Polymorphisms of growth hormone (GH) and insulin-like growth factor I (IGF-I) genes in prolific Lezhi Black Goat: possible association with litter size
2014-02-17ZIXiangDongMUXiaoKunLUJianyuanMALiWANGYong
ZI Xiang-Dong, MU Xiao-Kun, LU Jian-yuan, MA Li, WANG Yong
(1.School of Life Science and Technology, Southwest University for Nationalities, Chengdu 610041, P.R.C.;2. Biology Department, Zhaoqing College, Zhaoqing 526061, P.R.C. )
Polymorphisms of growth hormone (GH) and insulin-like growth factor I (IGF-I) genes in prolific Lezhi Black Goat: possible association with litter size
ZI Xiang-Dong1, MU Xiao-Kun2, LU Jian-yuan1, MA Li1, WANG Yong1
(1.School of Life Science and Technology, Southwest University for Nationalities, Chengdu 610041, P.R.C.;
2. Biology Department, Zhaoqing College, Zhaoqing 526061, P.R.C. )
The growth hormone (GH) and insulin-like growth factor I (IGF-I) play a key role both in the follicular development and in the follicular atresia. In this study, the polymorphisms of GH andIGF-Igenes in 157 Lezhi black does were detected by polymerase chain reaction–single strand conformation polymorphism (SSCP). For GH gene, two conformational patterns were found in each of 5' flanking region and exon 2, four in exon 3, three in each of exon 4 and 5. ForIGF-Igene, two conformational patterns were found in exon 2. In addition, 14 single nucleotide polymorphisms (SNPs)—T48C, A55G) (P1 locus), T781C (P2 locus), G1122A, A1161G, A1171G, C1179T and C1180G (P3 locus), T1481C, A1494G, G1549T, and C1590T (P4 locus), G1942A and G1957A (P5 locus) were identified byGHgene sequencing and PCR-SSCP analysis. One SNP G3270C (P6 locus) was identified inIGF-I. Association analysis showed that only in exon 3 ofGHgene, Lezhi black does with genotype BB had 0.32 (p<0.05) more kids than those with genotype AA. This study preliminarily showed polymorphisms of GH gene and possible association with litter size in Lezhi black goat.
GH gene;IGF-Igene; PCR-SSCP; Litter size; Lezhi black goat
Reproductive rate, especially fecundity, is one of the most economically important traits in animal production and is regulated by genetic and environmental factors. Ovulation rate can be a major determinant of reproductive efficiency. The processes of recruitment and selection lead to the development of numbers of ovulatory follicles that vary among species and breeds[1]. Since development of primordial to early antral follicles is not dependent on gonadotrophins[2], the role of locally produced growth factors in ovarian folliculogenesis has been extensively studied in mammals, especially rodents. There is a growing body of evidence that growth hormone (GH)[3-5]and members of the insulin-like growth factor (IGF) family system play a key role both in the development of preantral to preovulatory follicles and in the process of follicular atresia[5-8].
Recently, correlations between production traits and polymorphisms at the GH[9-11]and IGF-I[12-13]genes have been observed. In Gulin Ma goats, two polymorphisms at 5′ flanking regulatory region of GH gene were found to affect litter size traits, but these polymorphisms did not affect litter size in Chuandong White goats and Guizhou White goats[12]. Polymorphisms of 5′ regulatory region of exons of IGF-I gene significantly affected lambing rate in sheep[13]. Therefore, GH and IGF genes are possible candidate genes for reproductive traits in goat. Lezhi black goat is a local Chinese breed famous for its good meat performance and high fecundity[14]. The main purposes of this work were firstly to evaluate the genetic variability of GH and IGF genes and their association with litter size in Lezhi black goat by PCR-SSCP method.
1 Materials and Methods
1.1 Animals, Sample Collection and DNA Isolation
Blood samples of 5–6 ml were collected from 157 does of Lezhi black goats along with data on litter size in the Lezhi Black Goat Breeding Farm in Sichuan Province (30°30′N, 105°02′E and 596.3 m altitude), P.R. China. Venous blood was collected using EDTA as an anticoagulant. DNA extraction was performed within 24 h of blood collection, according to Sambrook and Russell[15]. After checking the quality and quantity, DNA was diluted to a final concentration of 50 ng/μl in water and stored at 4°C for immediate use while for long term kept at –20°C.
