隐蔽型真菌毒素的形成及降解方法的研究进展
2014-03-08王彦苏周佳宇谢星光丁成龙戴传超
王彦苏,周佳宇,谢星光,丁成龙,戴传超,*
(1.南京师范大学生命科学学院,江苏省微生物资源产业化工程技术研究中心,江苏 南京 210023;2.江苏省农业科学院畜牧研究所,江 苏 南京 210014)
隐蔽型真菌毒素的形成及降解方法的研究进展
王彦苏1,周佳宇1,谢星光1,丁成龙2,戴传超1,*
(1.南京师范大学生命科学学院,江苏省微生物资源产业化工程技术研究中心,江苏 南京 210023;2.江苏省农业科学院畜牧研究所,江 苏 南京 210014)
真菌毒素广泛分布于霉变的或受霉菌污染的粮食、谷物、饲料中,对人类和家畜健康造成严重威胁。植物和微生物可以降解真菌毒素为弱毒性的毒素,但是这种降解并不彻底,隐蔽型的真菌毒素依然存在于植物体内或环境中。本文主要从真菌毒素的分布及危害、植物生长和储存过程中降解真菌毒素的不完全性、微生物转化或降解真菌毒素形成“隐蔽型真菌毒素”、隐蔽型真菌毒素降解过程这四个方面进行论述,希望为今后真菌毒素的完全降解研究提供参考。
真菌毒素降解;植物转化;微生物降解;隐蔽型真菌毒素
真菌毒素是丝状真菌产生的有毒次级代谢产物。庄稼收获的前后容易感染真菌,如果谷物储存条件潮湿并且温度有利于真菌生长,就会有大量真菌毒素的产生。主要产毒素的真菌属有:镰刀菌属(Fusarium)、青霉属(Penicillium)、曲霉属(Aspergillus)、链格孢霉属(Alternar ia)。储存谷物中也常常会有真菌毒素,污染了真菌毒素的食品或饲料对人体或动物有一定危害。许多方法可以减弱或消除真菌毒素:通过非营养吸附化合物如:活性碳、膨润土、沸石等,可以减少动物胃肠道对毒素的吸收,最终可以减少毒素转移到动物产品中[1];一些动植物或微生物可以通过改变真菌生长环境条件抑制真菌毒素的产生,来自植物宿主的次级代谢物质可以参与调节真菌毒素合成途径,减少毒素积累;植物在生长和储存过程中会降解一部分真菌毒素,微生物也可以通过吸附、水解、还原、糖苷化、甲酰化等方式,对真菌毒素起到解毒作用。但是用以上方法处理过的食品看似安全,实际上却存在着被植物或微生物代谢转化的隐蔽型真菌毒素。隐蔽型真菌毒素不容易被检测到,常常以共轭形式存在,如Deoxynivalenol(DON)的糖苷式(deoxynivalenol-3-D-glucopyranoside,D3G),Zearalenone(ZON)的硫化物Zearalenone-14-sulfate(Z14S),存在一定毒性,危害人体或动物健康,对于形成的隐蔽型真菌毒素的完全降解方 法也很少有研究。因此隐蔽型真菌毒素的检测技术、解毒机制和降解方法还需要进一步深入研究。
1 真菌毒素的分布
真菌毒素是丝状真菌产生的有毒次级代谢产物,主要的真菌毒素有:玉米赤霉烯酮、单端孢霉烯族毒素、黄曲霉毒素、赭曲霉毒素、麦角生物碱、伏马菌素、展青霉素等,容易污染玉米、麦类等谷物、食品、饲料。Richard等[2]在成熟玉米青贮样品中检测到7种真菌毒素,如黄曲霉毒素B1、橘霉素、脱氧雪腐镰刀菌烯醇、伏马菌素B1、胶霉毒素、赭曲霉素、玉米赤霉烯酮。小麦贮藏中比较常见的真菌污染有棒曲霉(Aspergillus alternata)(14.6%)、烟曲霉(Aspergillus fumigatus)(10.5%)、黑曲霉(Aspergillus niger)(8.3%)、橘青霉(Penicillium citrinum)(3.8%)[3]。大麦储存过程中,主要被镰刀菌(Fusarium)污染,镰刀菌所产生的毒素有伏马菌素、脱氧雪腐镰刀菌烯醇等。Sulyok等[4]在小麦和玉米中分离出39 种毒素,其中包括单端孢霉烯族毒素类、玉米烯酮、伏马菌素、麦角生物碱等,如果食用了感染丝状真菌的粮食和饲料,会对人体或动物造成一定危害。DON是单端孢霉烯族类毒素B中最主要的一种,能引起胃肠障碍、造血机能障碍、脏器出血,并具有致癌、致突变作用。DON的糖基化衍生物DON-3-glucoside(DON3Glc)被发现存在于谷物和啤酒中[5-6]。Shephard等[7]发现在家庭酿造的大麦啤酒可以检测出伏马菌素B1(fumonisins,FB1)、B2(FB2)和B3(FB3)。Siegel等[8]在啤酒和饮料中检测到交链孢菌酮酸(tenuazonic acid,TA),平均含量达到11 μg/kg,最高达到了175 μg/kg。在饲料中,特别是青贮饲料中,如果青贮原料感染真菌或青贮过程不严格厌氧,造成真菌繁殖,会使青贮饲料中存在真菌毒素,对家畜健康造成危害[9]。Lepom[10]研究发现,如果使用感染了大刀镰刀霉(Fusarium culmorum)的玉米进行青贮,11 d后检测不到F. culmorum,但是该菌所产生的玉米烯酮(zearalenone)含量却在整个12 周内没有变化。
2 植物对真菌毒素的代谢转化以及隐蔽型真菌毒素的存在
真菌毒素和其他异源物质一样,可以被活体植物代谢,形成共轭的真菌毒素。病原菌在感染植物过程中,病原菌细胞壁降解酶基因表达上调,降解植物细胞壁侵染植物细胞[11]。Hariprasad等[12]研究发现,绿色蔬菜可吸收并通过疏水组织运输真菌毒素到地上部分。植物种子可以通过被 动运输,在水通道蛋白的参与下,吸收真菌毒素,并通过植物蒸腾作用吸收到叶部,也可积累到植物根内皮层[13]。真菌毒素 作为异源生物在植物体内被识别需要经历3 个化学修饰阶段,第一个阶段是还原、氧化和乙酰化,这个阶段可能导致激活衍生型分子的毒性变高;第二个阶段是酶转化活性基团如进行结合、糖苷化、硫基化作用,形成亲水性化合物,更好地消除真菌毒素、减弱毒性;第三个阶段是隔离真菌毒素,使毒素区域化,如将真菌毒素包裹在植物细胞液泡内,或结合在植物细胞壁上[14]。
