Im proved quantification accuracy for dup lex real-time PCR detection of genetically modified soybean and maize in heat processed foods
2013-06-01CHENGFangSHENPingZHANGDabingLIJianyueYANGLitao
CHENG Fang,SHEN Ping,ZHANG Dabing,LIJianyue,YANG Litao*
(1.College of Life and Environment Sciences,Shanghai Normal University,Shanghai 200234,China;2.Development Center for Science and Technology,Ministry of Agriculture,Beijing 100026,China;3.School of Life Science and Biotechnology,Shanghai Jiao Tong University,Shanghai 200240,China)
Im proved quantification accuracy for dup lex real-time PCR detection of genetically modified soybean and maize in heat processed foods
CHENG Fang1,SHEN Ping2,ZHANG Dabing3,LIJianyue1,YANG Litao3*
(1.College of Life and Environment Sciences,Shanghai Normal University,Shanghai 200234,China;2.Development Center for Science and Technology,Ministry of Agriculture,Beijing 100026,China;3.School of Life Science and Biotechnology,Shanghai Jiao Tong University,Shanghai 200240,China)
Real-time PCR technique has been widely used in quantitative GMO detection in recent years.The accuracy of GMOs quantification based on the real-time PCR methods is still a difficult problem,especially for the quantification of high processed samples.To develop the suitable and accurate real-time PCR system for high processed GM samples,wemade ameliorations to several realtime PCR parameters,including re-designed shorter target DNA fragment,similar lengths of amplified endogenous and exogenous gene targets,similar GC contents and melting temperatures of PCR primers and TaqMan probes.Also,one Heat-Treatment Processing Model(HTPM)was established using soybean flour samples containing GM soybean GTS 40-3-2 to validate the effectiveness of the improved real-time PCR system.Tested results showed that the quantitative bias of GM content in heat processed samples were lowered using the new PCR system.The improved duplex real-time PCR was further validated using processed foods derived from GM soybean,andmore accurate GM content values in these foodswas also achieved.These results demonstrated that the improved duplex real-time PCR would be quite suitable in quantitative detection of high processed food products.
geneticallymodified organisms;soybean;maize;processed foods;real-time PCR
1 Introduction
Global areas of cultivated GM crops have increased rapidly from 1.7 million hectares in 1996 to 160 million hectares in 2012[1].To address public concerns over GMO safety issues and protect consumer rights,many countries have implemented strict labeling policies for GMO ingredients in foods,for example,the threshold for GMO labeling is 0.9%in European Union(EU),3%in Korea,and 5%in Japan[2].
To accurately label GMO contents in accordance with specific thresholds,development of quantitative detection methods for GMOs are obviously in great need.Among different detection techniques,quantitative realtime PCR has been demonstrated to be both robust and reliable in quantitative GMO detection[3].One type of real-time PCR techniques,the TaqMan technology,has been widely used in clinicalmedicine and GMO detection field for its high accuracy and easy operation.Many real-time PCR assays have been well developed to quantify GM maize,rice,papaya,canola,and soybean etc[4-8].However,most of these assays were developed using raw materials such as seeds,whereas GM soybean,maize and others are usually processed to be ingredients of various kinds of products.The processing proceduresmight result in target DNA degradation that could affect quantification accuracy,especially in deeply processed food productswhich usually go through high temperature and high pressure conditions[9].There are only few reports about how to decrease quantification bias in processed food products[10].Many factorsmay contribute to GMO quantification bias in processed foods,such as target DNA degradation,inconsistent PCR efficiencies between endogenous and exogenous genes,length of amplified PCR fragments.In this study,we improved the design of PCR primers,TaqMan probes,and lengths of both endogenous and exogenous gene amplicons in duplex real-time PCR assays.The new duplex real-time PCR assay was employed to quantify GM soybean in heat-treating-process-modeling(HTPM)samples.The new PCR system could be applied effectively in more accurate quantitative detection of GM contents in heat processed food products.
2 Materials and methods
2.1 GM Seeds,HTPM sam ples and processed foods
Seeds of GM Roundup Ready Soybean GTS 40-3-2(RRS)and Event176 maize were developed by Monsanto Co.(St.Louis,MO,USA)and Novartis Seeds Inc.,respectively.Non-GM soybean and maize seedswere purchased from local supermarket in Shanghai,China.Mixed seed samples containing RRS seeds or Event176 seeds at concentrations of 1%and 5%of total seed weightwere prepared in our laboratory.