1.2 DNA Amplification by PCR
Polymerase chain reaction (PCR) was carried out on about 50–100 ng genomic DNA in a 25 μl reaction volume. Six pairs of primers were designed to amplify the 5' flanking region and exons 1–5; seven pairs of primers were designed to amplify the 5' flanking region, exons 1–6 of IGF-I; twelve pairs of primers were designed to amplify exons 2–10 of IGF-II. The primers were synthesized by Shanghai Invitrogen Biotechnology, Ltd., Shanghai, P.R. China. In the PCR products of 25 pairs of primers, only six listed in Table 1 had polymorphisms, and other primers with no polymorphism detected in their amplification regions are not listed. The 25 μl PCR contained 50 ng of genomic DNA, 12.5 μl of 2×reaction mix (including 500 μM dNTP each, 20 mM Tris-HCl, pH 9, 100 mM KCl, 3 mM MgCl2), 0.5 μM of each primer and 0.5U Taq DNA polymerase (Tiangen Biotech, Beijing, P.R. China). The cycling protocol was 4 min at 95°C followed by 35 cycles of denaturing at 94°C for 30 s, annealing at X°C (Table 1) for 30 s, extending at 72°C for 30 s, with a final extension at 72°C for 7 min on Eppendorf Mastercycler gradient (Eppendorf AG 22331, Hamburg, Germany). The PCR products were separated by electrophoresis on 1.0 % agarose gels in parallel with DNA marker I (Tiangen, Beijing, P.R. China).
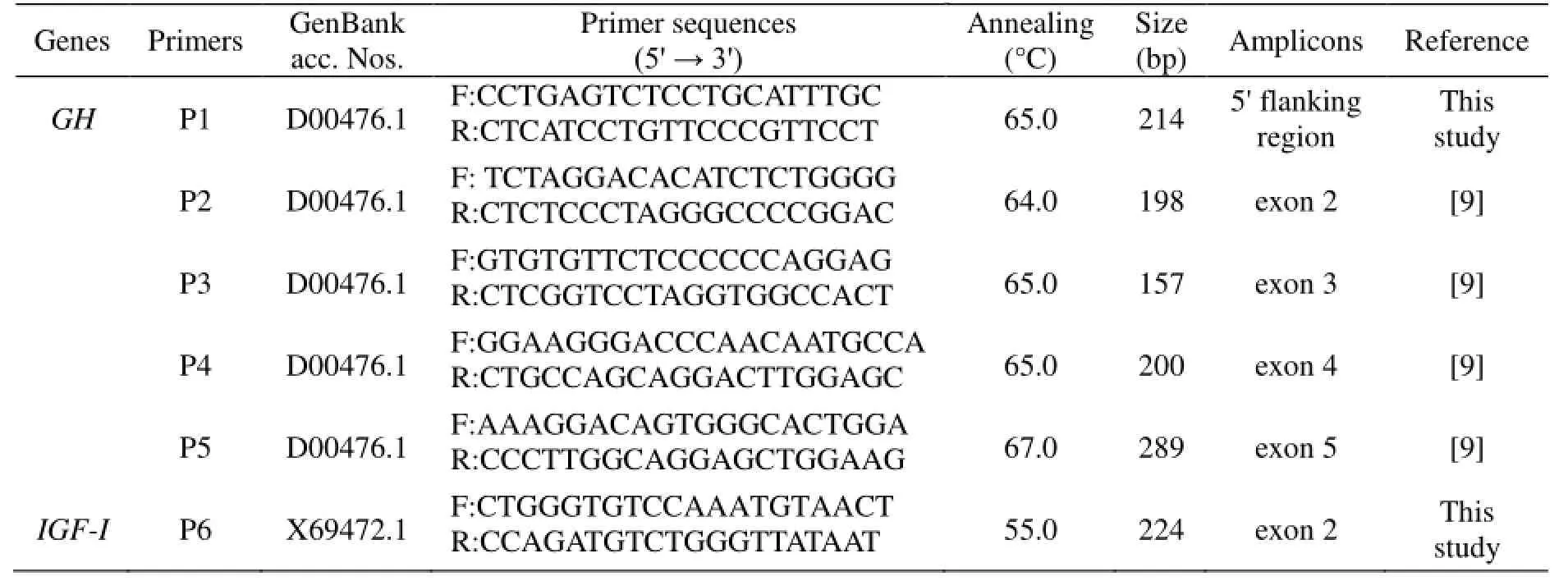
Table 1 Primer sequences and information of the goat GH and IGF-I genes
1.3 Single Strand Conformation Polymorphism (SSCP) and DNA Sequencing
For SSCP analysis, 2 μl of each amplification products were mixed with 1 μl 6×loading buffer containing 30 mM EDTA, 36% glycerol, 0.035% xylene cyanol FF, and 0.05% bromophenol blue (TaKaRa, Dalian, P.R. China). The mixtures were denatured at 98°C for 10 min, and immediately placed on ice for 6 min. The products were then subjected to 8–10% PAGE (polyacrylamide gel electrophoresis) at a constant temperature (4°C) under voltage of 250 V for 5 min, and 180 V for 2 h, and then gels were stained with 0.1% silver nitrate[16]. After the polymorphisms were detected, each of the DNA bands on the SSCP gel was extracted and the PCR products of the different electrophoresis patterns were sent for sequencing in both directions (repeated three times) in an ABI 377 DNA analyzer (Applied Biosystems) and the sequences were determined with DNAstar software (version 7.1) and blast in NCBI (National Center for Biotechnology Information).
