甲真菌病概述及其诊断最新进展
2012-03-28徐建平HeatherYoell
徐建平,Heather Yoell
(麦克马斯特大学生物学系,汉密尔顿,安大略,L8S 4K1,加拿大)
甲真菌病概述及其诊断最新进展
徐建平,Heather Yoell
(麦克马斯特大学生物学系,汉密尔顿,安大略,L8S 4K1,加拿大)
甲真菌病(Onychomycosis)是指真菌所致的指、趾甲感染,真菌包括皮肤癣菌、酵母菌及非皮肤癣菌的霉菌。甲真菌病虽然很少致命,但越来越成为重要的医疗问题,全球人口患病率约占5%。许多种真菌可引起甲真菌病,由于不同种类真菌的发病机制及对抗真菌药物敏感性的不同,治疗成功有赖于快速而准确的病原真菌诊断。因此,研究人员一直努力研究准确、快速而价廉的诊断技术,制药公司也在积极开发靶向抗真菌药物和高效的药物剂型。本文对甲真菌病进行简要概述,重点论述甲真菌病诊断的最新进展。
甲感染;毛癣菌;假丝酵母菌;诊断;物种识别
Funding:Research in my lab on medical fungi has been supported by McMaster University,Ontario Ministry of Research and Innovation,and Natural Sciences and Engineering Research Council of Canada.
1 Introduction
Onychomycosis refers to any fungal infection that affects the toenails or the fingernails.The infection may involve any component of the nail,including the nailmatrix,nail bed,and nail plate.Because of the unique structure and development of the nails,their infections are difficult to treat and eradicate and often require long-term therapy.The overall prevalence of onychomycosis is about 5%but ranges from 2-13%among geographic populations[1,2].For example,in a survey of 15,000 patients in four different clinics in Ontario,Canada,approximately 17%of the patientshad nails which were abnormal in appearance,and 8%had mycological evidence of onychomycosis[2]。People of all ages can have nail infections.However,the prevalence of onychomycosis can differ greatly in people of different age,sex,geographic origin,and occupation.Decades of epidemiological surveys have identified many risk factors for onychomycosis,including but not limited to:old age;compromised immunity from HIV infection,organ transplantation,and cancer treatments;wearing tight non-breathable footwear for extended periods of time;increased use of damp spaces such as locker rooms and gymnasiums by large groups of people;and working in certain occupations such as mining,lodging,and the military[1,3,4].
While typically not life threatening,onychomycosis can cause noticeable disfigurement of the nail and entire fingers and toes,as well as overall discomfort and pain.It can impact mobility and may result in occupational limitations.In addition,because nail infectionsmay be easily visible to others,significant psychosocial and emotional effects can develop and impact the overall quality of life for those suffering the infection[1,4].
2 Types ofOnychomycosis
Onychomycosis is traditionally classified into the following four types:distal subungual onychomycosis(DSO),white superficial onychomycosis(WSO),proximal subungual onychomycosis(PSO),and candidal onychomycosis (CO)[4].Patients may have a combination of these types.Total dystrophic onychomycosis refers to the most advanced infection of any of the four types.As the names indicate,different types differ in several aspects:the specific site of the nail that is infected,the route of infection,the overall pathogenesis,and in the case of CO,the infectious agents.
DSO is the most common form of onychomycosis.In DSO,the fungus spreads from plantar skin and invades the nail bed via the hyponychium.Mild inflammation develops,causing focal parakeratosis and subungual hyperkeratosis,both of which ultimately can result in onycholysis (detachment of the nail plate from the nail bed)and thickening of the subungual region.DSO is usually caused by the dermatophyte Trichophyton rubrum,although T.mentagrophytes,T.tonsurans,and E.floccosum also are known to be causative agents.
The second type,PSO,is less common than DSO.It occurs when fungi penetrate the nailmatrix via the proximal nail fold and colonize the newly formed portion of the proximal nail plate.The clinical presentation includes subungual hyperkeratosis,proximal onycholysis,leukonychia,and destruction of the proximal nail plate.The dominant agent for this type is T.rubrum,as for DSO.
The third type,WSO,occurs when certain fungi invade the superficial layers of the nail plate directly,causing distinctive opaque “white islands” on the external nail plate.The “white islands” can coalesce and spread as the disease progresses.The WSO-infected nail progressively becomes rough,soft,and crumbly.However,the underlying tissue is typically not infected.The most common etiologic agent for WSO is T.mentagrophytes.Several non-dermatophytemolds such as Aspergillus terreus and Fusarium oxysporum can also causeWSO.
