4种昆虫基因组中的线粒体假基因
2012-12-09杨明茹周志军常岩林张艳霞
杨明茹,周志军,常岩林,张艳霞
(河北大学生命科学学院,河北保定 071002)
4种昆虫基因组中的线粒体假基因
杨明茹,周志军,常岩林,张艳霞
(河北大学生命科学学院,河北保定 071002)
为了进一步了解昆虫核基因组中线粒体假基因(Numts)序列分布情况,避免Numts序列对基于线粒体DNA(mt DNA)进行系统发育关系研究结果的误导,利用Blast N对GenBank中已完成核基因组和mtDNA测序的4种昆虫核基因组中的Numts序列进行检索,结果表明:冈比亚按蚊Anophelesgambiae中没有Numts序列;黑腹果蝇Drosophilamelanogaster中仅有少量Numts序列;赤拟谷盗Triboliumcastaneum和意大利蜜蜂Apismellifera基因组中Numts序列超过100条,尤其是意大利蜜蜂中的Numts序列涵盖全部mtDNA.ND2,ND4,ND5,COⅠ与lrRNA向核内转移频率高于其他mt DNA基因片段,因此,在使用其进行系统发育关系研究时需加倍谨慎.
mt DNA;Numts;基因组
线粒体DNA(mt DNA)具有结构简单、进化速率快和母系遗传等特征,是研究基因组结构和基因表达的良好模型,同时也是在分子进化和系统发育研究中应用最广的分子标记[1].随着聚合酶链式反应(PCR)和DNA测序技术的发展,mtDNA作为分子标记被广泛用于不同类群的系统进化和群体遗传结构研究[2-4].
线粒体假基因(nuclear mitochondrial pseudogenes,Numts)是在核基因组中发现的、与mtDNA高度相似的DNA片段[5],通常被认为是mtDNA向核基因组转移的结果,在真菌、无脊椎动物、脊椎动物和植物中都有Numts存在[6-8].由于核基因组中的Numts序列进化速率较mt DNA慢,更接近于mtDNA的祖先序列[9-12],因此,在使用通用引物进行扩增时,核基因组中的Numts极易被优先扩增出来或与mt DNA靶序列一起扩增出来[13-15].Numts序列的存在不仅增加了获得mt DNA靶序列的难度,而且如果将Numts序列误作为mtDNA靶序列使用,甚至会得出错误的结论[14-17].Sorenson等[18]分别将在斑胸树鸭Dendrocygnaarcuata中发现的COⅠ-Numts序列和mtDNA基因序列用于树鸭属系统发育关系分析,发现其分支拓扑结构完全不同.同时,Numts也可为系统进化研究提供有益的线索[16,19],与内含子、卫星序列和转位因子等冗余DNA一样,Numts不受进化的选择作用,保留着更多的祖先特征,可以为物种的分子进化研究提供大量的信息[20],比如,人们已把Numts作为分子标记,对人[21]和拟南芥[22]进行了研究.
mtDNA向核内转移现象非常普遍.2001年,Bensasson等[19]统计发现在80余种真核生物(包括20余种昆虫)中存在Numts序列.此后,Numts在弄蝶[23]、蜜蜂[24]、蚂蚁[25]、蝉[26]、石蛃[27]等类群中被报道.在Genbank中,以“Insecta Numt”作为关键词,进行Numts序列检索,共获得881条Numts序列,隶属于昆虫纲6个目,分别为直翅目、鳞翅目、膜翅目、石蛃目、双翅目和半翅目;涵盖7种基因类型,分别为COⅠ(部分)、ND5(部分)、COⅠ(部分)-trn L-COⅡ(部分)、trn L-COⅡ(部分)、ND2(部分)-COⅠ(部分)、COⅢ(部分)、ATP6(部分)[23,25-27,28-35].
此前关于Numts的报道,都是基于单一物种基因组中的Numts分析,且不同研究所使用的参数设置也不尽相同,难以进行比较.为了进一步深入了解昆虫核基因组中Numts序列分布情况,对基因组序列组装基本完成(Y染色体除外)的4种昆虫中的Numts序列的大小、数目及分布特点等进行了研究.
1 材料与方法
1.1 实验数据来源
从公共数据库NCBI中下载物种的基因组数据,见表1.