植物代谢物质,如一些植物挥发性物质也可以减少毒素积累,甚至会促进植物降解真菌毒素。丁香、山百里香等植物挥发油可以抑制黄曲霉毒素(aflatoxin B1,AFB1)的积累[15]。3 种药用植物(Thy mus daenensis、Satureja khozistanica和Satureja macrosiphonia)挥发油、乙醇提取物可以抑制曲霉属(Aspergillus)的生长,但水萃取物可以不同程度降解AFB1[16-18]。玉米烯酮是一种镰刀菌毒素, Berthiller等[19]使用玉米烯酮溶液处理拟南芥(Arabidopsis thaliana)植物,液相色谱-质谱联用检测后发现有17 种代谢物质,主要有糖苷、丙糖苷、玉米烯酮双已糖和已糖-戊糖二糖,α-和β-玉米烯酮等。真菌毒素的降解并不完全,会形成共轭真菌毒素并积累在植物体内或表面。
真菌毒素的发生、生物代谢还不是很清楚,并且用于分析真菌毒素的仪器大都不能检测到共轭的真菌毒素。还有一个令人头疼的问题是减毒后的共轭真菌毒素被人类或动物消化后可以重新激活,也有人把这种真菌毒素称为隐蔽型真菌毒素[20]。以DON和ZON为例,在粮食储存和啤酒发酵过程中,会容易形成共轭的DON和ZON,危害人类健康。
2.1 DON
Tran和Smith在澳大利亚多个地区对DON进行的一项调查中显示。2009—2011年污染样品中都检测到以共轭形式存在的隐蔽型DON,污染率分别达到61%、87%、68%。共轭DON对作物存在潜在影响[21]。啤酒是比较受人们欢迎的饮品,麦芽是其基本的原料之一,在酿造过程中特别容易污染镰刀菌(Fusarium)毒素,如DON、3-Acetyldeoxynivalenol(3ADON)、D3G。Varga等[22]发现啤酒所含D3G和DON平均质量浓度分别是6.9 μg/L和8.4 μg/L。
2.2 ZON
1990年,Gareis等[23]在谷物样品中发现玉米烯酮的糖苷式Zearalenone-glycoside,用含有zearalenone-4-p-D-glucopyranoside的饲料饲喂猪14 d后,在消化过程中,糖苷被水解掉,又会形成ZON和α-Zearalenol。这种 不容易被检测到,但对人体或动物存在潜在危害的真菌毒素被Gareis为“masked mycotoxin”。Vendla等[24]发现以谷物为基础的产品中发现大量毒素,如Deoxynivalenol、Zearalenone,还存在它们的代谢物质D3G、3ADON l、Zearalenol-4-glucopyranoside、α-Zearalenol、β-Searalenol、α-Zearalenol-4-glucopyranoside、β-Zearalenol-4-glucopyranoside和Zearalenone-4-sulfate。其中,丰富度最高的是Zearalenone-4-sulfate。麦麸中有较高的毒素含量,其中DON 254 ng/g、Zearalenone-4-sulfate 6 ng/g和ZON 44 ng/g。
3 微生物对真菌毒素的不完全降解
3.1 乳酸菌的抑菌作用
生物防治病原菌是抑制食品腐败提高食品保质期的有效方法,乳酸菌作为益生菌可以抑制食品中的腐败菌,提高食品保质期[25]。乳酸菌抑制腐败菌生长的作用机理分为几个原因:乳酸菌产生细菌素(蛋白类物质抗菌多肽)、乳酸、乙酸、苯基乳酸、脂肪酸、酚类化合物[26],许多乳酸菌可以产生黄素蛋白氧化酶类但不产生过氧化氢酶,在氧气存在的条件下产生H2O2,使具有强氧化作用的H2O2可以破坏细菌细胞蛋白。乳酸菌(Lactobacillus paracasei ssp. tolerans)对真菌和霉菌(如Fusarium proliferatum M5689、M5991、Fusarium graminearum R4053)有一定的抑制作用[27]。Magnusson从环境中分离37株抑制真菌活性的乳酸菌,能够抑制A. fumigatus、Aspergillus nidulans、Penicillium commune和Fusarium sporotrichioides的生长[28]。表1显示了乳酸菌能够抑制的一些病原菌或腐败菌类型。
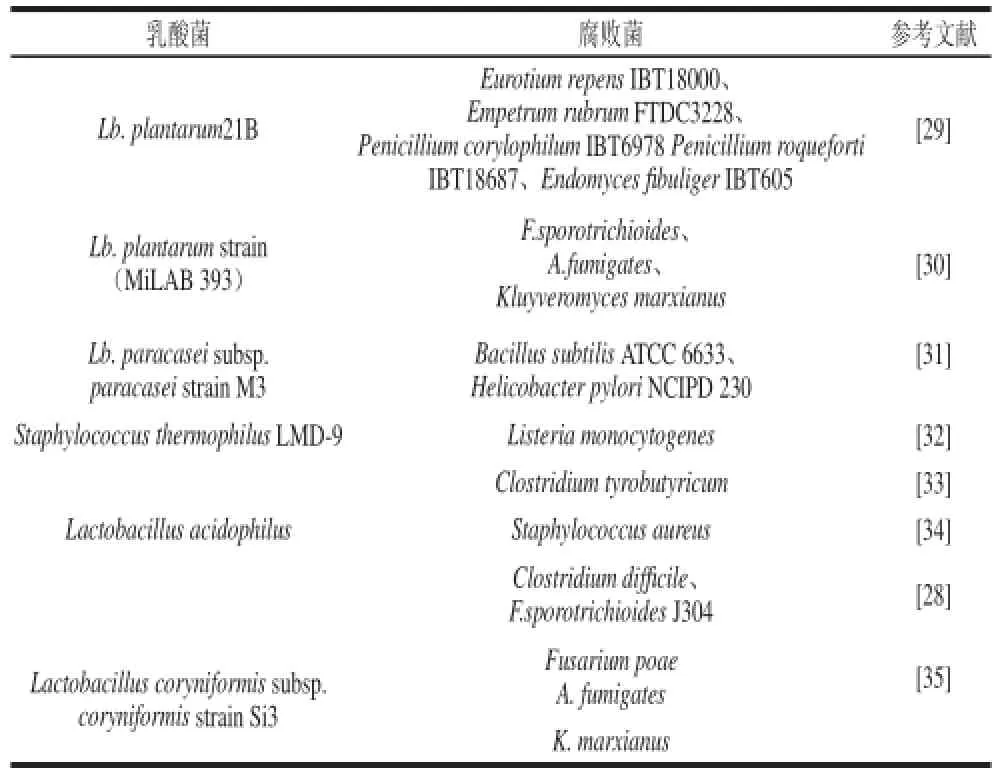
表1 乳酸菌抑制菌种Table1 Strains against lactic acid bacteria
3.2 乳酸菌对真菌毒素的结合及降解作用
乳酸菌细胞壁与真菌毒素可以通过氢键、离子键、疏水作用力进行结合,减弱样品毒性[36]。Wu等[37]发现酵母和乳酸菌作为黄曲霉霉素的生物吸附剂可以阻止其进入人类和动物肠道。乳酸菌(Lactobacillus casei、Lb. plantarum和Lactobacillus fermentum)等一些传统的益生菌可以结合黄曲霉毒素,起到对食品进行解毒的作用。被镰刀菌毒素污染的玉米青贮中可以分离到8 株乳酸杆菌属(Lactobacilli)和3 株明串珠菌属(Leuconostoc),研究发现这几株菌可以将ZON转化成 α-Zearalenol,但是不能转化DON和伏马菌素(fumonisins),许多菌可以和这些毒素结合[38]。Fazeli等[38]从酸面包和奶制品中分离得到乳酸菌(Lb. casei、Lb. plantarum和Lb. fermentum),发现所有菌株都有能力去除AFB1,Lb. fermentum和Lb. plantarum可以快速去除AFB1,去除率达到61%~65%。根据以上结果,乳酸菌与AFB1可以有效结合,但是,Niderkorn等发现[39]结合不是很紧密,经过离心后,会有AFB1脱离菌体,不同菌体与不用毒素的结合能力不同,可能与毒素结构和菌体表面结构不同。Franco等[40]发现不管是活体乳酸菌细胞还是热灭活的乳酸菌细胞,乳酸菌在体外可以去除DON,pH值和培养基浓度都不会影响去除能力,并且发现热灭活的细胞有更强的去除真菌毒素的能力,DON减少率35%~67%,活体细胞只达到16%~56%。Niderkorn等[41]发现乳酸菌与毒素结合反应涉及到乳酸菌中的肽聚糖和TCA循环,特别是细胞壁肽聚糖中部分肽结构在与伏马菌素B1和B2结合的过程中起到明显作用,热激或酸处理后的细胞结合真菌毒素的能力增强,可能是热激和酸处理后细胞壁表面结合位点暴露。乳酸菌在植物加工或贮存过程中,是可以减弱真菌毒素污染的良好菌剂,也可以提高食物风味,可以作为粮食储存保护剂或修复剂[42]。在啤酒发酵过程中,麦芽是不可缺少的原料,在麦芽浸泡过程中,乳酸菌数量达到108CFU/g,其中异型发酵明串珠菌属占优势地位[43-44],乳酸菌在麦芽发酵过程中,可以产生细菌素,抑制腐败菌生长[45]。
