and tumors, and the cell cycle proteins required for cell survival and proliferation are up-regulated or are continuously activated74. In recent years, considerable insight into the mechanisms of aerodigestive tract carcinogenesis has been gained. The aforementioned studies indicate that the manipulation of gene expression as a means to restore cell cycle regulation and induce apoptosis is feasible in esophageal cancer cells in vitro, as well as in clinical seings. Furthermore, recent laboratory experiments have demonstrated that novel compounds that inhibit survival signaling markedly enhance the efficacy of conventional therapeutic regimens for esophageal cancer. These data clearly support the evaluation of these combination treatment regimens using well-designed clinical protocols. We are entering a new era as regards the treatment and prevention of cancer, but whether the targeted molecular therapies can inhibit the pathogenesis and clinical progression of esophageal malignancies remains to be proven75.
Curative treatment of malignant tumors with ionizing radiation (IR) was introduced 80 years ago76. The conditions of the tumor microenvironment that favor tumor cell survival after IR include hypoxia and secretion of radiation-protective cytokines and growth factors that promote the growth and survival of tumor tissues. Tumor radioresistance serves an important function in treatment failure for esophageal cancer. Therefore, the mechanisms involved in tumor radioresistance must be determined to improve prognosis77. Moreover, a number of genes have been implicated in the response mechanism of eukaryotic cells to IR. These genes include a number of cell cycle, checkpoint, and DNA repair genes, as well as mediators of apoptosis, such as p53, bax, and Bcl-2. The expression or repression of related genes is associated with cell survival or cell death in simple model systems, but shed no light on intercellular events in in-situ tumors or in the clinical outcome of radiotherapy. In addition, studies have shown that miRNA expression fi ngerprints correlate with the clinical and biological characteristics of tumors, including tissue type, differentiation, aggression, response to therapy, and prognosis78. For example, analysis of MDM2 overexpression in relation to p53 gene status has revealed significant associations between p53 missense mutations and the lack of detectable MDM2 protein expression79. Inhibition of MDM2 can restore p53 activity in cancers with wt p53, resulting in anti-tumor effects with apoptosis and growth inhibition18. The silencing of HDM2 mRNA directly enhances MCF-7 cell apoptosis and decreases cell proliferation. These results provide strong evidence that the siRNA technology can be an e ff ective method for inhibiting oncogene expression and activating apoptotic and tumor suppressor genes80. MDM2 is a critical component of the responses to both ionizing and UV radiation81. The decreased levels of MDM2 sensitize cells upon IR. Thus, MDM2 is a potential target for therapeutic intervention because its inhibition may radiosensitize the subset of human tumors expressing wt p53, making radiotherapy more e ff ective81.
Non-specific cytotoxic agents that affect both normal and cancer cells in targeted therapies and personalized medicine are considered as novel cancer therapeutics. Targeted therapies are directed at the unique molecular signatures of cancer cells for enhanced e ffi cacy with low toxicity18. Cancer is a multistep genetic and epigenetic disease with a complex etiology, and cancer cells have been characterized by several defects, such as mutations, down-regulation, over-expression, and deletions of oncogenes and tumor suppressor genes74. Expression array technology has become an important method for many applications, including the identi fi cation of disease-related and treatment-responsive genes, as well as the determination of carcinogenicity, toxicity, and safety of drugs82.ese techniques have the capability to identify genes with expressions that correlate with ESCC because these genes could be potentialcandidates as molecular markers for the prevention and early detection of ESCC. Moreover, identification of genes that are di ff erentially expressed between radiosensitive and radioresistant cancer cells is important for predicting the clinical e ff ectiveness of radiotherapy83.erefore, activation of the cellular apoptotic program is a current strategy for the treatment of human cancers. Studies have demonstrated that radiation and standard chemotherapeutic drugs kill some tumor cells through the induction of apoptosis84. Most chemotherapeutic agents and radiation therapy target the DNA. Non-DNA targets may be effective in killing the cell or modifying the cell in such a way that it becomes more susceptible to cell killing after radiationinduced damage85. Recent advances in the molecular biology of esophageal cancer have documented the function of genetic alterations in tumorigenesis and have facilitated the development of potential new therapeutic approaches designed to target such genetic alterations11. Further study is needed to elucidate the markers or combinations of markers that best enhance the effects of radiotherapy in ESCC. Such markers might prove valuable, not only as clinical predictors, but also as targets for ESCC treatment. For example, the treatment might result in an increased sensitivity if these abnormal functions and expressions return to normal86.