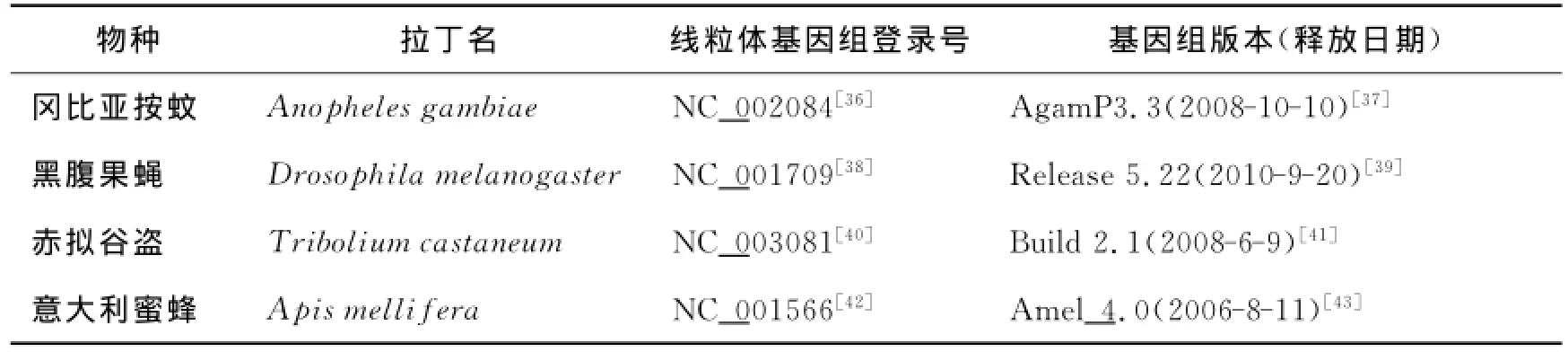
表1 实验物种及基因组数据来源Tab.1 Specimens and data collection of the genome
1.2 实验方法
分别以mt DNA编码的37个基因(13个蛋白质编码基因、22个tRNA基因和2个r RNA基因)为诱饵序列,在NCBI中逐个对其核基因组序列进行Blast N同源性序列比对检索,分别统计4种昆虫基因组不同染色体上的Numts序列.在进行Numts序列统计时,为避免假阳性和减少遗漏早期进化的Numts序列,选择了较保守的期望值(E值为10-4)[6,44-46],没有统计序列长度≤50 bp的检索结果,同时,对相邻位点的Numts序列进行核查以确定他们是否属于同一Numts序列.
2 结果
2.1 4种昆虫核基因组中的Numts分布
NCBI-Blast N同源性序列分析结果(如表2):冈比亚按蚊基因组中没有检测到Numts;黑腹果蝇中检测到25条Numts序列,总长度为2.634 kb,其中,最长的Numts序列长度为541 bp,对应于mt DNA的ND2(部分)和COⅠ(部分)及它们之间的3个tRNA基因簇(trn W-trnC-trn Y).赤拟谷盗中检测到123条Numts序列,总长度为18.485 kb,其中,最长的Numts序列长度为951 bp,对应于mtDNA的ND4(部分);意大利蜜蜂中的Numts含量最多,共检测到994条Numts序列,总长度达327.102 kb,其Numts序列已涵盖mt DNA的全部序列.其中,最长的Numts序列长度为1 267 bp,对应于mtDNA的4个tRNA基因(trn M-trn Q-trn A-trnI)和整个ND2.所检测到的Numts序列长短不一,但大多数小于300 bp(表2).