1.4 Statistical Analysis
The genotypic frequencies, heterozygosity (He) and polymorphism content (PIC) were calculated using Cluster-analysis software (version 1.2), and Hardy-Weinberg equilibrium was analyzed using χ2–test, which was performed by SPSS software (version 16.0). The software SPSS (version 16.0) was also used to determine the relationship between genotypes and litter size. Data on litter size in the first and the second parities from 157 does born in two successive years were used in the present study. Years were regarded as two kidding years. Least square variance analysis was conducted for six primer pairs, respectively. Therefore, the following statistical model was fitted to compare difference of litter size between genotypes. Yijkl= μ + Li+ Pj+ Gk+ eijkl, where Yijklis phenotypic value of litter size; μ is population mean; Liis the fixed effect of the ith (i = 1,2) kidding year; Pjis the fixed effect of the jth (j =
1,2) parity; Gkis the fixed effect of the kth (k=1,2,3) genotype for gene; and eijklis random error effect of each observation.
2 Results
2.1 Polymorphisms of GH and IGF-I Genes
Polymerase chain reaction (PCR) was carried out on about 50–100 ng genomic DNA in a 25 μl reaction volume, using the primers described in Table 1. In PCR products of five pairs of primers ofGHgene, according to Gupta et al.[10]about the naming of SSCP patterns, in P1 locus, different SSCP patterns were named AB and BB genotypes; in P2 locus, different SSCP patterns were named AB and BB genotypes; in P3, different SSCP patterns were named AA, AB, BB, and BC genotypes; in P4 locus, different SSCP patterns were named AA, BB, and CC genotypes; in P5 locus, different SSCP patterns were named AC, BC, and CC genotypes (Fig. 1). In P6 locus ofIGF-Igene, different SSCP patterns were named AB and BB genotypes (Fig. 1). Allele and Genotype frequencies, He, and PIC are shown in Table 2. However, some genotypes were not observed; AA was not observed in P1 and P2 loci ofGHgene; AB, BC and AC were not observed in P4 locus of GH gene; AA and BB were not observed in P5 locus ofGHgene; AA was not observed in P6 locus ofIGF-Igene. The single nucleotide polymorphisms (SNPs) loci were in Hardy-Weinberg disequilibrium in this goat population (p<0.05).