The siRNA complexes silence gene expression in vitro or in vivo with excellent specificity in cells bearing the receptor recognized by the antibody and can target cells, such as primary lymphocytes, which are refractory to lipid-mediated transfection. RNAi possesses high specificity and high efficiency in downregulating gene expressions, making it a potential therapeutic strategy against human cancer. Several molecules involved in cell-cycle regulation have been targeted for RNAi intervention in an effort to suppress cancer cell growth. Two cell-cycle regulators, namely, pRb and p53, are of special importance in cancer therapy and worthy of discussion87. RNAi has facilitated the identi fi cation and study of the components of apoptosis and survival pathways, thus enabling the identification of specific gene targets for improving the e ff ectiveness of cancer therapies.
Conclusion
Significant advances in understanding the molecular biology of esophageal cancer have resulted in the application of gene therapeutic methods, in which genetic material is transferred into human cells and expressed in those cells for a therapeutic purpose. Many of the genes described above may contribute to the resistance to chemotherapy or irradiation. For example, the expression of antiapoptotic proteins by cancer cells is an important mechanism by which cancer cells resist chemotherapy or irradiation. Using RNAi to target antiapoptotic proteins may be a promising strategy for use in conjunction with chemotherapy and radiotherapy for cancer treatment. RNAi therapy can potentially be used in conjunction with chemotherapy, radiotherapy, and/or immunotherapy. The same types of tumors, even with similar clinical phases, differ in terms of radiosensitivity. In recent years, many genes related to the radiosensitivity of tumor cells have been found. Studies focus on controlling the radiosensitive genes and adjusting the fraction dose and interval of radiation, as well as on how to realize the individualization of radiotherapy, change tumor cell radiosensitivity, and reduce the normal tissue damage to achieve the optimum therapeutic e ff ects. Safety and e ff ectiveness are the key factors in gene therapy, and its control mechanism affects its targets. siRNA technology has several major advantages over other post-transcriptional gene silencing techniques, such as the antisense and gene knockout technologies. siRNA technology is easier to deliver, requires only small doses of siRNA to produce its silencing effect, and can directly inactivate a gene at almost any stage in development with biological molecules through a number of theoretically possible reactions.e gene therapy in the fi eld of esophageal cancer is in the primitive stage compared with those in lung, head and neck, as well as brain cancers.
Conflict of interest statement
No potential con fl icts of interest are disclosed.
1. Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin 2005;55:74-108.
2. Socialstyrelsen. Cancer incidence in sweden 2003. In:e National Board of Health and Welfare; Centre for Epidemiology. O ffi cial Statistics of Sweden. Stockholm: 2005.
3. Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med 2003;349:2241-2252.
4. Stoner GD, Gupta A. Etiology and chemoprevention of esophageal squamous cell carcinoma. Carcinogenesis 2001;22:1737-1746.
7. Dinwoodie WR, Bartolucci AA, Lyman GH, Velez-Garcia E, Martelo OJ, Sarma PR. Phase II evaluation of cisplatin, bleomycin,and vindesine in advanced squamous cell carcinoma of the esophagus: a Southeastern Cancer Study Group Trial. Cancer Treat Rep 1986;70:267-270.
8. Bleiberg H, Conroy T, Paillot B, Lacave AJ, Blijham G, Jacob JH, et al. Randomised phase II study of cisplatin and 5- fl uorouracil (5-FU) versus cisplatin alone in advanced squamous cell oesophageal cancer. Eur J Cancer 1997;33:1216-1220.
9. Wadler S, Haynes H, Beitler JJ, Hu X, Fell S, Camacho M, et al. Phase II clinical trial with 5- fl uorouracil, recombinant interferonalpha-2b, and cisplatin for patients with metastatic or regionally advanced carcinoma of the esophagus. Cancer 1996;78:30-34.
11. Park JC, Moon C. Gene therapy for esophageal cancer. Cancerer 2008;6:35-46.
12. Nakanoko T, Saeki H, Morita M, Nakashima Y, Ando K, Oki E, et al. Rad51 expression is a useful predictive factor for the e ffi cacy of neoadjuvant chemoradiotherapy in squamous cell carcinoma of the esophagus. Ann Surg Oncol 2014;21:597-604.