表2 4种昆虫基因组中的Numts大小分布(Blast N检索E值为10-4)Tab.2 Size distribution of Numts in four insect genomes detected by Blast N at a threshold of 10-4
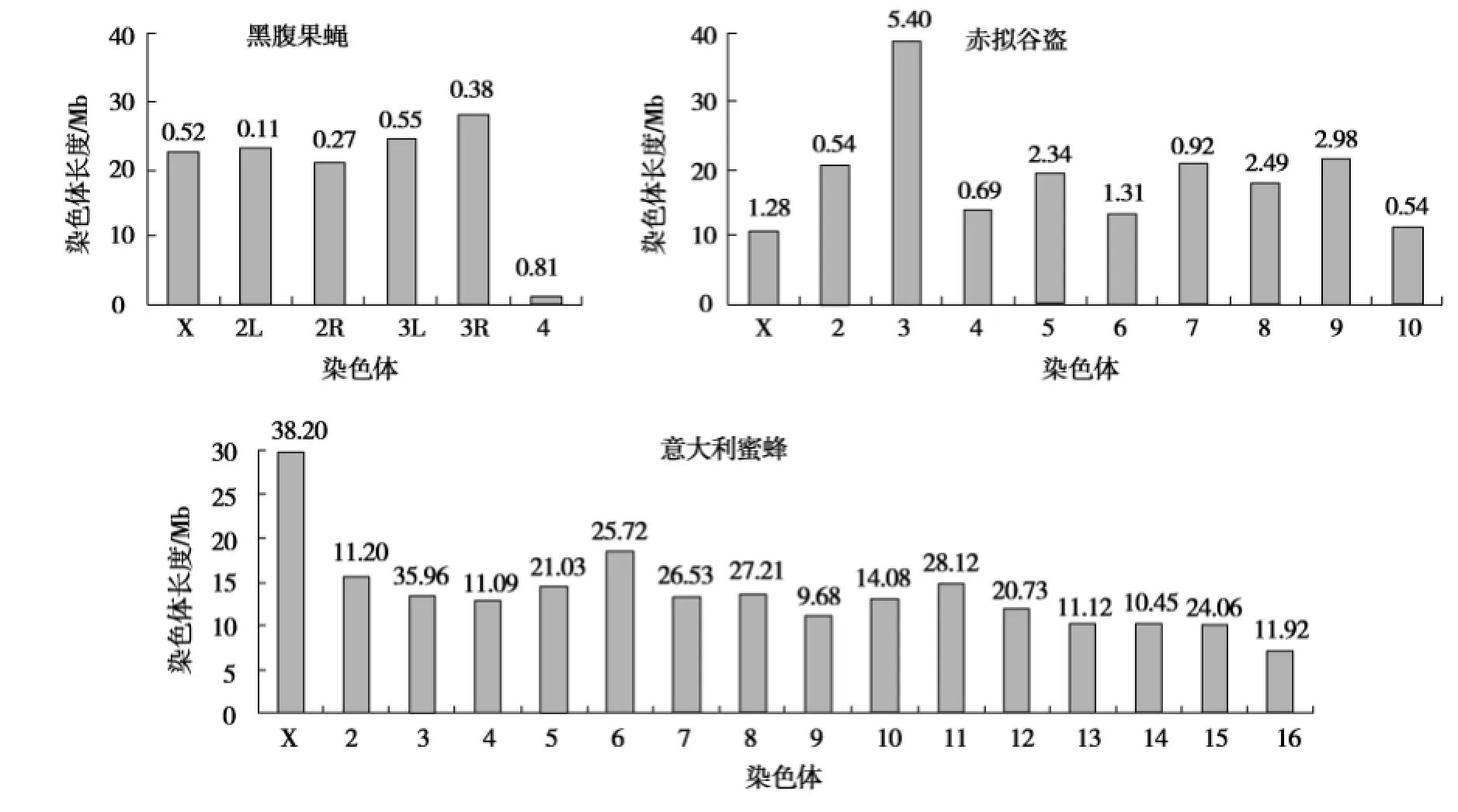
图1 4种昆虫基因组中染色体长度及Numts序列分布情况Fig.1 Chromosome length and the distribution of Numts in four insect genomes
2.2 不同染色体中的Numts分布
4种昆虫基因组中,不同染色体上的Numts分布情况如图1所示,黑腹果蝇的5个常染色体和1个X染色体,赤拟谷盗的9个常染色体和1个X染色体及意大利蜜蜂的15个常染色体和1个X染色体中都有Numts分布.其中,黑腹果蝇的4号染色体、赤拟谷盗的3号染色体,意大利蜜蜂的X染色体检测到的Numts序列最多,总长度分别为0.81,5.40,38.20 kb;而黑腹果蝇的2L号染色体,赤拟谷盗的2号和10号染色体,意大利蜜蜂的9号染色体检测到的Numts序列最少,长度分别仅为0.11,0.54,9.68 kb.意大利蜜蜂的Numts含量明显高于赤拟谷盗与黑腹果蝇,仅Numts序列含量最少的9号染色体的Numts数目都远高于黑腹果蝇基因组的整体Numts量.
2.3 不同线粒体基因向核内转移频率差异分布
mtDNA中,不同的基因片段向核内转移的频率不同,其中,重排发生频繁、长度较短的tRNA基因的转移频率较低,而位置相对固定,长度较长的蛋白质编码基因和rRNA基因的转移频率较高,ND2,ND4,ND5,COⅠ与lrRNA基因的转移的次数较高(如图2).