研究发现乳酸菌可以有效降解真菌毒素,在酸面团发酵中,乳酸菌与酵母混合接菌可以降解赭曲霉素(ochratoxin A)[46]。被毒素污染的玉米与乳酸菌共培养处理组相比于未用乳酸菌处理组,对SNO细胞(人类食道癌细胞系)毒性较弱[47]。Roig从肠道分离得到9 株乳酸菌(Bif i dobacterium longum、Bif i dobacterium bif i dum、Bifidobacterium breve、Bifidobacterium adolescentes、Lactobacillus rhamnosus、Lactobacillus casei-casei、Streptococcus termofilus、Lactobacillus ruminis、Lb. casei)和22株酿酒酵母(Saccharomyces cerevisiae),所有菌株在体外培养基中降解范围为5%~99%,在小麦粉食物系统中Enniations(ENs)的降解1.3%~49.2%[48]。
食品安全是食品行业和消费者越来越关注的问题,乳酸菌可以抑制病原菌的生长、吸附或降解真菌毒素。因此,今后应加强乳酸菌与病原菌拮抗机理的研究,探讨乳酸菌在食品生产加工过程微生物群落中生态作用,使乳酸菌菌剂可以应用于食物防腐工业中。乳酸菌会产生一些代谢产物,如细菌素、有机酸等,对病原菌具有良好的抑制作用,如果能大规模生产使用,在粮食储存、食品加工过程中有广阔的应用前景。但是以上研究显示,乳酸菌只能部分降解或结合真菌毒素,并没有完全消除真菌毒素的危害,对于乳酸菌是否能完全彻底的降解真菌毒素还需要进一步研究。
3.3 其他降解菌对真菌毒素的降解作用
研究发现许多从土壤、水体等自然环境中获得真菌毒素降解菌[49-50],也有许多研究发现从人体或动物胃肠道获得的微生物可以抑制真菌生长或降解真菌毒素。Kusumaningtyas等[51]研究将酿酒酵母(S. cerevisiae)和寡孢根霉菌(Rhizopus oligosporus)分别接种和组合接种被黄曲霉毒素污染的鸡饲料中,在处理第5天后,AFB1达到最大降解率,R. oligosporus接种的处理组降解AFB1活性最高。被赫曲霉素污染的大麦固体发酵物,在接种白腐真菌Pleurotus ostreatus 4周后,OTA降解到23%,OTB降解到3%,在降解过程中OTB水解为Ochratoxin β[52]。Zuo等[53]发现Lb. casei、枯草芽孢杆菌(B. subtilis)和异常毕赤酵母(Pichia anomala)混合培养可以有效地降解AFB1,在12 h内可去除59.3%,在48 h内可去除87.04%。非病原菌嗜吡啶红球菌(Rhodococcus pyridinivorans)K408与ZON孵育5 d后可以达到87.21%的降解率[54]。
以上研究发现微生物对真菌毒素具有良好的降解或转化能力,可能是微生物产生一些物质(如酶类)作用于腐败真菌,使菌体生长减弱,或作用于真菌毒素,使毒素降解或使毒性减弱。Meca等[55]研究发现酿酒酵母(S. c erevisiae LO9、YE5、A34和A17)混合粗酶液,加到白僵菌毒素(beauvericin)标准溶液中,毒素降解率为83%~100%,以污染白僵菌毒素的玉米粉为基质,降解率为66%~91%。细菌和黑酵母降解伏马菌素的途径,首先是伏马菌素羧酸脂酶水解两个三羧酸侧链,第二步是在黑色酵母中胺氧化酶作用水解后的伏马菌素B1脱氨基作用,或是在细菌中氨基转移酶作用转氨基。Hartinger等[56-60]发现鞘脂单胞菌(Sphingopyxis sp. MTA144)编码氨基转移酶FumⅠ,FumⅠ能够催化水解了的伏马菌素B1脱氨基作用,FumⅠ和伏马菌素羧酸脂酶Fum D联合作用,使伏马菌素解毒成为可能。

图1 黄曲霉毒素BB1水解过程Fig.1 Hydrolysis of aflatoxin B1
AFB1通过环氧化物水解酶水解为AFB1-8-9-二氢二醇(AFB1-8,9-dihydrodiol)(图1),Wang等[61]对来自白腐真菌Phanerochaete sordid YK-624的锰过氧化物酶可以有效消除AFB1,5 nkat的MnP与AFB1共培养48 h AFB1的最大消除达到86.0%,锰过氧化物酶将ABF1环氧化成AFB18,9-epoxide,再水解为AFB1-8,9-二氢二醇(AFB1-8,9-dihydrodio l)。羧肽酶A 、胰蛋白酶、α-糜蛋白酶和组织蛋白酶C能在体外条件下水解赭曲霉毒素OTA为弱毒性的OTα[62-63](图2)。

图2 赭曲霉毒素A水解过程Fig.2 Hydrolysis of ochratoxin A
不同种类的微生物对真菌毒素的降解途径不同,以微生物降解DON为例,革兰氏阳性菌和革兰氏阴性菌对DON具有不同的降解作用,革兰氏阳 性菌可以吸收DON,将DON作为碳源利用,而革兰氏阴性菌则不能吸收利用,需要在含有DON培养一段时间后才能起到降解DON的作用。革兰氏阳性菌与革兰氏阴性菌存在不同DON降解途径,在这两种途径中有不同的降解酶基因表达调控机制,但Sato等[64]发现两种菌的降解产物均为3-Epi-deoxynivalenol。

图3 脱氧雪腐镰刀菌烯醇结构式Fig.3 Structure of deoxynivalenol
DON毒性主要是中间环氧键的存在(图3)[65]。许多胃肠道微生物可以将DON还原去环氧基团,一些来自动物消化系统的微生物代谢可以降解DON为弱毒性的Deepoxy-deoxynivalenol (DOM-1)[66]。Gratz等[67]发现人类粪便微生物可以解毒DON,形成DOM-1。Yu等[68]从鸡肠道中分离得到DON降解细菌,将DON转化成DOM-1。许多研究都是胃肠道微生物在厌氧环境下通过还原去环氧化作用解毒DON[69-71]。Islam等[72]用农田土壤制备培养基,在有氧条件下可以降解DON、去环氧化。