13. Posner M, Forastiere A, Minsky B. Cancer of the esophagus. In: DeVita VT, Jr, Hellman S, Rosenberg SA. eds. Cancer principles & practices of oncology. 7th ed. Philadelphia: LippincoWilliams & Wilkins; 2005: 861-909.
15. Leu CM, Chang C, Hu C. Epidermal growth factor (EGF) suppresses staurosporine-induced apoptosis by inducing mcl-1 via the mitogen-activated protein kinase pathway. Oncogene 2000;19:1665-1675.
16. Akimoto T, Nonaka T, Harashima K, Ishikawa H, Sakurai H, Mitsuhashi N. Selective inhibition of survival signal transduction pathways enhanced radiosensitivity in human esophageal cancer cell lines in vitro. Anticancer Res 2004;24:811-819.
17. Okamoto H, Fujishima F, Nakamura Y, Zuguchi M, Miyata G, Kamei T, et al. Murine double minute 2 and its association with chemoradioresistance of esophageal squamous cell carcinoma. Anticancer Res 2013;33:1463-1471.
18. Yuan Y, Liao YM, Hsueh CT, Mirshahidi HR. Novel targeted therapeutics: inhibitors of MDM2, ALK and PARP. J Hematol Oncol 2011;4:16.
19. Lam AK. Molecular biology of esophageal squamous cell carcinoma. Crit Rev Oncol Hematol 2000;33:71-90.
20. Mandard AM, Hainaut P, Hollstein M. Genetic steps in the development of squamous cell carcinoma of the esophagus. Mutat Res 2000;462:335-342.
21. Song Y, Li L, Ou Y, Gao Z, Li E, Li X, et al. Identi fi cation of genomic alterations in oesophageal squamous cell cancer. Nature 2014;509:91-95.
22. Okamoto H, Fujishima F, Nakamura Y, Zuguchi M, Ozawa Y, Takahashi Y, et al. Signi fi cance of CD133 expression in esophageal squamous cell carcinoma. World J Surg Oncol 2013;11:51.
23. Nam TK, Lee JS, Kim HR, Ahn SJ, Song JY, Yoon MS. Molecular prognostic factors in rectal cancer treated by preoperative chemoradiotherapy. Oncol Le2010;1:23-29.
24. Hollstein MC, Peri L, Mandard AM, Welsh JA, Montesano R, Metcalf, et al. Genetic analysis of human esophageal tumors from two high incidence geographic areas: frequent p53 base substitutions and absence of ras mutations. Cancer Res 1991;51:4102-4106.
25. Gao H, Wang LD, Zhou Q, Hong JY, Huang TY, Yang CS. p53 tumor suppressor gene mutation in early esophageal precancerous lesions and carcinoma among high-risk populations in Henan, China. Cancer Res 1994;54:4342-4346.
26. Xing EP, Nie Y, Wang LD, Yang GY, Yang CS. Aberrant methylation of p16INK4a and deletion of p15INK4b are frequent events in human esophageal cancer in Linxian, China. Carcinogenesis 1999;20:77-84.
27. Xu XC, Liu X, Tahara E, Lippman SM, Lotan R. Expression and up-regulation of retinoic acid receptor-beta is associated with retinoid sensitivity and colony formation in esophageal cancer cell lines. Cancer Res 1999;59:2477-2483.
28. Jiang W, Zhang YJ, Kahn SM, Hollstein MC, Santella RM, Lu SH, et al. Altered expression of the cyclin D1 and retinoblastoma genes in human esophageal cancer. Proc Natl Acad Sci U S A 1993;90:9026-9030.
29. Hollstein MC, Smits AM, Galiana C, Yamasaki H, Bos JL, Mandard A, et al. Ampli fi cation of epidermal growth factor receptor gene but no evidence of ras mutations in primary human esophageal cancers. Cancer Res 1988;48:5119-5123.
30. Lu SH, Hsieh LL, Luo FC, Weinstein IB. Ampli fi cation of the EGF receptor and c-myc genes in human esophageal cancers. Int J Cancer 1988;42:502-505.
31. Guo YJ. Ampli fi cation of int-2 gene in primary esophageal carcinoma and fetal esophageal carcinoma induced by N-methyl-N-benzylnitrosamine (NMBzA). Zhonghua Zhong Liu Za Zhi 1993;15:91-93.
32. Lord RV, Salonga D, Danenberg KD, Peters JH, DeMeester TR, Park JM, et al. Telomerase reverse transcriptase expression is increased early in the Barre’s metaplasia, dysplasia, adenocarcinoma sequence. J Gastrointest Surg 2000;4:135-142.