Two soybean floursamples containing RRA soybean at 1%and 5%of total weight were prepared in the lab and subjected to heat-treating-process-modeling(HTPM),in which the sampleswere autoclaved at121℃for 0, 10, 30,and 60 minutes,respectively.Processed foods including tofu,soy milk,corn chip and popcorn were prepared.
2.2 DNA extraction
Genomic DNA from seeds,HTPM samples and processed foods were extracted and purified using Plant DNA Extraction Kit developed by ShanghaiRuifeng Agro-tech Co.Ltd(Shanghai,China).500mg startingmaterial was used for each DNA extraction and purification,and 1μL purified DNA was used in real-time PCR.
2.3 Oligonucleotide primers and TaqM an probes
Sequences of oligonucleotide PCR primers and TaqMan probes were designed using Primer Express software Version 3.0(Applied Biosystems,Foster City,CA)and listed in Table 1.Endogenous gene probewas labeled at the 5′end with fluorescent reporter 5-hexachloro-fluorescein(HEX),and exogenous gene probe was labeled at the 5′end with fluorescent reporter 6-carboxy-fluorescein(FAM).Fluorescent quencher 6-carboxytetramethylrhodamine(TAMRA)was labeled at the 3′end of the probes.For detection of 5-enolpyruvylshikimate-3-phosphate synthase(CP4-EPSPS)gene and nopaline synthase terminator(NOS)in RRS GM soybean,two groups of primers and probes were designed based on published gene sequence[11].The CryIA(b)transgene of Event176 maize was amplified using primers E1/E2 and probe E-p[12].All primers and TaqMan probeswere synthesized and purified by Shanghai Invitrogen Co.Ltd.(Shanghai,China).
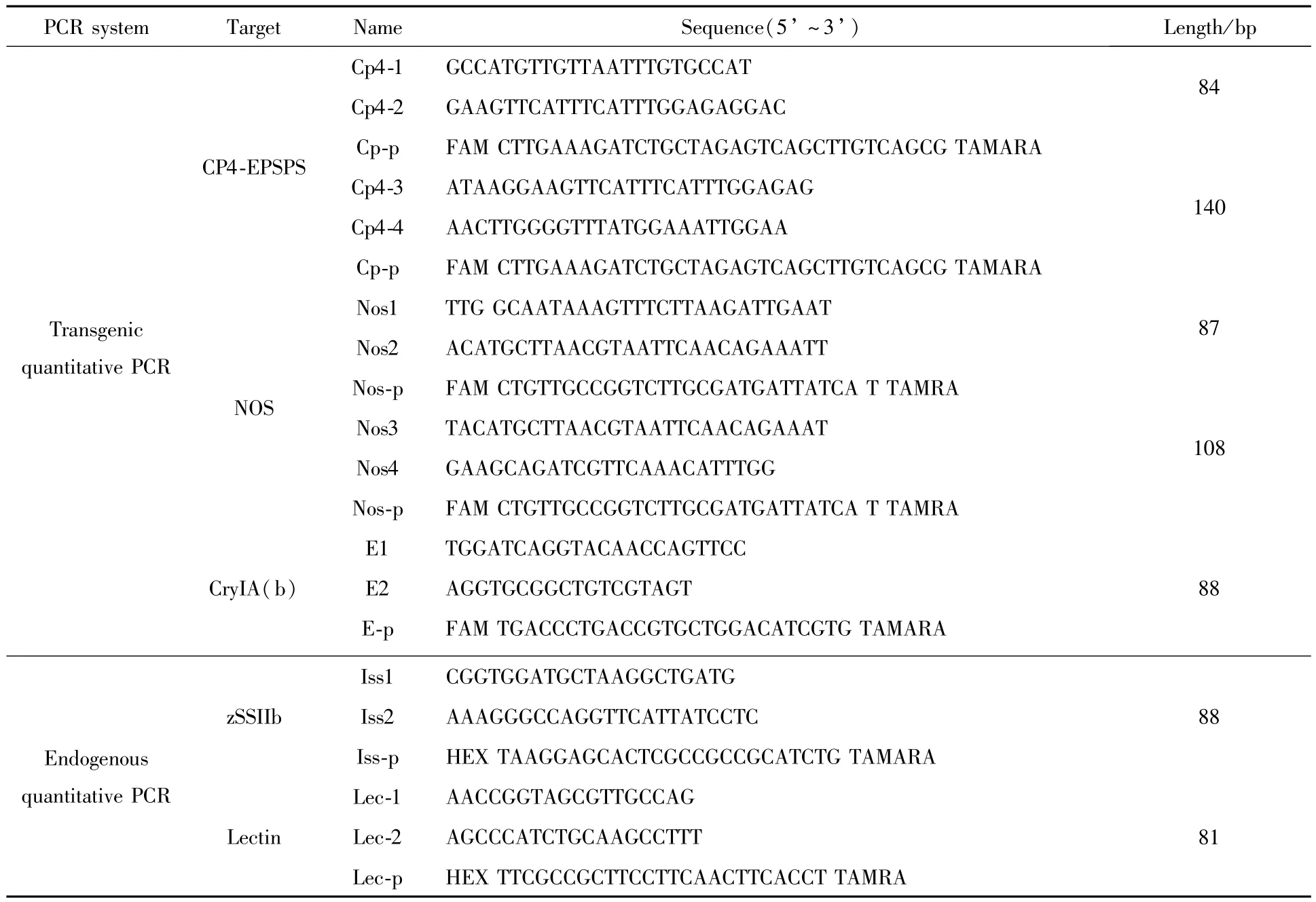
Table1 List of primers and TaqMan probes for real-time PCR
2.4 Dup lex real-time PCR
Real-time PCR reactions were carried out using a Rotor-Gene 3000 fluorescent thermal cycler(Corbett Research,Australia)in a reaction volume of 25μL.Each PCR reaction contained the following components:12.5μL PCR Buffer(TaqManⓇUniversal Master Mix,Applied Biosystems,USA);25 nmol/L endogenous primers;100 nmol/L exogenous primers;200 nmol/L endogenous and 400 nmol/L exogenous probes;and 5μL DNA template.Real-time PCR program was as following:50℃for 2 minu;denaturation at 95℃for 10 min;50 cycles of 15 at94℃,60 at60℃.Fluorescent signalwasmonitored in every PCR cycle at the annealing step.