图4 脱氧雪腐镰刀菌烯醇乙酰化衍生物结构式Fig.4 Structure of acetylated derivatives of deoxynivalenol
DON存在下常常伴随着两个乙酰化衍生物:3-ADON和15-ADON(图4)。Pinton等[73]通过对细胞增殖、胃肠道屏障功能和肠道形态结构等观察研究,比较体内体外条件下DON、3-ADON、15-ADON毒性,发现15-DON因为能激活MAPK途径使其具有更高的毒性。Pinton等[74]对DON、3-DON、15-DON毒性进行了分析,它们的毒性大小依次为3-ADON≤DON<15-ADON。但是DON虽然被转化和降解,但是降解并不彻底,其衍生物依然有弱毒性,危害人类健康。
3.4 微生物对真菌毒素降解的不完全性
真 菌毒素的降解取决于环境条件,如:微生物的种类、数量、化合物结构等。往往微生物对真菌毒素具有一定 的降解作用都具有不完全性,虽然降解率较高,但降解产物具有不彻底性,只能减弱毒性,被人体或动物吸收后,经过胃肠道微生物作用和修饰,降解产物又会回复到原来的毒性较强的化合物,很少会转变成无机矿物质、H2O、CO2、。
4 真菌毒素检测与降解存在的问题
对于真菌毒素的检测存在许多种方法薄层层析法、酶联免疫分析方法、气相色谱法、高效液相色谱法等。这几种方法各有优缺点。由于真菌毒素有不同的化学式和物理化学性质,特殊的提取、净化、合适的检 测器,只能有效的检测一种或一类真菌毒素。Tanaka等[76]论述了水稻真菌毒素的检测方法,脱氧雪腐镰刀菌烯醇(deoxynivalenol,DON)、镰刀菌烯酮和雪腐镰刀菌醇一般使用气相色谱-质谱联用检测,单 端孢霉烯族化合物使用气相色谱-电子捕获检测(gas chromatography and electron capture detection,GC-ECD)确定。杂色曲霉素(sterigmatocystin,ST)是曲霉属或其他一些真菌产生的致癌性化合物,气相色谱-质谱联用(gas chromatography-mass spectrometry,GC-MS)、液相色谱-质谱联用(liquid chromatography-mass spectrometry(LC-MS)、LC-MS/MS)和液相紫外检测(liquid ultraviolet detection,LC-UV) 都可以用于STE的检测,但是LC-UV的检测效果比较好。但是对隐蔽型真菌毒素的检测没有做具体论述。
多种方法并用,可以更有效地检测难以检测的隐蔽型真菌毒素。LC-MS/MS由于其普遍性、选择性、敏感性的优点,近几年应用比较广泛。Vendl等[77]用LC-MS/MS从谷物食物中检测到DON和ZON还有8 种它们的代谢产物如:Deoxynivalenol-3-glucoside (D3G)、3-Acetyl-deoxynivalenol(3ADON)、Zearalenone-4-glucoside(Z4G)、α-Zearalenol (α-ZOL)、β-Zearalenol(β-ZOL)、α-Zearalenol-4-glucoside (α-ZG)、β-Zearalenol-4-glucoside(β-ZG)、Zearalenone-4-sulfate(Z4S)。Nakagawa等[78]利用高分辨率LC-轨道阱质谱检测到单端孢霉烯族化合物A型的糖基化衍生物:Neosolaniol-glucoside(NESGlc)和Diacetoxyscirpenolglucoside(DASGlc)、单端孢霉烯族化合物B型化合物的糖基化衍生物D3G。Vidal等[79]论述了生物传感器在检测真菌毒素方面的应用,但是多种毒素检测、生物标志物和蒙面真菌毒素的检测方面还需进一步研究。
虽然在检测共轭真菌毒素技术上得到提高,但是对于共轭真菌毒素的完全解毒还需要进一步研究。通过糖基化作用来“伪装”的真菌毒素,如DON的糖基化DON-3-Glc可以水解又回到DON,研究发现DON-3-Glc可以抵抗酸性环境,因此在胃液的酸性环境中水解DON-3-Glc是非常困难的。Berthiller等[80]发现人类胞质中的β-葡糖苷酶对D3G水解效果较差,但是纤维素酶和纤维二糖酶对其的最大水解率分别达到13%和73%,这表明D3G被瘤胃动物吸收后,在瘤胃纤维素分解菌的作用下使D3G回到DON。同时他们又发现胃肠道细菌如Bifidobacterium adolescentis、Enterococcus durans、Enterococcus mundtii、Lb. plantarum 8 h对D3G的水解率分别为:17%~25%、14%~27%、38%、62%。虽然植物将DON解毒为D3G,但是在肠道微生物作用下D3G又水解为DON。
转基因植物中过表达几种植物或细菌基因可以加强植物修复系统,在汞污染的修复过程中,Heaton等[81]将植物转基因表达细菌的merA基因或merB基因,这两种基因来自细菌的mer操纵子,merA基因编码NADPH依 赖的汞离子还原酶,可以转化离子汞(Hg(Ⅱ))转化成(Hg(0)),merB基因编码有机汞裂合酶,可以降解MeHg为甲烷和Hg(Ⅱ),有效的修复汞污染。以此借鉴,细菌中水解酶基因转移到植物中,植物可以更好地降解土壤以及吸收后的真菌毒素,使真菌毒素钝化,利用转基因植物导入细菌或真菌编码水解或还原真菌毒素基因的方法,希望可以有效缓解被黄曲霉毒素污染现状。对于环境中防治真菌毒素,高温度、高湿度会黄曲霉大量繁殖,RNA沉默技术减少黄曲霉生长和其毒素的合成,从而减弱毒性[82]。
5 结 语