33. Tanaka H, Kijima H, Tokunaga T, Tajima T, Himeno S, Kenmochi T, et al. Frequent expression of inducible nitric oxide synthase in esophageal squamous cell carcinomas. Int J Oncol 1999;14:1069-1073.
34. Hiyama T, Yokozaki H, Kitadai Y, Haruma K, Yasui W, Kajiyama G,et al. Overexpression of human telomerase RNA is an early event in oesophageal carcinogenesis. Virchows Arch 1999;434:483-487.
35. Raida M, Sarbia M, Clement JH, Adam S, Gabbert HE, Höen K. Expression, regulation and clinical signi fi cance of bone morphogenetic protein 6 in esophageal squamous-cell carcinoma. Int J Cancer 1999;83:38-44.
36. Zimmermann KC, Sarbia M, Weber AA, Borchard F, Gabbert HE, Schrör K. Cyclooxygenase-2 expression in human esophageal carcinoma. Cancer Res 1999;59:198-204.
37. Kimura Y, Shiozaki H, Doki Y, Yamamoto M, Utsunomiya T, Kawanishi K, et al. Cytoplasmic beta-catenin in esophageal cancers. Int J Cancer 1999;84:174-178.
38. Ishii H, Ba ff a R, Numata SI, Murakumo Y, Raan S, Inoue H, et al.e FEZ1 gene at chromosome 8p22 encodes a leucine-zipper protein, and its expression is altered in multiple human tumors. Proc Natl Acad Sci U S A 1999;96:3928-3933.
39. Daigo Y, Nishiwaki T, Kawasoe T, Tamari M, Tsuchiya E, Nakamura Y. Molecular cloning of a candidate tumor suppressor gene, DLC1, from chromosome 3p21.3. Cancer Res 1999;59:1966-1972.
40. Futami T, Miyagishi M, Seki M, Taira K. Induction of apoptosis in HeLa cells with siRNA expression vector targeted against bcl-2. Nucleic Acids Res Suppl 2002;(2):251-252.
41. Jackson PK, Eldridge AG, Freed E, Furstenthal L, Hsu JY, Kaiser BK, et al.e lore of the RINGs: substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol 2000;10:429-439.
42. Toledo F, Wahl GM. MDM2 and MDM4: p53 regulators as targets in anticancer therapy. Int J Biochem Cell Biol 2007;39:1476-82.
44. Mirzayans R, Andrais B, ScoA, Wang YW, Murray D. Ionizing radiation-induced responses in human cells with di ff ering TP53 status. Int J Mol Sci 2013;14:22409-22435.
45. Ozaki T, Nakagawara A. p53: the aractive tumor suppressor in the cancer research fi eld. J Biomed Biotechnol 2011;2011:603925.
46. Shen H, Maki CG. Pharmacologic activation of p53 by smallmolecule MDM2 antagonists. Curr Pharm Des 2011;17:560-568.
47. van Leeuwen IM, Higgins M, Campbell J, Brown CJ, McCarthy AR, Pirrie L, et al. Mechanism-speci fi c signatures for smallmolecule p53 activators. Cell Cycle 2011;10:1590-1598.
48. Wang B, Fang L, Zhao H, Xiang T, Wang D. MDM2 inhibitor Nutlin-3a suppresses proliferation and promotes apoptosis in osteosarcoma cells. Acta Biochim Biophys Sin (Shanghai) 2012;44:685-91.
49. Eads CA, Lord RV, Wickramasinghe K, Long TI, Kurumboor SK, Bernstein L, et al. Epigenetic paerns in the progression of esophageal adenocarcinoma. Cancer Res 2001;61:3410-3418.
50. Hibi K, Nakamura H, Hirai A, Fujikake Y, Kasai Y, Akiyama S, et al. Loss of H19 imprinting in esophageal cancer. Cancer Res 1996;56:480-482.
51. Mori M, Inoue H, Shiraishi T, Mimori K, Shibuta K, Nakashima H, et al. Relaxation of insulin-like growth factor 2 gene imprinting in esophageal cancer. Int J Cancer 1996;68:441-446.
52. Inoue H, Mori M, Li J, Mimori K, Honda M, Nakashima H, et al. Human esophageal carcinomas frequently express the tumorrejection antigens of MAGE genes. Int J Cancer 1995;63:523-526.