2.5 Construction of real-tim e PCR standard curves
To construct PCR standard curves,series of10-fold diluted genomic DNAs(range:100~0.01ng/μL)of the RRS soybean and Event176 maize were used.PCR efficiency was calculated from each linear regression using the equation:Efficiency=10-slope-1.PCR standard curves weremade and Ct values were determined to quantify GM contents.
2.6 Determ ination of the lim it of detection
Limit of detection(LOD)was determined using serial dilutions of DNAs extracted from 100%RRS soybean or Event176 maize.DNA concentrations ranging from 10000 to 1 copy of haploid genome were prepared based on the average size of soybean ormaize genomes and used in LOD determination[13].Each DNA concentration was assayed in triplicate per PCR reaction.
2.7 Quantification of HTPM sam ples and processed foods
Based on the constructed endogenous gene and exogenous gene PCR standard curves,Ct values were de-termined and used to quantify the amount of total DNA and GM DNA in each sample or processed food.GM contents of soybean ormaize were expressed as the ratio of GM DNA to total DNA.
3 Results and discussion
3.1 Different PCR Efficiencies in two duplex real-time PCR systems

Table 2 PCR efficiencies of the two duplex real-time PCR systems(A and B)
Inaccuracies of real-time PCR quantification resultswere observed in heat processed samples containing genetically modified(GM)soybean ormaize.To discover the causes of increased bias in quantification of GM soybean in heat processed samples,duplex real-time PCR efficiency of system A and system B was evaluated using mixed seed samples,respectively(Table 2).To improve the PCR accuracy,new prime/probewere designed In system A,in which all primer/probe had similar GC%and Tm values,and the amplified targetswere shorter and simiar in size.In system A,PCR efficiencies of endogenous gene(Lectin)and exogenous gene(CP4-EPSPS or NOS)were similar.In system B,PCR efficiencies of exogenous gene assays (CP4-EPSPS or NOS)were slightly lower than that of endogenous Lectin gene assay.In system A,the Tm values of all primer/probe and sizes of amplified exogenous gene and endogenous genewere similar;whereas in system B,the Tm values and amplicon sizeswere remarkable difference.