真菌毒素广泛存在于粮食、饲料、食品中,会对人类和动物健康造成威胁。许多研究发现,外界环境可以影响真菌毒素的合成,甚至可以调节毒力因子基因的表达,一些产毒素真菌还会受其宿主植物信号分子的影响,从而调控真菌毒素基因表达。粮食作物在加工或储存过程中可能会有效地减弱真菌毒素的毒性,但作用并不完全。乳酸菌作为益生菌可以调节人类胃肠道菌群,提高食品或饲料的口感、营养。其细胞壁特殊肽段对真菌毒素具有吸附作用,还有研究表明乳酸菌可以对真菌毒素起到降解作用。许多环境微生物对真菌毒素具有降解作用,降解的机理有水解作用、还原作用、乙酰化作用等,但是降解作用并不彻底,形成真菌毒素的中间衍生物,没有完全降解成H2O和CO2等,依然对环境有害。因此对于隐蔽型真菌毒素的降解还需进一步研究,归结为以下几点。1)来自宿主植物的信号分子可以减少真菌毒素积累,植物挥发油发现可以促进降解真菌毒素,这就为减少真菌毒素污染提供了新的思路;2)研究发现来自环境和肠道微生物具有降解真菌毒素的功能,新型、安全的真菌毒素降解菌还需要进一步筛选;3)在以往的检测中,往往忽略了隐蔽型真菌毒素的存在及其危害性,因此需进一步提高检测技术,检测粮食或饲料中隐蔽型真菌毒素;4)大部分植物还是微生物对真菌毒素只是起到吸附、不完全降解或减弱毒性的作用,并没有将真菌毒素完全降解成H2O或CO2,隐蔽型真菌毒素的完全降解还需深入研究。
[1] RAMOS A J, FINK-GREMMELS J, HERNANDEZ E. Prevention of toxic effects of mycotoxins by means of nonnutritive adsorbent compounds[J]. Journal of Food Protection, 1996, 59(6): 631-641.
[2] RICHARD E, HEUTTE N, BOUCHART V, et al. Evaluation of fungal contamination and mycotoxin production in maize silage[J]. Animal Feed Science and Technology, 2009, 148(2): 309-320.
[3] MATHEW S, THOMAS G, AHMAD T. An evaluation of the fungi isolated from sub-epidermal region of post-harvested stored wheat grains[J]. Nepal Journal of Biotechnology, 2011, 1(1): 9-13.
[4] SULYOK M, BERTHILLER F, KRSKA R, et al. Development and validation of a liquid chromatography/tandem mass spectrometric method for the determination of 39 mycotoxins in wheat and maize[J]. Rapid Communications in Mass Spectrometry, 2006, 20(18): 2649-2659.
[5] LANCOVA K, HAJSLOVA J, POUSTKA J, et al. Transfer of Fusarium mycotoxins and ‘masked’ deoxynivalenol (deoxynivalenol-3-glucoside) from field barley through malt to beer[J]. Food Additives and Contaminants, 2008, 25(6): 732-744.
[6] MALACHOVA A, DZUMAN Z, VEPRIKOVA Z, et al. Deoxynivalenol, deoxynivalenol-3-glucoside, and enniatins: the major mycotoxins found in cereal-based products on the Czech market[J]. Journal of Agricultural and Food Chemistry, 2011, 59(24): 12990-12997.
[7] SHEPHARD G S, van der WESTHUIZEN L, GATYENI P M, et al. Fumonisin mycotoxins in traditional Xhosa maize beer in South Africa[J]. Journal of Agricultural and Food Chemistry, 2005, 53(24): 9634-9637.
[8] SIEGEL D, MERKEL S, KOCH M, et al. Quantification of the Alternaria mycotoxin tenuazonic acid in beer[J]. Food Chemistry, 2010, 120(3): 902-906.
[9] YEKELER H, BITMIS K, OZCELIK N, et al. Analysis of toxic effects of Alternaria toxins on esophagus of mice by light and electron microscopy[J]. Toxicologic Pathology, 2001, 29(4): 492-497.
[ 10] LEPOM P. Occurrence of Fusarium species and their mycotoxins in maize: 1. Method of determining zearalenone in maize and maize silage by means of high performance liquid chromatography (HPLC) using nuorescence detection[J]. Archives of Animal Nutrition, 1988, 38: 799-806.