The fourth type is CO.As the name suggests,themain difference between CO and the other three subtypes is the causal organism involved.Although species in several yeast genera (Table 1)can cause CO,themain agents are Candida albicans and Candida parapsilosis.Based on the site and symptoms ofinfection,CO can be further classified into the following four forms:(i)candida paronychia,marked by erythematous swelling of the proximal and lateral folds; (ii)candida granuloma,characterized by a thickened nail plate and paronychia;(iii)candida onycholysis,distinguished by separation of the nail plate from the nail bed;and(iv)total dystrophic onychomycosis(TDO),characterized by a thickened and dystrophic nail unit.As discussed earlier,TDO can be caused by dermatophytes and non-dermatophyte molds.
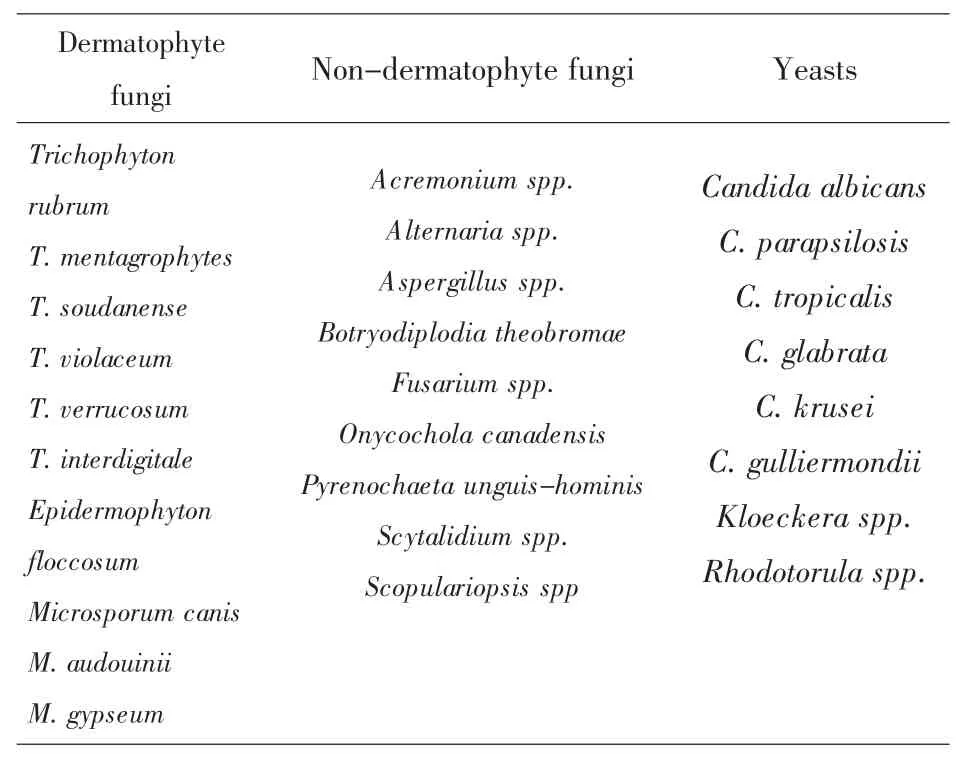
Table1 Common fungal species causing onychomycosis
3 CausalAgents ofOnychomycosis
Many fungal species can cause onychomycosis.These fungi are traditionally grouped into three types:dermatophytes,yeasts,and non-dermatophyte molds.Table 1 summarizes the commonly found specieswithin each of these groups.
At present,among the three groups of fungi,dermatophytes are responsible for the majority of onychomycosis in North America and Europe while non-dermatophyte molds and yeasts play relatively minor roles[1,4].For example,of the~1200 patients with mycological evidence of onychomycosis identified in the survey in Ontario,Canada,90.5%had infections caused by dermatophytes,whereas 7.8%had infections caused by non-dermatophyte molds and 1.7%by Candida species[2].Globally,T.rubrum is themost important cause of onychomycosis.However,there are significant differences in species distributions among geographic regions and study populations[3].For example,in two large-scale studies of patients with suspected or confirmed onychomycosis in China,both the spectrum of fungi and the prevalence of individual fungal species differed from each other and from those in North America and Europe.In a countrywide survey by Wu et al.[5]from May 2002 to November 2003,739 strains of fungal pathogens were isolated from the infected nails of 1084 patients.The species diversity was high and the relative frequencies of pathogens were T.rubrum(56.4%),C.albicans(10%),non-dermatophytemolds(mostly Aspergillus spp,~6%),C.parapsilosis(5.8%),T.mentagrophytes (5.5%),C.glabrata(5.1%),C.tropicalis(4.6%),M.gypseum(2.6%),C.krusei(1.9%),E.floccosum(1.5%),M.canis(0.3%),T.violaceum(0.1%),with other yeasts and dermatophytes accounting for the remaining 0.3%.In contrast,in a study by Chen et al.[6],631 fungal strains were isolated from 1036 clinically suspected cases of onychomycosis.Among these fungi,about 50%were yeasts belonging to genera Candida,Kloeckera,and Rhodotorula;29%were non-dermatophyte molds;and less than 22%were dermatophytes.The five most common organisms were T.mentagrophytes(11.25%),C.albicans (10.14%),Rhodotorula spp.(9.98%),T.rubrum (9.51%)and Aspergillus spp.(8.87%).