3.2 LOD of the two duplex real-time PCR systems
To test the LODs of these two real-time PCR systems,CP4-EPSPS gene and NOS terminator assays were evaluated.Both genes could be detected in 9 replicated amplification reactions using DNA template as low as 10 copies of haploid genome in system A.In system B,the lowest detection level was 20 copies of haploid genome(Table 3).Concluded from these results,the LOD of PCR system A was lower than system B,indicating system A wasmore sensitive than system B.
3.3 Quantitative analysis of HTPM sam ples using two duplex real-time PCR systems
Inquantitative analysis of HTPM samples containing 1%RRS soybean,the detected GM contents ranged from 0.969%to 1.123%using CP4-EPSPS gene as exogenous gene target in PCR system A,and the range from 0.803%to 1.046%were calculated in PCR system B.Using NOS terminator as exogenous gene target,the GM content resultswere from 0.998%to 1.049%in system A;and from 0.637%to 0.925%in system B (Table 4).When HTPM samples containing 5%RRS soybean were analyzed in system A,GM soybean was determined to be from 4.904%to 5.400%using CP4-EPSPS transgene as exogenous target;and from 4.734%to 5.395%when using NOS terminator as exogenous target.In system B,the results were from 3.806%to 6.016%when targeting CP4-EPSPSgene;and from 5.132%to 6.031%targeting NOS terminator(Table 4).By comparing two duplex real-time PCR systems(A and B)in quantitative analysis of HTMP RRS samples,we concluded that quantification accuracy could be increased by similar GC%and Tm values of primer/probe,short amplified target length.Test results of the improved duplex real-time PCR system A on RRS soybean detection in heat treated foods validated the effectiveness of this system for detection of GM soybean contents in deeply processed food products.