[11] CARAPITO R, VORWERK S, JELTSCH J M, et al. Genome-wide transcriptional responses of Fusarium graminearum to plant cell wall substrates[J]. FEMS Microbiology Letters, 2013. 340: 129-134.
[12] HARIPRASAD P, DURIVADIVEL P, VENKATESWARAN G. Natural occurrence of a atoxin in green leafy vegetables[J]. Food Chemistry, 2013, 138: 1908-1913.
[13] SNIGDHA M, HARIPRASAD P, VENKATESWARAN G. Mechanism of aflatoxin uptake in roots of intact groundnut (Arachis hypogaea L.) seedlings[J]. Environmental Science and Pollution Research, 2013, 20(12): 1-9.
[14] BERTHILLER F, LEMMENS M, WERNER U, et al. Short review: metabolism of the Fusarium mycotoxins deoxynivalenol and zearalenone in plants[J]. Mycotoxin Research, 2007, 23(2): 68-72.
[15] BIUMA R, AMAIDEN M R, ETCHEVERRY M. Screening of Argentine plant extracts: impact on growth parameters and aflatoxin B1accumulation by Aspergillus section Flavi[J] . International Journal of Food Microbiology, 2008, 122: 114-125.
[16] AKBAR G, MOHSEN F, MAHMUD S, et al. A atoxin B1-reduction of Aspergillus fl avus by three medicinal plants (Lamiaceae)[J]. Food Control, 2013, 31: 218-223.
[17] DEABES M M, ELSOUD N H A, ELKASSEM L T A. in vitro Inhibition of growth and aflatoxin B1production of Aspergillus fl avus strain (ATCC 16872) by various medicinal plant essential oils[J]. Macedonian Journal of Medical Sciences, 2011, 4(4): 345-350.
[18] OMIDBEYGI M, BARZEGAR M, HAMIDI Z, et al. Antifungal activity of thyme, summer savory and clove essential oils against Aspergillus fl avus in liquid medium and tomato paste[J]. Food Control, 2007, 18(12): 1518-1523.
[19] BERTHILLER F, WERNER U, SULYOK M, et al. Liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) determination of phaseⅡ metabolites of the mycotoxin zearalenone in the model plant Arabidopsis thaliana[J]. Food Additives and Contaminants, 2006, 23(11): 1194-1200.
[20] BERTHILLER F, SULYOK M, KRSKA R, et al. Chromatographic methods for the simultaneous determination of mycotoxins and their conjugates in cereals[J]. International Journal of Food Microbiology, 2007, 119(1): 33-37.
[21] TRAN S T, SMITH T K. A survey of free and conjugated deoxynivalenol in the 2009, 2010 and 2011 cereal crops in Australia[J]. Animal Production Science, 2013, 53(5): 40 7-412.
[22] VARGA E, MALACHOVA A, SCHWARTZ H, et al. Survey of deoxynivalenol and its conjugates deoxynivalenol-3-glucoside and 3-acetyl-deoxynivalenol in 374 beer samples[J]. Food Additives & Contaminants: Part A, 2013, 30(1): 137-146.
[23] GAREIS M, BAUER J, THIEM J, et al. Cleavage of zearalenoneglycoside, a “masked” mycotoxin, during digestion in swine[J]. Journal of Veterinary Medicine, Series B, 1990, 37: 236-240.
[24] VENDL O, CREWS C, MACDONALD S, et al. Occurrence of free and conjugated Fusarium mycotoxins in cereal-based food[J]. Food Additives and Contaminants, 2010, 27(8): 1148-1152.
[25] STILES M E. Biopreservation by lactic acid bacteria[J]. Antonie van Leeuwenhoek, 1996, 70: 331-345.
[26] SCHNURER J, MAGNUSSON J. Antifungal lactic acid bacteria as biopreservatives[J]. Trends in Food Science & Technology, 2005, 16: 70-78.
[27] HASSAN Y I, BULLERMAN L B. Antifungal activity of Lactobacillus paracasei ssp. tolerans isolated from a sourdough bread culture[J]. International Journal of Food Microbiology, 2008, 121(1): 112-115.
[28] MAGNUSSON J, STROM K, ROOS S, et al. Broad and complex antifungal activi ty among environmental isolates of lactic acid bacteria[J]. FEMS Microbiology Letters, 2003, 219(1): 129-135.
[29] LAVERMICOCCA P, VALERIO F, EVIDENTE A, et al. Purification and characterization of novel antifungal compounds from the sourdough Lactobacillus plantarum strain 21B[J]. Applied and Environmental Microbiology, 2000, 66(9): 4084-4090.
[30] STROM K, SJOGREN J, BROBERG A, et al. Lactobacillus plantarum MiLAB 393 produces the antifungal cyclic dipeptides cyclo(L-Phe-LPro) and Cyclo( L-P he-trans-4-OH-L-Pro) and 3-phenyllactic acid[J]. Applied and Environmental Microbiology, 2002, 68(9): 4322-4327.
[31] ATANASSOVA M, CHOISET Y, DALGALARRONDO M, et al. Isolation and partial biochemical characterizati on of a proteinaceous anti-bacteria and anti-yeast compound produced by Lactobacillus paracasei su bsp. paracasei strain M3[J]. International Journal of Food Microbiology, 2003, 87: 63-73.
[32] FONTAINE L, HOLS P. The inhibitory spectrum of ther mophilin 9 from Streptococcus thermophilus LMD-9 depends on the production of multiple peptides and the activity of BlpGSt, a thiol-disulfide oxidase[J]. Applied and Environmental Microbiology, 2008, 74(4): 1102-1110.
[33] MATHOT A G, BELIARD E, THUAULT D. Streptococcus thermophilus 580 produces a bacteriocin potentially suitable for inhibition of Clostridium tyrobutyricum in hard cheese[J]. Journal o f Dairy Science, 2003, 86(10): 3068-3074.
[34] MKRTCHYAN H, GIBBONS S, HEIDELBERGER S, et al. Purification, characterisation and identification of acidocin LCHV, an antimicrobial peptide produced by Lactobacillus acidophilus n.v. Er 317/402 strain Narine[J]. International Journal of Antimicrobial Agents, 2010, 35(3): 255-260.
[35] MAGNUSSON J, SCHNURER J. Lactobacillus cory niformis subsp. coryniformis strain Si 3 produces a broad-spectrum proteinaceous antifungal compound[J]. Applied and Environmental Microbiology, 2001, 67(1): 1-5.