53. Mashino K, Sadanaga N, Tanaka F, Yamaguchi H, Nagashima H, Inoue H, et al. Expression of multiple cancer-testis antigen genes in gastrointestinal and breast carcinomas. Br J Cancer 2001;85:713-720.
54. Zendman AJ, Ruiter DJ, Van Muijen GN. Cancer/testis-associated genes: identi fi cation, expression pro fi le, and putative function. J Cell Physiol 2003;194:272-288.
55. Maley CC, Galipeau PC, Li X, Sanchez CA, Paulson TG, Reid BJ. Selectively advantageous mutations and hitchhikers in neoplasms: p16 lesions are selected in Barre’s esophagus. Cancer Res 2004;64:3414-3427.
56. Kuroki T, Trapasso F, Yendamuri S, Matsuyama A, Alder H, Mori M, et al. Promoter hypermethylation ofSSF1A in esophageal squamous cell carcinoma. Clin Cancer Res 2003;9:1441-1445.
57. Wang Y, Fang MZ, Liao J, Yang GY, Nie Y, Song Y, et al. Hypermethylation-associated inactivation of retinoic acid receptor beta in human esophageal squamous cell carcinoma. Clin Cancer Res 2003;9:5257-5263.
58. Si HX, Tsao SW, Lam KY, Srivastava G, Liu Y, Wong YC, et al. E-cadherin expression is commonly downregulated by CpG island hypermethylation in esophageal carcinoma cells. Cancer Le2001;173:71-78.
59. Hasan R, Chauhan SS, Sharma R, Ralhan R. siRNA-mediated downregulation of TC21 sensitizes esophageal cancer cells to cisplatin. World J Gastroenterol 2012;18:4127-4135.
60. Eads CA, Lord RV, Kurumboor SK, Wickramasinghe K, Skinner ML, Long TI, et al. Fields of aberrant CpG island hypermethylation in Barre’s esophagus and associated adenocarcinoma. Cancer Res 2000;60:5021-5026.
61. Zambon A, Mandruzzato S, Parenti A, Macino B, Dalerba P, Ruol A, et al. MAGE, BAGE, and GAGE gene expression in patients with esophageal squamous cell carcinoma and adenocarcinoma of the gastric cardia. Cancer 2001;91:1882-1888.
62. Akcakanat A, Kanda T, Koyama Y, Watanabe M, Kimura E, Yoshida Y, et al. NY-ESO-1 expression and its serum immunoreactivity in esophageal cancer. Cancer Chemother Pharmacol 2004;54:95-100.
63. Schrump DS, Chen GA, Consuli U, Jin X, Roth JA. Inhibition of esophageal cancer proliferation by adenovirally mediated delivery of p16INK4. Cancer Geneer 1996;3:357-364.
64. Ishii H, Dumon KR, Vecchione A, Trapasso F, Mimori K, Alder H, et al. E ff ect of adenoviral transduction of the fragile histidine triad gene into esophageal cancer cells. Cancer Res 2001;61:1578-1584.
65. Noguchi T, Takeno S, Kimura Y, Uchida Y, Daa T, Yokoyama S, et al. FHIT expression and hypermethylation in esophageal squamous cell carcinoma. Int J Mol Med 2003;11:441-447.
66. Brock MV, Gou M, Akiyama Y, Muller A, Wu, Montgomery E, et al. Prognostic importance of promoter hypermethylation of multiple genes in esophageal adenocarcinoma. Clin Cancer Res 2003;9:2912-2919.
67. Yakushiji T, Uzawa K, Shibahara T, Noma H, Tanzawa H. Overexpression of DNA methyltransferases and CDKN2A gene methylation status in squamous cell carcinoma of the oral cavity. Int J Oncol 2003;22:1201-1207.
68. Toh Y, Yamamoto M, Endo K, Ikeda Y, Baba H, Kohnoe S, et al. Histone H4 acetylation and histone deacetylase 1 expression in esophageal squamous cell carcinoma. Oncol Rep 2003;10:333-338.
69. Schrump DS, Nguyen DM. Targeting the epigenome for the treatment and prevention of lung cancer. Semin Oncol 2005;32:488-502.
70. Yu X, Guo ZS, Marcu MG, Neckers L, Nguyen DM, Chen GA, et al. Modulation of p53, ErbB1, ErbB2, and Raf-1 expression in lung cancer cells by depsipeptide FR901228. J Natl Cancer Inst 2002;94:504-513.