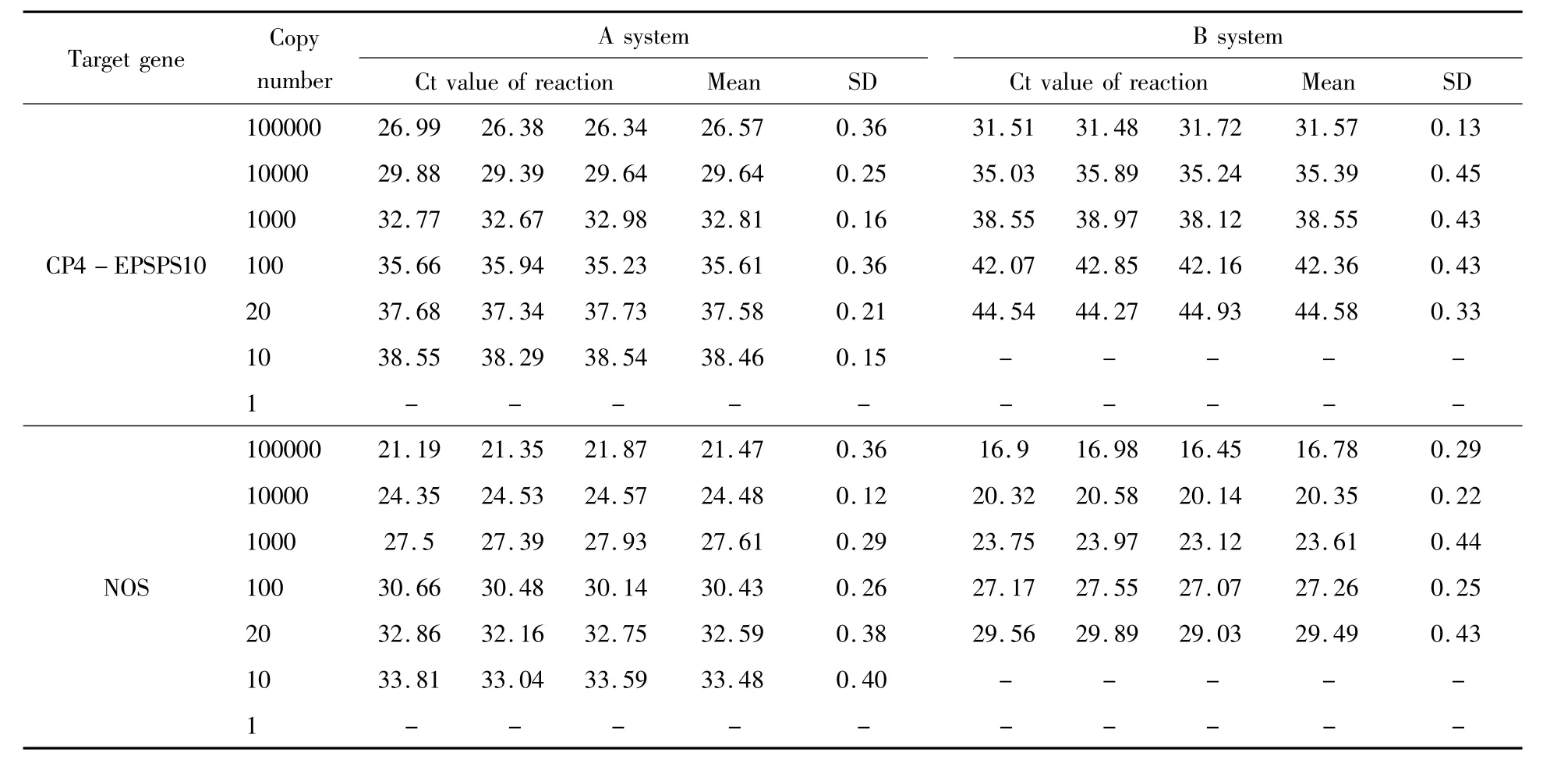
Table 3 LOD evaluation of CP4-EPSPS gene and NOS terminator assays of systems A and B
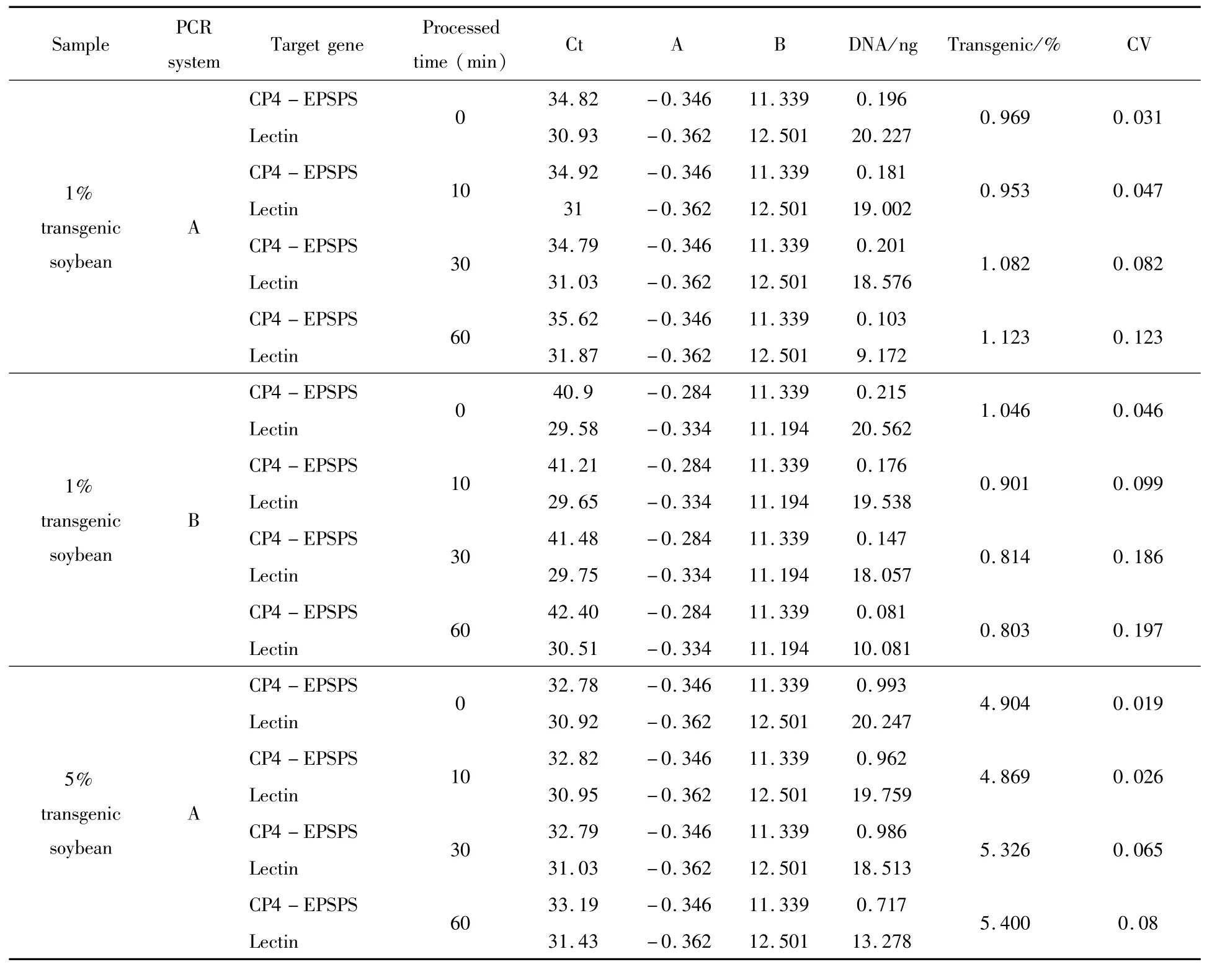
Table 4 Determination of GM contents in HTPM RRS soybean samples using A and B systems
3.4 Com parison of the quantitative results of system s A and B
Thebias of real-time PCR quantification was denoted by coefficient of variation(CV)value,which wascalculated with the formula of CV=1-Calculated GM contents/Known GM contents.The quantitative results of HTPM RRS samples in system A or B were showed in Table 4.CV valueswere plotted over the time of heat treatment(autoclaving)in Figure 1.In samples containing 1%of RRS soybean,CV values in system A increased from 0.019 to 0.123 for CP4-EPSPS gene as the autoclaving time increased from 0 to 60 minutes;For NOS terminator assay,CV values increased from 0.002 to 0.105.In system B,the CV values increased from 0.046 to 0.239 for CP4-EPSPSgene quantification and from 0.026 to 0.363 for NOS terminator as the autoclaving time increased from 0 to 60 minutes.
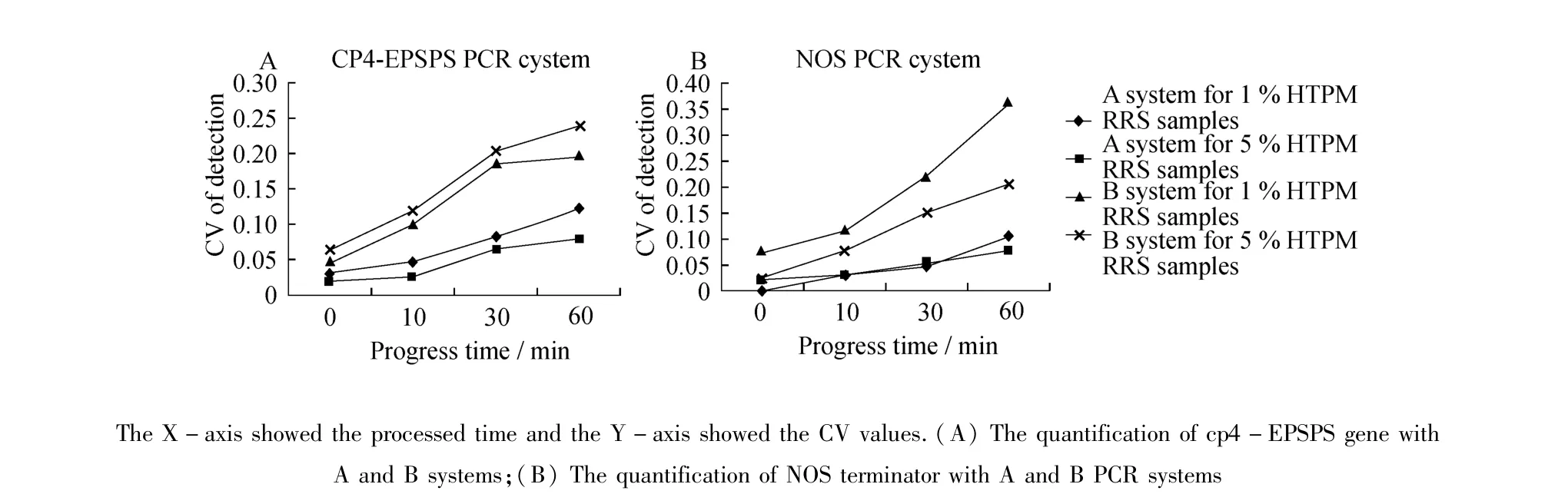
Figure 1 The coefficient of variation(CV)of HTMPRRS samples in CP4-EPSPS gene and NOS terminator detection using A and B systems
Comparing the calculated CV values of the HTPM RRS samples in A and B systems,we found that corresponding CV values in A and B systemswere similar and differ by less than 0.10 when the RRS sampleswere not heat treated,indicating that there was no obvious difference to use either A or B system for raw RRS seeds or flourmaterials.However,as the HTMP sampleswere autoclaved in extended time,the quantification