[36] 韩鹏飞, 贺稚非, 李洪军, 等.微生物细胞壁结构及结合真菌毒素的研究进展[J]. 食品科学, 2012, 33(11): 294-298.
[37] WU Q, JEZKOVA A, YUAN Z, et al. Biological degradation of aflatoxins[J]. Drug Metabolism Reviews, 2009, 41(1): 1-7.
[38] FAZELI M R, HAJIMOHAMMADALI M, AZAMOSSADAT M, et al. Aflatoxin B1binding capacity of autochthonous strains of lactic acid bacteria[J]. Journal of Food Protection, 2009, 72(1): 189-192.
[39] NIDERKOM V, MORGAVI D P, PUJOS E, et al. Screening of fermentative bacteria for their ability to bind and biotransform deo xynivalenol, zearalenone and fumonisins in an in vitro simulated corn silage model[J]. Food Additives and Contaminants, 2007, 24(4): 406-415.
[40] FRANCO T S, GARCIA S, HIROOKA E Y, et al. Lactic acid bacteria in the inhibition of Fusarium graminearum and deoxynivalenol detoxification[J]. Journal of Applied Microbiology, 2011, 111(3): 739-748.
[41] NIDERKON V, MORGAVI D P, ABOAB B, et al. Cell wall component and mycotoxin moieties involved in the binding of fumonisin B1 and B2 by lactic acid bacteria[J]. Journal of Applied Microbiology, 2009, 106(3): 977-985.
[42] OLIVEIRA P M, ZANNINI E, ARENDT E K. Cereal fungal infection, mycotoxins, and lactic acid bacteria mediated bioprotection: from crop farming to cereal products[J]. Food Microbiology, 2013, 37: 78-95.
[43] BOOYSEN C, DICKS L M T, MEIJERING I, et al. Isolation, identification and changes in the composition of lactic acid bacteria dur ing the malting of two different barley cultivars[J]. International Journal of Food Microbiology, 2002, 76(1): 63-73.
[44] JUSTE A, MALFLIET S, LENAERTS M, et al. Microflora during malting of barley: overview and impact on malt quality[J]. Brewing Science, 2011, 64: 22-31.
[45] LAITILA A, KOTAVIITA E, PELTOLA P, et al. Indigenous microbial commun ity of barley greatly influences grain germination and malt quality[J]. Journal of the Institute of Brewing, 2007, 113( 1): 9-20.
[46] PIOTROWSKA M, ŻAKOWSKA Z. The biodegradation of ochratoxin A in food products by lactic acid bacteria and baker’s yeast[J]. Progress in Biotechnology, 2000, 17: 307-310.
[47] MOKOENA M P, CHELULE P K, GQALENI N, Reduction of fumonisin B1 and zearalenone by lactic acid bacteria in fermented maize meal[J]. Journal of Food Protection, 2005, 68: 2095-2099.
[48] ROIG M, MECA G, FERRER E, et al. Reduction of the enniatins A, A1, B, B1 by an in vitro degradation employing different strains of probiotic bacteria: identification of degradat ion products by LC-MSLIT[J]. Toxicon, 2013, 70: 44-53.
[49] HE P, YOUNG L G, FORSBERG C. Microbial transformation of deoxynivalenol (vomitoxin)[J]. Applied Environment Microbiology, 1992, 58: 3857-3863.
[50] SHIMA J, TAKASE S, TAKAHASHI Y, et al. Novel detoxification of the trichothecene mycotoxin deoxynivalenol by a soil bacterium isolated by enrichment culture[J]. Applied Environment Microbiology, 1997, 63(10): 3825-3830.
[51] KUSUMANINGTYAS E, WIDIASTUTI R, MARYAM R. Reduction of aflatoxin B1in chicken feed by using Saccharomyces cerevisiae, Rhizopus oligosporus and their combination[J]. Mycopathologia, 2006, 162(4): 307-311.
[52] ENGELHARDT G. Degradation of ochratoxin A and B by the white rot fungus Pleurotus ostreatus[J]. Mycotoxin Research, 2002,18: 37-42.
[53] ZUO Ruiyu, CHANG Juan, YIN Qingqiang, et al. Inhibiting Aspergillus flavus growth and degrading aflatoxin B1by combined beneficial microbes[J]. African Journal of Biotechnology, 2012, 11(65): 12903-12909.
[54] KRISZT R, KRIFATON C, SZOBOSZLAY S, et al. A new zearalenone biodegradation strategy using non-pathogenic Rhodococcus pyridinivorans K408 Strain[J]. PLoS One, 2012, 7(9): e43608.
[55] MECA G, RITIENI A, ZHOU T, et al. Degradation of the minorFusarium mycotoxin beauvericin by intracellular enzymes of Saccharomyces cerevisiae[J]. Food Control, 2013, 33(2): 352-358.
[56] HARTINGER D, SCHWARTZ H, HAMETNER C, et al. Enzyme characteristics of aminotransferase FumI of Sphingopyxis sp. MTA144 for deamination of hydrolyzed fumonisin B1[J]. Applied Microbiology and Biotechnology, 2011, 91(3): 757-768.
[57] BLACKWELL B A, GILLIAM J T, SAVARD M E, et al. Oxidative deamination of hydrolyzed fumonisin B1 (AP 1) by cultures of Exophiala spinifera[J]. Natural Toxins, 1999, 7: 31-38.
[58] DUVICK J, MADDOX J, GILLIAM J, et al. Compositions and methods for fumonisin detoxification: U.S. Patent 6,534,291[P]. 2003-03-18.
[59] HEINL S, HARTINGER D, THAMHESL M, et al. Degradation of fumonisin B1 by the consecutive action of two bacterial enzymes[J]. Journal of Biotechnology, 2010, 145(2): 120-129.
[60] HEINL S, HARTINGER D, THAMHESL M, et al. An aminotransferase from bacterium ATCC 55552 deaminates hydrolyzed fumonisin B1[J]. Biodegradation, 2011, 22(1): 25-30.
[61] WANG J, OGATA M, HIRAI H, et al. Detoxification of aflatoxin B1by manganese peroxidase from the whiterot fungus Phanerochaete sordida YK-624[J]. FEMS Microbiology Letters, 2011, 314(2): 164-169.
[62] DOSTER R C, SINNHUBER R O. Comparative rates of hydrolysis of ochratoxins A and B in vitro[J]. Food and Cosmetics Toxicology, 1972, 10(3): 389-394.