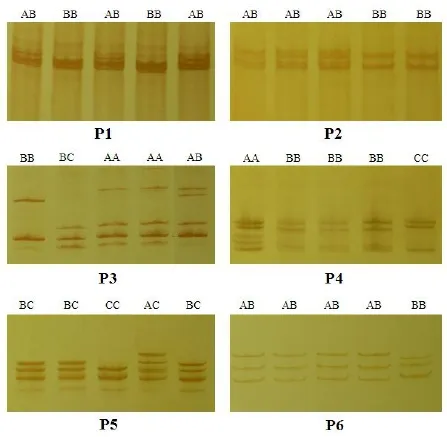
Fig. 1 SSCP analysis of PCR products using GH 5' flanking region primer (P1), GH exon 2 primer (P2), GH exon 3 primer (P3), GH exon 4 primer (P4), GH exon 5 primer (P5), and IGF-I exon 2 primer (P6) of Lezhi black goat.
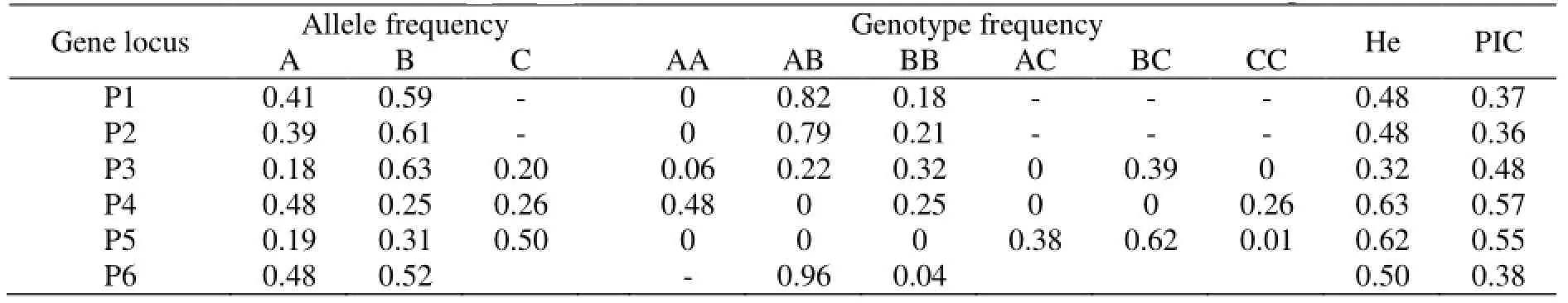
Table 2 Allele and genotype frequencies of GH and IGF-I genes in 157 Lezhi black goats
2.2 Sequencing of Different Genotypes
Different SSCP patterns of GH and IGF-I genes shown in Fig. 1 were sequenced, and sequence comparisons were
carried out among the genotypes at the same locus. In P1 locus ofGHgene, two base substitutions (T48C and A55G) were detected; in P2 locus, one base substitution (T781C) was detected; in P3 locus, five base substitutions (G1122A, A1161G, A1171G, C1179T and C1180G) were detected; in P4 locus, four base substitutions (T1481C, A1494G, G1549T and C1590T) were detected; and in P5 locus, two base substitutions (G1942A and G1957A) were detected. In P6 locus of IGF-I gene, one base substitution (G3270C) was detected (Table 3).