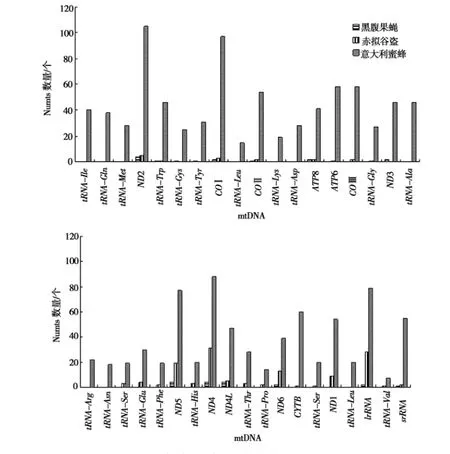
图2 不同mt DNA编码基因对应的Numts数量Fig.2 Numbers of Numts of different mt DNA encoding genes
3 讨论
3.1 4种昆虫基因组中Numts序列比较
生物在长期进化过程中,mtDNA向核内转移已被证实,并且很可能仍在持续进行之中,已经报道的不同物种mtDNA向核内转移的数量差异很大[6-7].
基于相同的参数设置,对4种昆虫基因组中的Numts序列进行研究发现:2种双翅目昆虫基因组中没有或仅含有少量Numts,鞘翅目的赤拟谷盗基因组中Numts数量较多,膜翅目意大利蜜蜂基因组中的Numts数量最多,与之前的报道基本一致[6-7,24,47].
本研究发现冈比亚按蚊、黑腹果蝇、赤拟谷盗和意大利蜜蜂基因组的Numts序列总长度分别为0,2.6,18.5,327 kb.之前报道黑腹果蝇基因组的Numts序列总长度分别为0.5[6],10.3[7]和0.8 kb[24],赤拟谷盗中分别为31.2 kb[7]和8.8 kb[24],意大利蜜蜂中分别为0[48],172[7],237 kb[24]和272 kb[47].Pamilo等[24]发现赤拟谷盗中ND4与ND5的转移频率很高,约占总量的1/2;笔者发现lrRNA的转移频率同样很高,三者约占总量的2/3.Behura等[47]发现意大利蜜蜂的10号染色体Numts含量最少,COⅠ转移频率最高;而本课题组发现9号染色体Numts序列最少,ND2转移频率高于COⅠ.造成上述差异的主要原因可能是由于实验参数设置和基于的基因组组装版本差异.所检测到的Numts序列长度多数小于300 bp(表2),表明较小的基因片段具有更高的向核内转移效率.
此前报道在蚱蜢[28]、蚜虫[49]、伊蚊[30]和沙漠蝗[50]的一些基因具有很高的转移频率,而在一些虎甲中Numts却十分罕见[51].由于蚱蜢和蚜虫是不完全变态昆虫,而冈比亚按蚊、黑腹果蝇、意大利蜜蜂、赤拟谷盗及虎甲都是完全变态昆虫.赤拟谷盗和意大利蜜蜂基因组中检测到大量Numts序列,推翻了此前关于完全变态昆虫中缺乏Numts的观点[51].
本研究涉及的4种昆虫基因组大小差异不大,因此,不能用“基因组越大Numts越多”的观点[7]解释这些物种间Numts的数量差异.Hazkani-Covo等[52]发现人类基因组中的Numts序列仅30%来自mt DNA直接转移,其余70%则由转移后的mt DNA片段在核内复制、转座而成.Tourmen等[53]发现一些重复的Numts的侧翼区域都是长末端重复序列(LTR),表明核基因组中存在Numts序列复制、转座的机制.此前,有报道认为意大利蜜蜂基因组的重组率远高于黑腹果蝇和冈比亚按蚊[54],那么基因组中Numts数量变化是否与基因组的重组率具有一定相关性?仍有待于进一步的研究.
3.2 Numts序列对分子进化研究的影响
mtDNA向核内转移的频率决定于细胞内的竞争强度、父系基因渗透概率和物种种群数量[55].随着在更多物种中的Numts序列发现,如何处理其对分子进化研究的影响成为我们不得不面对的问题.