error in system B increased more rapidly than in system A.Quantification bias in system A was lower than in system B in almost each HTMP sample,especially when autoclaving time of HTPM sampleswas extended to 60 minutes. These results indicated that system A wasmore accurate and effective than system B in HTPM samples quantification.There could be several reasons behind the differences in quantification bias between A and B systems. First,genomic DNAs in HTPM RRS samples could have been damaged under extended autoclaving time.Since system A had shorter amplicon sizes for both exogenous and endogenous genes,itwould be less affected by this DNA damage compared to system B.Secondly the higher and similar PCR efficiencies between endogenous and exogenous genes in system A would help to decrease chances of selective target amplification.
3.5 Quantitative analysis of processed foods using re-designed real-time PCR system s
Based on the comparison between system A and B in analysis of HTPM RRS soybean samples,real-time PCR primers should have similar GC contents and Tm values,and the sizes of amplified DNA targets should be shortand similar in accurate real-time PCR system.Upon these considerations,we designed real-time PCR primers and probes for Event 176 maize targeting CryIA(b)exogenous gene and zSSIIb endogenous gene to detect Event176 content in processed foods(Table 1).The duplex real-time PCR systems employing improved primer pair/probe and shorter target length were used in quantification of RRS soybean and Event176 maize in several processed foods,including bean curd,soy milk,popcorns and corn chips.Each of the processed food contained 1%or 5%RRS soybean or Event 176 maize.Quantification values from 1.08%to 1.42%were obtained in processed foods containing 1%RRS or Event 176 maize;values from 5.22%to 5.91%were observed in processed foods containing 5%RRS soybean or Event176 maize(As shown in Figure 2A and 2B).