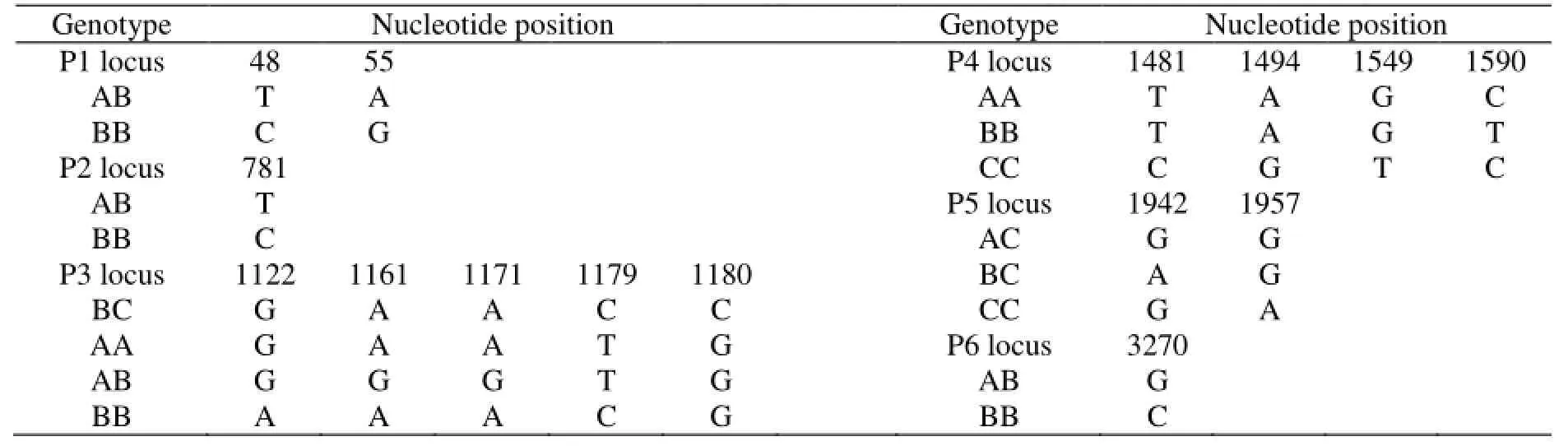
Table 3 The nucleotide mutations between genotypes of GH and IGF-I genes in 157 Lezhi black goats
2.3 Association of GH and IGF-I Genotypes with Litter Size
Table 4 presents the least square means obtained for litter size. At the P3 locus ofGHgene, the Lezhi black does with genotype BB had 0.32 (p<0.05) more kids than those with genotype AA. However, at P1, P2, P4, P5 loci of GH gene and P6 of IGF-I gene, the records of litter size did not show any significant correlation with genotypes.

Table 4 Least squares mean and standard error for litter size of different GH and IGF-I genotypes in Lezhi black goat
The data are expressed as least square means ± standard errors. Values with different superscripts in the same primer differ significantly at p<0.05.
3 Discussion
The polymorphisms of 5' flanking region, exons 2–5 ofGHgene, and exon 2 ofIGF-Igene were detected in 157 Lezhi black goats, nevertheless, some genotypes were not observed. Consistent with previous studies in other goat breeds[9-10,17-18], theGHandIGF-Igenes had polymorphisms in the Lezhi black goat population and certain genotypes are absent. The results can be explained by the following two reasons: (1) there is a lower frequency for missing genotypes, and the samples were small[9-12,17,18]and (2) the missing genotypes of theGHandIGF-Igene loci have negative effects on individual performance, so individuals with the missing genotypes have been eliminated in breeding[19-20]. Due to the important roles of GH and IGF-I in the regulation of a multitude of physiological functions, including growth[22-23], reproduction[5-8,21], lactation[9,24]and metabolism[25], it is an interesting phenomenon worthy of further investigation on mutations that influence production traits.
The associations between polymorphisms of GH andIGF-Igenes and production traits have been analyzed in some breeds of goat. Their SNPs had significant effects on growth traits and milk traits suggesting that both of these genes are strong candidate genes for these traits[9,17,18,26-29]. Recently, the possible associations of polymorphisms ofGHandIGF-Igenes with reproductive traits have also been studied in some livestock species. Lan et al.[27]identified T1651G mutation in exon 5 ofGHgene, which was associated with milk performance and litter size in seven goat populations reared in China. Korwin-Kossakowska et al.[30]studied the CA repeat ofIGF-Igene and found that the sows with 130/132 bp genotype had larger litter size (total number of piglets born and number of piglets born alive) and higher litter weight (litter weight at birth). In Gulin Ma goats, two deletions (CA) in theIGF-Imicrosatellite and two
mutations (A1637G, T1640C) in 5′ flanking regulatory region of IGF-I were found to affect litter size traits, but these polymorphisms did not affect litter size in Chuandong White goats and Guizhou White goats[12]. Two mutations (C1511G and A1513G) in 5′ regulatory region of IGF-I gene were identified in Small Tail Han sheep, and the ewes with genotype BB or AB had 0.96 or 0.38 lambs more than those with genotype AA[13]. In the present study, the polymorphisms of 5' flanking region, exons 2–5 of GH gene, and exon 2 of IGF-I gene were firstly detected in Lezhi black goats. Interesting results appeared from analysis which revealed the association between exon 3 of GH different genotypes and litter size in this research; the Lezhi black does with genotype BB had 0.32 (p<0.05) more kids than those with genotype AA. The other polymorphisms did not significantly affect litter size, but some of these polymorphisms may be associated with growth traits and milk traits. However, we did not carry out such association analyses due to the lack of performance records in this study. These associations are worthy of being further verified.