首先,在使用通用引物对mt DNA基因进行扩增时,应尽量选择线粒体含量较丰富的组织或器官[56];其次,可以通过设计特异性引物、以长距PCR产物为模板进行二次PCR,RT-PCR等方法避免Numts序列干扰[19,56-58];最后,可以通过Numts序列特征识别所获得数据中的假基因.蛋白质编码基因对应的Numts序列,通常会由于碱基替换或插入/缺失导致移码突变或终止突变.例如,周志军等[35]利用线粒体COⅠ基因片段进行暗褐蝈螽不同地理种群间的遗传分化研究时,发现5条COⅠ-Numts序列,其长度略短于mtDNACOⅠ,存在2处碱基缺失(3 bp和21 bp),在另外2条COⅠ-Numts中还存在额外的1 bp缺失.除了这些明显的Numts特征外,也可以通过翻译后的氨基酸序列是否发生保守位点突变加以判断.Sorenson等[59]在熊斑树鸭核基因组发现COⅠ-Numts与mtDNA-COⅠ相比,不存在碱基插入/缺失,异常的终止密码子,但其氨基酸序列有7个保守位点发生了变化.对于tRNA和rRNA基因,Numts序列识别显得更复杂,有些学者提出可以通过比较序列的二级结构来识别Numts[60].
核内Numts序列在给基于mtDNA的分子进化研究带来困难的同时,也提供了一个新的有力工具.首先,可以根据Numts中保留的基因信息确定物种之间的亲缘关系和进化距离.通过分别构建mt DNA和Numts的进化关系树,可以确定mt DNA和Numts,不同Numts之间的分歧时间.例如,张凤英等采用上述方法发现在拟穴青蟹的同一个体中存在2类Numts序列,分别代表了2次核整合事件[61].其次,可以作为系统发育研究的外群.例如,在人类起源问题的研究中,Zischler等[62]使用人类核基因组中的一段D-loop区的Numts序列作为外群,证实了现代人类起源于非洲的假说,结束了很久以来关于现代人类起源地的争论.Hay等[63]用cytb-Numts作为外群,构建系统发育树分析发现爬行类的Sphenodonpunctatus仅仅是Sphenodonguntheri的同物异名.第三,Numts序列作为分子标记.同一段Numts序列在近源种间是否存在及其变异程度,可以反映物种间的亲缘关系远近.Sato等[64]分别使用Numts序列和mtDNA序列,重建了达尔文雀系统发育关系,结果显示,2种序列建立的系统发育树分支拓扑结构基本一致.Hazkani-Covo利用Numts序列构建了5种灵长类动物系统发育树,结果支持人类-黑猩猩的姊妹群关系[65].
综上所述,Numts序列给基于mt DNA的分子进化研究带来挑战的同时,也带来了新的契机.相信随着更多物种基因组测序计划的完成,对Numts序列研究的逐步深入,Numts序列必将在分子进化研究中展现更大的价值.
[1]BOORE J L.Animal mitochondrial genomes[J].Nucleic Acids Res,1999,27(8):1767-1780.
[2]AVISE J C,AMOLD J,BALL R M,et al.Intraspecific phylogeography:the mitochondrial DNA bridge between population genetics and systematics[J].Annu Rev Ecol Syst,1987,18:489-522.
[3]徐其放,陈嘉昌,朱世杰,等.动物线粒体DNA的特异结构及应用分子系统学分析的方法[J].中国比较医学杂志,2005,15(5):315-319.
XU Qifang,CHEN Jiachang,ZHU Shijie,et al.Structural characteristics of animal's mitochondrial DNA and its application on molecular systematics[J].Chinese Journal of Comparative Medicine,2005,15(5):315-319.
[4]BOORE J L,BROWN W M.Big trees from little genomes:mitochondrial gene order as a phylogenetic tool[J].Curr Opin Genet Dev,1998,8(6):668-674.
[5]LOPEZ J V.Numt,a recent transfer and tandem amplification of mitochondrial DNA to the nuclear genome of the domestic cat[J].J Mol Evol,1994,39:174-190.
[6]RICHLY E,LEISTER D.Numts in sequenced eukaryotic genomes[J].Mol Biol Evol,2004,21(6):1081-1084.
[7]HAZKANI-COVO E,ZELLER R M,MARTIN W.Molecular poltergeists:mitochondrial DNA copies(numts)in sequenced nuclear genomes[J].PLoS Genet,2010,6(2):e1000834.
[8]SACERDOT C,CASAREGOLA S,LAFONTAINE I,et al.Promiscuous DNA in the nuclear genomes of hemiascomycetous yeasts[J].FEMS Yeast Res,2008,8:846-857.
[9]BROWN W M,GEORGE M,WILSON A C.Rapid evolution of animal mitochondrial DNA[J].Proc Natl Acad Sci USA,1979,76(4):1967-1971.
[10]TAMURA K.The rate and pattern of nucleotide substitution in Drosophila mitochondrial DNA[J].Mol Bid Ed,1992,9(5):814-825.