Figure 2 The quantified GM contents of trial processed foods employing improved PCR systems
4 Conclusion
Real-time PCR technique has been widely used in quantitative detection of GM foods because of its high accuracy and easy operation.We discovered that similar GC%and Tm values of primer/probe,short and similar amplified target length could increase quantification accuracy in processed samples.The duplex real-time PCR system developed in this study has provided an effective GM quantification system for at leastGM soybean and maize in heat processed foods,ormaybe other types of processed foods thatwould be investigated in further studies.
Acknow ledgem ents
Thiswork was funded by the Ministry of Science and Technology of China(2011BAK10B03),and Shanghai Ring Star project(11QA1403300).
[1] ISMAIL IA,ABDEL-MONIEM A SH,EL-SHAZLY E A,etal.Biodetrimental effects of the Corn hybrids,Neemazal-T/Sand Chlorphan 48%on the pink corn borer Sesamia cretica Led[J].Archives of Phytopathology And, 2012,45(17):2014-2025.
[2] CARTER C A,GRUERE G P.International approaches to the labeling of geneticallymodified foods[J].Choices, 2003,18(2):1-4.
[3] STUDER E,RHYNER C,LUTHY J,et al.Quantitative competitive PCR for the detection of geneticallymodified soybean and maize[J].Z Lebensm Unters F A, 1998,207(3):207-213.
[4] YANG L T,GUO JC,ZHANG H B,et al.Qualitative and quantitative event-specific PCR detection methods for oxy-235 canola based on the 3'integration flanking sequence[J].JAgric Food Chem, 2008,56(6):1804-1809.