In conclusion, the polymorphisms we have identified in exon 3 of GH gene (P3 locus) could be potential genetic markers for litter size in goats. However, more tests are needed to confirm the associated effects of the genetic markers on other breeds, and the exact mechanism remains to be elucidated.
[1] HANRAHAN JP, QUIRKE JF. Contribution of variation in ovulation rate and embryo survival to within breed variation in litter size. In: Land RB, Robinson DW (eds.), Genetics of Reproduction in Sheep. London: Butterworths, 1985:193-201.
[2] FORTUNE JE. The early stages of follicular development: activation of primordial follicles and growth of preantral follicles[J]. Anim Reprod Sci, 2003,78:135-163.
[3] ZHAO J, TAVERNE MA, VAN DEWEIJDEN GC, et al. Immunohistochemical localisation of growth hormone (GH), GH receptor (GHR), insulin-like growth factor I (IGF-I) and type I IGF-I receptor, and gene expression of GH and GHR in rat pre-antral follicles[J]. Zygote, 2002,10:85-94.
[4] SHIMIZU T, MURAYAMA C, SUDO N, et al. Involvement of insulin and growth hormone (GH) during follicular development in the bovine ovary[J]. Anim Reprod Sci, 2008, 106: 143-152.
[5] SILVA JRV, FIGUEIREDO JR, VAN DEN HURK R. Involvement of growth hormone (GH) and insulin-like growth factor (IGF) system in ovarian folliculogenesis[J]. Theriogenology, 2009, 71: 1193-1208.
[6] ZHAO J, TAVERNE MA, VAN DEWEIJDEN GC, et al. Insulin-like growth factor-I (IGF-I) stimulates the development of cultured rat pre-antral follicles[J]. Mol Reprod Dev, 2001,58:287-296.
[7] HASTIE PM, HARESIGN W. Expression of mRNAs encoding insulin-like growth factor (IGF) ligands, IGF receptors and IGF binding proteins during follicular growth and atresia in the ovine ovary throughout the oestrous cycle[J]. Anim Reprod Sci, 2006, 92: 284-299.
[8] LI L, FU YC, XU JJ, et al. Caloric restriction promotes the reproductive capacity of female rats via modulating the level of insulinlike growth factor-1 (IGF-1) [J]. Gen Comp Endocrinol, 2011,174:232-237.
[9] MALVEIRO E, PEREIRA M, MARQUES PX, et al. Polymorphisms at the five exons of the growth hormone gene in the algarvia goat: possible association with milk traits[J]. Small Rumin Res, 2001, 41: 163-170.
[10] GUPTA N, PANDEY A, MALIK G, et al. Single nucleotide polymorphism (SNP) in growth hormone gene of Jakhrana, a prominent milk goat breed in India[J]. Small Rumin Res, 2009, 81: 35-41.
[11] TAHMOORESPUR M, TAHERI A, GHOLAMI H, et al. PCR-SSCP variation of GH and STAT5A genes and their association with estimated breeding values of growth traits in Baluchi sheep[J]. Anim Biotechnol, 2011,22:37-43.
[12] WANG PQ, TAN Y, ZHANG BY, et al. DNA Polymorphisms of 5´ flanking region of insulin-like growth factor 1 gene and their association with reproduction traits in goats[J]. Agri Sci China, 2011,10:1609-1617.