[11]ARCTANDER P.Comparison of a mitochondrial gene and a corresponding nuclear pseudogene[J].Proc R Soc Lond Biol,1995,262:13-19.
[12]PERNA N T,KOCHER T D.Mitochondrial DNA:molecular fossils in the nucleus[J].Current Biology,1996,6(2):128-129.
[13]KUYL A C,KUIKEN C L,DEKKER J T,et al.Nuclear counterparts of the cytoplasmic mitochondrial 12Sr RNA gene:a problem of ancient DNA and molecular phylogenies[J].J Mol Evol,1995,40(6):652-657.
[14]ZH ANG Dexing,HEWITT G M.Nuclear integrations:challenges for mitochondrial DNA markers[J].Trends in Ecology and Evolution,1996,11(6):247-251.
[15]SONG H,BUHAY J E,WHITING M F,et al.Many species in one:DNA barcoding overestimates the number of species when nuclear mitochondrial pseudogenes are coamplified[J].PNAS,2008,105(36):13486-13491.
[16]ZH ANG Dexing,HEWITT G M.Nuclear DNA analyses in genetic studies of populations:practice,problems and prospect[J].Molecular Ecology,2003,12:563-584.
[17]LEISTER D.Origin,evolution and genetic effects of nuclear insertions of organelle DNA[J].Trends Genetics,2005,21(12):655-663.
[18]SORENSON M D,QUINN T W.Numt:A challenge for avian systematics and population biology[J].The Auk,1998,115(1):214-221.
[19]BENSASSON D,ZHANG Dexing,HARTL D L,et al.Mitochondrial pseudogenes:evolution's misplaced witnesses[J].Trends Ecol Evol,2001,16(6):314-321.
[20]ZUCKERKANDL E.Why so many noncoding nucleotides?The eukaryote genome as an epigenetic machine[J].Genetica,2002,115(1):105-129.
[21]THOMAS R,ZISCHLER H,P A··A··BO S,et al.Novel mitochondrial DNA insertion polymorphism and its usefulness for human population studies[J].Hum Biol,1996,68(6):847-854.
[22]ULLRICH H,LATTIG K,BRENNICKE A,et al.Mitochondrial DNA variations and nuclear RFLPs reflect different genetic similarities among 23 Arabidopsis thaliana ecotypes[J].Plant Mol Biol,1997,33:37-45.
[23]HEBERT P D,PENTON E H,BURNS J M,et al.Ten species in one:DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator[J].Proc Natl Acad Sci USA,2004,101(41):14812-14817.
[24]PAMILO P,VILJAKAINEN L,VIHAVAINEN A.Exceptionally high density of Numts in the honeybee genome[J].Mol Biol and Evol,2007,24(6):1340-1346.
[25]MARTINS J Jr,SOLOMON S E,MIKHEYEV A S,et al.Nuclear mitochondrial-like sequences in ants:evidence fromAtta cephalotes(Formicidae:Attini)[J].Insect Mol Biol,2007,16(6):777-784.
[26]LEE Y J,HILL K B R.Systematic revision of the genus Psithyristria St l(Hemiptera:Cicadidae)with seven new species and a molecular phylogeny of the genus and higher taxa[J].Syst Entomol,2010,35(2):277-305.
[27]BALDO L,DE QUEIROZ A,HEDIN M,et al.Nuclear-mitochondrial sequences as witnesses of past interbreeding and population diversity in the jumping bristletail mesomachilis[J].Mol Biol Evol,2011,28(1):195-210.
[28]BENSASSON D,ZHANG Dexing,HEWITT G M.Frequent assimilation of mitochondrial DNA by grasshopper nuclear genomes[J].Mol Biol Evol,2000,17(3):406-415.
[29]SAWAMURA K,KOGANEBUCHI K,SATO H,et al.Potential gene flow in natural populations of theDrosophila ananassaespecies cluster inferred from a nuclear mitochondrial pseudogene[J].Mol Phylogenet Evol,2008,48(3):1087-1093.
[30]HLAING T,TUN-LIN W,SOMBOON P,et al.Mitochondrial pseudogenes in the nuclear genome ofAedesaegyptimosquitoes:implications for past and future population genetic studies[J].BMC Genetics,2009,10:11.
[31]MOULTON M J,SONG H,WHITING M F.Assessing the effects of primer specificity on eliminating numt coamplification in DNA barcoding:a case study from Orthoptera(Arthropoda:Insecta)[J].Mol Ecol Resour,2010,10(4):615-627.