[5] WANG S,LIX,YANG L T,etal.Developmentand in-house validation of a referencemolecule pMIR604 for simplex and duplex event-specific identification and quantification of GM maize MIR604[J].Eur Food Res Technol, 2009,230(2):239-248.
[6] JIANG L X,YANG L T,RAO J,et al.Developmentand in-house validation of the event-specific qualitative and quantitative PCR detection methods for geneticallymodified cotton MON15985[J].JSci Food Agric, 2010,90(3):402-408.
[7] GUO J,CHEN L,LIU X,et al.A multiplex degenerate PCR analytical approach targeting to eight genes for screening GMOs[J].Food Chem, 2012,132(3):1566-1573.
[8] YANG L T,GUO JC,PAN A H,et al.Event-specific quantitative detection of nine genetically modified maizes usingone novel standard referencemolecule[J].JAgric Food Chem, 2007,55:15-24.
[9] BURNSM,SHANAHAN D,VALDIVIA H,et al.Quantitative event-specific multiplex PCR detection of Roundup Ready soya using LabChip technology[J].Eur Food Res Technol, 2003,216(5):428-433.
[10] CORBISIER P,TRAPMANN S,GANCBERG D,et al.Quantitative determination of Roundup Ready soybean(Glycine max)extracted from highly processed flour[J].Anal Bioanal Chem, 2005,383(2):282-290.
[11] JAMESD,SCHMIDT A,WALL E,et al.Reliable detection and identification of genetically modified maize,soybean,and canola bymultiplex PCR analysis[J].JAgric Food Chem, 2003,51(20):5829-5834.
[12] GARCIA-CANASV,CIFUENTESA,GONZÁLEZ R.Quantitation of transgenic Bt event-176 maize using double quantitative competitive polymerase chain reaction and capillary gel electrophorsesis laser-induced fluorescence[J].Anal Chem, 2004,76(8):2306-2313.
[13] TAVERNIERS I,WINDELSP,VAITILINGOM M,et al.Event-specific plasmid standards and real-time PCR methods for transgenic Bt 11,Bt 176,and GA21 maize and transgenic GT73 canola[J].J Agric Food Chem, 2005,53(8):3041 -3052.
(责任编辑:顾浩然)
转基因大豆和玉米加工产品的双重精确定量PCR检测方法
程 芳1,沈 平2,张大兵3,李建粤1,杨立桃3*
(1.上海师范大学生命与环境科学学院,上海200234;2.农业部科学和技术发展中心,北京100026;3.上海交通大学生命科学技术学院,上海200240)
在很多国家,转基因生物(GMOs)及其衍生产品必须标有精确的转基因含量.最近研究中,实时定量PCR技术广泛应用于转基因成分的检测.然而,转基因生物的实时定量PCR方法的精确度仍然是一个难以解决的问题,尤其是对于高温处理过的样品.为了更好地准确定量高温处理样品中转基因的含量,对普通的实时定量PCR体系做了一些改进,包括重新设计内源基因和外源基因的引物,使得扩增较短并且大小接近的目标DNA片段,同时引物的GC含量和溶解温度也都相近.此外,采用热处理加工模型(HTPM)的方法,制备了含有转基因大豆GTS 40-3-2的样品,并验证了改进后的实时定量PCR系统.实验结果表明:使用改进后的实时定量PCR体系测定热处理过的样品,发现其中的转基因含量的定量偏差明显降低.同时使用改进的双重实时定量PCR进一步验证转基因大豆的加工食品,结果也显示,转基因含量的定量结果更准确.这些结果表明:改进的双重实时定量PCR将适用于热加工产品的定量检测.
转基因生物;大豆;玉米;加工食品;荧光定量PCR
Q 37
A
1000-5137(2013)02-0197-09
Received date:2013-03-25
Foundation item:Thiswork was supported by the National Transgenic Plant Special Fund,China(2011ZX08012-003),and Shanghai Ring Star project(11QA1403300).
Biography:CHENG Fang(1988-),female,graduate student,College of Life and Environment Sciences,ShanghaiNormal U-niversity;YANG Litao(1981-),male,associate professor,School of Life Science and Biotechnology,Shanghai Jiao Tong University.
*Corresponding Author