[63] RINGOT D, CHANGO A, SCHNEIDER Y J, et al. Toxicokinetics and toxicodynamics of ochratoxin A, an update[J]. Chemico-Biological Interactions, 2006, 159(1): 18-46.
[64] SATO I, ITO M, ISHIZAKA M, et al. Thirteen novel deoxynivalenoldegrading bacteri a are classified within two genera with distinct degradation mechanisms[J]. FEMS Microbiology Letters, 2012, 327(2): 110-117.
[65] ERIKSEN G S, PETTERSSON H, LUNDH T. Comparative cytotoxicity of deoxynivalenol, nivalenol, their acetylated derivatives and de-epoxy metabolites[J]. Food Chemistry Toxicology, 2004, 42: 619-624.
[66] GUAN S, HE J, YOUNG J C, et al. Transformation of trichothecene mycotoxins by microorganisms from fish digesta[J]. Aquaculture, 2009, 290(3): 290-295.
[67] GRATZ S W, DUNCAN G, RICHARDSON A J. The human fecal microbiota metabolizes deoxynivalenol and deoxynivalenol-3-glucoside and may be responsible for urinary deepoxydeoxynivalenol[J]. Applied and Environmental Microbiology, 2013, 79(6): 1821-1825.
[68] YU Hai, ZHOU Ting, GONG Jianhua, et al. Isolation of deoxynivalenol-transforming bacteria from the chicken intestines using the approach of PCR-DGGE guided microbial selection[J]. BMC Microbiology, 2010, 10(1): doi: 10.1186/1471-2180-10-182.
[69] KING R R, MCQUEEN R E, LEVESQUE D, et al. Transformation of deoxynivalenol (vomitoxin) by rumen microorganisms[J]. Journal of Agricultural and Food Chemistry, 1984, 32(5): 1181-1183.
[70] WESTLAKE K, MACKIE R I, DUTTON M F. in vitro metabolism of mycotoxins by bacterial, protozoal and ovine ruminal fluid preparations[J]. Animal Feed Science Technology, 1989, 25: 169-178.
[71] WORRELL N R, MALLETT A K, COOK W M, et al. The role of gut micro-organisms in the metabolism of deoxynivalenol administered to rats[J]. Xenobiotica, 1989, 19: 25-32.
[72] ISLAM R, ZHOU T, YOUNG J C, et al. Aerobic and anaerobic deepoxydation of mycotoxin deoxynivalenol by bacteria originating from agricultural soil[J]. World Journal of Microbiology and Biotechnology, 2012, 28(1): 7-13.
[73] PINTON P, TSYBULSKYY D, LUCIOLI J, et al. Toxicity of deoxynivalenol and its acetylated derivatives on the intestine: differential effects on morphology, barrier function, tight junction proteins, and mitogen-activated protein kinases[J]. Toxicological Sciences, 2012, 130(1): 180-190.
[74] PINTON P, TSYBULSKYYD, LAFFITTE J, et al. The intestine, a target for deoxynivalenol and its derivatives[J]. Journées de la Recherche Porcine en France, 2012, 44: 85-90.
[75] HARITASH A K, KAUSHIK C P. Biodegradation aspects of polycyclic aromatic hydrocarbons (PAHs): a review[J]. Journal of Hazardous Materials, 2009, 169(1): 1-15.
[76] TANAKA K, SAGO Y, ZHENG Y, et al. M ycotoxins in rice[J]. International Journal of Food Microbiology, 2007, 119(1): 59-66.
[77] VENDL O, BERTHILLER F, CREWS C, et al. Simultaneous determination of deoxynivalenol, zearalenone, and their major masked metabolites in cereal-based food by LC-MS-MS[J]. Analytical and Bioanalytical Chemistry, 2009, 395(5): 1347-1354.
[78] NAKAGAWA H, SAKAMOTO S, SAGO Y, et al. D etection of masked mycotoxins derived from type A trichothecenes in corn by high-resolution LC-Orbitrap mass spectrometer[J]. Food Additives & Contaminants: Part A, 2013, 30(8): 1407-1414.
[79] VIDAL J C, BONEL L, EZQUERRA A, et al. Electrochemical affinity biosensors for detection of mycotoxins: a review[J]. Biosensors and Bioelectronics, 2013, 49: 146-158.
[80] BERTHILLER F, KRSKA R, DOMIG K J, et al. Hydrolytic fate of deoxynivalenol-3-glucoside during digestion[J]. Toxicology Letters, 2011, 206(3): 264-267.
[81] HEATON A C P, RUGH C L, WANG N, et al. Phytoremediation of mercury-and methylmercury-polluted soils using genetically engineered plants[J]. Journal of Soil Contamination, 1998, 7(4): 497-509.
[82] ALAKONYA A E, MONDA E O. A n ew approach in aflatoxin management in Africa: targeting aflatoxin/sterigmatocystin biosynthesis in Aspergillus species by RNA silencing technique[J]. Agricultural and Biological Sciences, 2013, doi: 10.5772/51440.
Advances in the Formation and Degradation of Masked Mycotoxins
WANG Yan-su1, ZHOU Jia-yu1, XIE Xing-guang1, DING Cheng-long2, DAI Chuan-chao1,*
(1. Jiangsu Engineering and Technology Research Center for Industrialization of Microbial Resources, College of Life Science, Nanjing Normal University, Nanjing 210023, China; 2. Institute of Animal Science, Jiangsu Academy of Agricultural Sciences, Nanjing 210014, China)
Mycotoxin-producing fungi are ubiquitous in moldy ce reals and animal feeds and pose a serious threat to human and animal health. Mycotoxins can be partly detoxified through incomplete degradation by plants or microbes into masked mycotoxins which, however, still exist in plants or the environment. This paper is focused on the following aspects of mycotoxins: distribution and hazards, incomplete degradation during the growth and postharvest storage of plants, and the formation of masked mycotoxins by microbial conversion or degradation as well as their further degradation. Hopefully this paper can provide guidelines for future studies on complete degradation of mycotoxins.
mycotoxins degradation; phytotransformation; microbial degradation; masked mycotoxins
Q939.97
A
1002-6630(2014)21-0326-08
10.7506/spkx1002-6630-201421062
2013-12-08
江苏省农业自主创新项目(CX(12)1002)
王彦苏(1989—),女,硕士研究生,主要从事微生物学研究。E-mail:wangyansu1990@163.com
*通信作者:戴传超(1970—),男,教授,博士,主要从事微生物生态学研究。E-mail:daichuanchao@njnu.edu.cn