[32]ANDRIANOV B V,GORYACHEVA I I,MUGUE N S,et al.Comparative analysis of the mitochondrial genomes inDrosophilavirilisspecies group(Diptera:Drosophilidae)[J].Trends Evol Biol,2010:e4.
[33]MAGNACCA K N,BROWN M J.Mitochondrial heteroplasmy and DNA barcoding in Hawaiian Hylaeus(Nesoprosopis)bees(Hymenoptera:Colletidae)[J].BMC Evol Biol,2010,10:174.
[34]BORING C A,SHARANOWSKI B J,SHARKEY M J.Maxfischeriinae:a new braconid subfamily(Hymenoptera)with highly specialized egg morphology[J].Systematic Entomology 2011,36:529-548.
[35]周志军,张艳霞,常岩林,等.暗褐蝈螽不同地理种群间的遗传分化[J].遗传,2011,33(1):75-80.
ZHOU Zhijun,ZHANG Yanxia,CHANG Yanlin,et al.Genetic differentiation among different geographic populations ofGampsocleissedakovii[J].Hereditas,2011,33(1):75-80.
[36]BEARD C B,HAMM D M,COLLINS F H.The mitochondrial genome of the mosquitoAnophelesgambiae:DNA sequence,genome organization,and comparisons with mitochondrial sequences of other insects sequences of other insects[J].Insect Mol Biol,1993,2(2):103-124.
[37]HOLT R A,MANI SUBRAMANIAN G,HALPERN A,et al.The genome sequence of the Malaria MosquitoAnopheles gambiae[J].Science,2002,298:129-149.
[38]LEWIS D L,FARR C L,KAGUNI L S.Drosophilamelanogastermitochondrial DNA:completion of the nucleotide sequence and evolutionary comparisons[J].Insect Mol Biol,1995,4(4):263-278.
[39]ADAMS M D,CELNIKER S E,HOLT R A,et al.The genome sequence ofDrosophilamelanogaster[J].Science,2000,287(5461):2185-2195.
[40]FRIEDRICH M,MUQIM N.Sequence and phylogenetic analysis of the complete mitochondrial genome of the flour beetleTriboliumcastanaeum[J].Mol Phylogenet Evol,2003,26(3):502-512.
[41]TRIBOLIUM GENOME SEQUENCING CONSORTIUM[TGSC].The genome of the model beetle and pestTribolium castaneum[J].Nature,2008,452:949-955.
[42]CROZIER R H,CROZIER Y C.The mitochondrial genome of the honeybeeApismellifera:complete sequence and genome organization sequence and genome organization[J].Genetics,1993,133(1):97-117.
[43]HONEYBEE GENOME SEQUENCING CONSORTIUM[HGSC].Insights into social insects from the genome of the honeybeeApismellifera[J].Nature,2006,443:931-949.
[44]APARICIO S,CHAPMAN J,STUPKA E,et al.Whole-genome shotgun assembly and analysis of the genome ofFugu rubripes[J].Science,2002,297:1301-1310.
[45]ROST B.Twilight zone of protein sequence alignments[J].Protein Eng,1999,12(2):85-94.
[46]ANTUNES A,RAMOS M J.Discovery of a large number of previously unrecognized mitochondrial pseudogenes in fish genomes[J].Genomics,2005,86:708-717.
[47]BEHURA S K.Analysis of nuclear copies of mitochondrial sequences in honey beeApismelliferagenome[J].Mol Biol and Evol,2007,24(7):1492-1505.
[48]PEREIRA S L,BAKER A J.Low number of mitochondrial pseudogenes in the chicken(Gallusgallus)nuclear genome: implications for molecular inference of population history and phylogenetics[J].BMC Evol Biol,2004,4:17.
[49]SUNNUCKS P,HALES D F.Numerous transposed sequences of mitochondrial cytochrome oxidase I-II in aphids of the genusSitobion(Hemiptera:Aphididae)[J].Mol Biol Evol,1996,13(3):510-524.
[50]ZHANG Dexing,HEWITT G M.Highly conserved nuclear copies of the mitochondrial control region in the desert locustSchistocercagregaria:some implications for population studies[J].Mol Ecol,1996,5:295-300.
[51]PONS J,VOGLER A P.Complex pattern of coalescence and fast evolution of a mitochondrialrRNApseudogene in a recent radiation of tiger beetles[J].Mol Biol Evol,2005,22:991-1000.