[13] HE JN, ZHANG BY, CHU MX, et al. Polymorphism of insulin-like growth factor 1 gene and its association with litter size in Small Tail Han sheep[J]. Mol Biol Rep, 2012,39: 9801-9807.
[14] ZI XD, XU HW, WANG Y. Variation in sequences and mRNA expression levels of inhibin subunits α (INHA) and βA (INHBA) genes between prolific and non-prolific goat breeds[J]. Mol Reprod Dev, 2012, 79: 238.
[15] SAMBROOK J, RUSSELL DW. Molecular cloning: a laboratory manual[M]. 3rd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 2001.
[16] JI YT, CAO BY. An optimal method of DNA silver staining in polyacrylamide gels[J]. Electrophoresis, 2007, 28: 1173-1175.
[17] HUA GH, CHEN SL, YU JN, et al. Polymorphism of the growth hormone gene and its association with growth traits in Boer goat bucks[J]. Meat Sci, 2009, 81: 391-395.
[18] AN XP, HOU JX, WANG LX, et al. Novel polymorphisms of the growth hormone gene and their effect on growth traits in Chinese goats[J]. Meat Sci, 2010, 86: 758-763.
[19] BEUZEN ND, STEAR MJ, CHANG KC. Molecular markers and their use in animal breeding[J]. Vet J, 2000, 160: 42-52.
[20] VAN MARLE-KOSTER E, NEL LH. Genetic markers and their application in livestock breeding in South Africa: A review[J]. South Afr J Anim Sci, 2003, 33: 1-10.
[21] ECHTERNKAMP SE, ROBERTS AJ, LUNSTRA DD, et al. Ovarian follicular development in cattle selected for twin ovulations and births[J]. J Anim Sci, 2004, 82: 459-471.
[22] BREIER BH. Regulation of protein and energy metabolism by the somatotropic axis[J]. Domest Anim Endocrinol, 1999,17: 209-218.
[23] DUAN CM, REN HX, GAO S. Insulin-like growth factors (IGFs), IGF receptors, and IGF-binding proteins: Roles in skeletal muscle growth and differentiation[J]. Gen Comp Endocrinol, 2010,167: 344-351.
[24] BALDI A. Manipulation of milk production and quality by use of somatotropin in dairy ruminants other than cow[J]. Domest Anim
Endocrinol, 1999, 17: 131-137.
[25] BAUMAN DE. Bovine somatotropin and lactation: from basic science to commercial application, Domest[J]. Anim Endocrinol, 1999, 17: 101-116.
[26] MIN LJ, LI MY, SUN GQ, et al. Relationship between polymorphism of growth hormone gene and production traits in goats[J]. Acta Genetica Sinica, 2005, 32: 650-654.
[27] LAN XY, CHEN H, PAN CY, et al. Polymorphism in growth hormone gene and its association with production traits in goats[J]. J Appl Anim Res, 2007, 32: 55-60.
[28] MARQUES PX, PEREIRA M, MARQUES MR, et al. Association of milk traits with SSCP polymorphisms at the growth hormone gene in the Serrana goat[J]. Small ruminant Res, 2003, 50: 177-185.
[29] ZHANG CX, ZHANG W, LUO HL, et al. A new single nucleotide polymorphism in the IGF-I gene and its association with growth traits in the Nanjiang huang goat[J]. Asian-Aust J Anim Sci, 2008, 21: 1073-1079.
[30] KORWIN-KOSSAKOWSKA A, SENDER G, KURYL J. Associations between the microsatellite DNA sequence in the IGF1 gene, polymorphism in the ESR gene and selected reproduction traits in F1 (Zlotnicka Spotted × Polish Large White) sows[J]. Anim Sci Pap Rep, 2004, 22: 215-226.
S814; S827
: A
: 1003-4271(2014)03-0344-06
2014-03-17
字向东(1963-), 男, 白族, 云南云龙人, 教授, E-mail: zixd@sina.com.
四川省应用基础项目(2013JY0043); 中央高校基本科研业务费专项资金项目(12NZYTH07)
10.3969/j.issn.1003-4271.2014.03.04