71. Neckers L, Schulte TW, Mimnaugh E. Geldanamycin as a potential anti-cancer agent: its molecular target and biochemical activity. Invest New Drugs 1999;17:361-373.
73. Kvols LK. Radiation sensitizers: a selective review of molecules targeting DNA and non-DNA targets. J Nucl Med 2005;46:187S-190S.
74. Abdelrahim M, Safe S, Baker C, Abudayyeh A. RNAi and cancer: Implications and applications. J RNAi Gene Silencing 2006;2:136-145.
75. Schrump DS, Nguyen DM. Novel molecular targeted therapy for esophageal cancer. J Surg Oncol 2005;92:257-261.
77. Ogawa R, Ishiguro H, Kuwabara Y, Kimura M, Mitsui A, Mori Y, et al. Identi fi cation of candidate genes involved in the radiosensitivity of esophageal cancer cells by microarray analysis. Dis Esophagus 2008;21:288-297.
78. Wang XC, Wang W, Zhang ZB, Zhao J, Tan XG, Luo JC. Overexpression of miRNA-21 promotes radiation-resistance of non-small cell lung cancer. Radiat Oncol 2013;8:146.
79. Arora S, Mathew R, Mathur M, Chaopadhayay TK, Ralhan R. Alterations in MDM2 expression in esophageal squamous cell carcinoma: relationship with p53 status. Pathol Oncol Res 2001;7:203-208.
80. Liu TG, Yin JQ, Shang BY, Min Z, He HW, Jiang JM, et al. Silencing of hdm2 oncogene by siRNA inhibits p53-dependent human breast cancer. Cancer Geneer 2004;11:748-756.
81. Perry ME. Mdm2 in the response to radiation. Mol Cancer Res 2004;2:9-19.
82. Farhadi M, Tahmasebi Z, Merat S, Kamangar F, Nasrollahzadeh D, Malekzadeh R. Human papillomavirus in squamous cell carcinoma of esophagus in a high-risk population. World J Gastroenterol 2005;11:1200-1203.
83. Matsuyama A, Inoue H, Shibuta K, Tanaka Y, Barnard GF, Sugimachi K, et al. Hepatoma-derived growth factor is associated with reduced sensitivity to irradiation in esophageal cancer. Cancer Res 2001;61:5714-5717.
84. Xie YE, Tang EJ, Zhang DR, Ren BX. Down-regulation of Bcl-XL by RNA interference suppresses cell growth and induces apoptosis in human esophageal cancer cells. World J Gastroenterol 2006;12:7472-7477.
85. Takahashi K, Miyashita M, Makino H, Akagi I, Orita H, Hagiwara N, et al. Expression of Akt and Mdm2 in human esophageal squamous cell carcinoma. Exp Mol Pathol 2009;87:42-47.
86. Zhou S, Ye W, Shao Q, Qi Y, Zhang M, Liang J. Prognostic signi fi cance of XIAP and NF-κB expression in esophageal carcinoma with postoperative radiotherapy. World J Surg Oncol 2013;11:288.
87. Pai SI, Lin YY, Macaes B, Meneshian A, Hung CF, Wu TC. Prospects of RNA interference therapy for cancer. Geneer 2006;13:464-477.
Cite this article as:Islamian JP, Mohammadi M, Baradaran B. Inhibition of human esophageal squamous cell carcinomas by targeted silencing of tumor enhancer genes: an overview. Cancer Biol Med 2014;11:78-85. doi: 10.7497/ j.issn.2095-3941.2014.02.002
Jalil Pirayesh Islamian
E-mail: pirayeshj@gmail.com
Received December 22, 2013; accepted April 8, 2014. Available at www.cancerbiomed.org
Copyright © 2014 by Cancer Biology & Medicine
杂志排行
Cancer Biology & Medicine的其它文章
- Staging the axilla in women with breast cancer: the utility of preoperative ultrasound-guided needle biopsy
- Research progress on the anticarcinogenic actions and mechanisms of ellagic acid
- Incidence and mortality of female breast cancer in the Asia-Paci fi c region
- Clinico-pathological signi fi cance of extra-nodal spread in special types of breast cancer
- Effects of postmastectomy radiotherapy on prognosis in different tumor stages of breast cancer patients with positive axillary lymph nodes
- Spindle cell carcinoma of the breast as complex cystic lesion: a case report