[52]HAZKANI-COVO E,SOREK R,GRAUR D.Evolutionary dynamics of large numts in the human genome,rarity of independent insertions and abundance of postinsertion duplications[J].J Mol Evol,2003,56:169-174.
[53]TOURMEN Y,BARIS O,DESSEN P,et al.Structure and chromosomal distribution of human mitochondrial pseudogenes[J].Genomics,2002,80(1):71-77.
[54]BEYE M,GATTERMEIER I,HASSELMANN M,et al.Exceptionally high levels of recombination across the honey bee genome[J].Genome Res,2006,16:1339-1344.
[55]YAMAUCHI A.Rate of gene transfer from mitochondria to nucleus:effects of cytoplasmic inheritance system and intensity of intracellular competition[J].Genetics,2005,171:1387-1396.
[56]王继文.动物线粒体假基因的识别及其在进化生物学中的应用[J].动物学杂志,2004,39(3):103-108.
WANG Jiwen.Numts identification and utility in evolutionary biology[J].Chinese Journal of Zoology,2004,39(3):103-108.
[57]COLLURA R V,AUERBACH M R,STEWART C B.A quick direct method that can differentiate expressed mitochondrial genes from their nuclear pseudogenes[J].Curr Biol,1996,6(10):1337-1339.
[58]CHENG S,HIGUCHI R,STONEKING M.Complete mitochondrial genome amplification[J].Nature Genetics,1994,7:350-351.
[59]SORENSON M D,FLEISCHER R C.Multiple independent transpositions of mitochondrial DNA control region sequences to the nucleus[J].Proc Natl Acad Sci USA,1996,93:15239-15243.
[60]OLSON L E,YODER A D.Using secondary structure to identify ribosomal numts:cautionary examples from the human genome[J].Mol Biol Evol,2002,19(1):93-100.
[61]张凤英,马凌波,乔振国,等.青蟹线粒体COⅠ假基因的分离和特征分析[J].遗传,2006,28(1):43-49.
ZH ANG Fengying,MA Lingbo,QIAO Zhenguo,et al.Separation and characterization of mitochondrialCOⅠpseudogenes inScyllaparamamosain[J].Hereditas,2006,28(1):43-49.
[62]ZISCHLER H,GEISERT H,HAESELER VON A,et al.A nuclear fossil of the mitochondrial D-loop and the origin of modern humans[J].Nature,1995,378:489-492.
[63]H AY J M,SARRE S D,DAUGHERTY C H.Nuclear mitochondrial pseudogenes as molecular outgroups for phylogenetically isolated taxa:a case study inSphenodon[J].Heredity,2004,93:468-475.
[64]SATO A,TICHY H,O'HUIGIN C,et al.On the origin of Darwin's Finches[J].Mol Biol Evol,2001,18(3):299-311.
[65]HAZKANI-COVO E.Mitochondrial insertions into primate nuclear genomes suggest the use of numts as a tool for phylogeny[J].Mol Biol Evol,2009,26(10):2175-2179.
Nuclear mitochondrial pseudogenes in four sequenced insect genomes
YANG Ming-ru,ZHOU Zhi-jun,CHANG Yan-lin,ZHANG Yan-xia
(College of Life Sciences,Hebei University,Baoding 071002,China)
The aim of this study was to have further information about the distribution of nuclear mitochondrial pseudogenes(Numts)in insect genomes,and to avoid misguidance of Numts sequences in the molecular phylogeny based on mt DNA.Sequences alignment(NCBI-Blast N)was carried out with mitochondrial and corresponding nuclear genome sequences in 4 insect species.The results showed:copy number ranges from none in the African malaria mosquitoAnophelesgambiae,few copies in the fruit flyDrosophilamelanogaster,to more than 100 in the flour beetleTriboliumcastaneumand the honeybeeApis mellifera,especially inA.mellifera,Numts spanned the whole mtDNA.The reconstructed frequency ofND2,ND4,ND5,COⅠandlr RNAintegrations into the nucleus were higher than other mitochondrial genes.So,it needed for doubly cautious when using it for the study of phylogenetic relationships.
mt DNA;Numts;g enome
Q96
A
1000-1565(2012)02-0165-08
2011-09-12
教育部高等学校博士学科点专项科研基金资助项目(20101301120006)
杨明茹(1984-),女,河北保定人,河北大学在读硕士研究生.
周志军(1980-),男,山西长治人,河北大学讲师,博士,主要从事直翅目线粒体基因组学研究.E-mail:zhijunzhou@163.com
赵藏赏)