The large diversity of fungi that can cause onychomycosis and their differences in susceptibility to antifungal drugs make it essential to develop rapid and efficientmethods to identify the causative agent(s)of onychomycosis.In the following sections,we describe the diagnostics of onychomycosis,starting with the collection of samples.
4 Obtaining Samples for Diagnosis of Onychomycosis
Like the diagnosis of all microbial infections,correct diagnosis of onychomycosis requires a series of critical steps to be taken correctly.These include the initial diagnosis by the clinician,appropriate sampling and transportation ofmaterials for laboratory examination,and timely identification and interpretation using reproducible methods.For onychomycosis,different subtypes of infection require different procedures for obtaining the correct samples for analysis.Similar to the skin and mucosal surfaces,nails are constantly exposed to environmental microbes.As a result,avoiding contaminants during sampling is key to robust diagnosis.The first step is thus a thorough cleaning of the nail area with 70%of ethanol to remove contaminants on the surface of nails.
For patients with DSO,because the fungus invades the nail bed rather than the nail plate,the specimen needs to be obtained from the nail bed.The nail should be clipped short and the specimentaken from the nail bed as close to the cuticle as possible.When necessary and possible,material should also be obtained from the underside of the nail plate,with emphasis placed on sampling from the advancing infected edge closest to the cuticle.This is because this area ismost likely to contain viable fungiand least likely to contain contaminants.
To obtain samples from patients with PSO,because the fungus invades under the cuticle before settling in the proximal nail bed while the overlying nail plate remains intact,the healthy nail plate should be gently pared away with a scalpel blade.Materials should be obtained from the infected proximal nail bed as close to the lunula as possible.
To obtain samples from patients with WSO,since the infection affects the nail plate surface,a sterile blade can be used to scrape the white area of the nail and remove the infected debris for analysis.
For patients with CO,material should be taken from the proximal and lateral nail edges for examination.If CO is suspected,the lifted nail bed and/or the undersurface of the nail plate should be scraped.
5 In Vitro Identification of Etiological Agent(s)of Onychom ycosis
Severalmethods have been developed for identifying etiological agents for onychomycosis[1,4].These methods differ in their degree of complexity,time requirement,cost effectiveness,and detail of results with regard to the specific agents.Listed in Table 2 are the six methods for diagnosing onychomycosis that have been used in clinical microbiology research and/or diagnostic service labs.The advantages and disadvantages of each method are briefly described.In clinical diagnostics(including both microbial infections and chronic diseases),there are two important criteria for determining the effectiveness of any methods:sensitivity and specificity.Sensitivity measures how often a test turns out positive for samples known to be positive (i.e.samples are known by othermethods to be infected by a specific agent or group of agents).On the other hand,specificity refers to how often the test turns out negative for samples that are known to be negative.In clinicalmicrobiology testing,methods with both high sensitivity and high specificity are constantly sought.At present,sensitivity has been estimated for various methods used for identifying agents of onychomycosis(Table 3;references 17-31 are cited in this table).However,there is little information about the specificity for thesemethods and most cannot identify the agent(s)to the species level.
Current diagnosis is primarily based on direct microscopic examination of nail scrapings as well as macroscopic and microscopic identification of the infectious fungi in culture assay.Because of the sharp contrast it offers,direct microscopic examination of fluorescently-stained cells is by far the most sensitive for detecting rare fungal hyphae and spores in dermatological samples.Relatively few fungal cells,live or dead,may be sufficient for observation.However,directmicroscopic examinations do not provide genus or species identification of the infecting fungus.In addition,results from both microscopy and culture assays are often difficult to interpret.This is mainly due to the presence of contaminants belonging to various non-dermatophyte molds in most healthy and diseased nails.The current criterion for confirming non-dermatophytes as causal agents of onychomycosis requires recurrent isolations of the same non-dermatophytemold from diseased nails[7].Surveys have shown that the frequency of positive dermatophyte cultures from nail samples is typically only about 30%of the positive samples as identified by direct nail mycological examinations.Most fungi cultured in the survey were non-dermatophytemolds[8].
Of the microscopy and culture-based methods,the current clinical standard primarily uses a combination of KOH treatment followed by microscopic examination and culture for identifying and confirming agents of onychomycosis.However,as seen in Table 3,there is considerable variability in the diagnostic sensitivity obtained by various investigators for each of these methods.Overall,PAS and molecular methods appear to havemore consistent sensitivity among the investigators.Like most other methods,asignificant drawback of PAS is that it cannot identify the causative agent(s)to the genus or species level.Correct identification of the infectious agent in nail infections is essential as Fusarium spp.and nondermatophytemolds are often tolerant or resistant to systemic terbinafine and azole treatments[9].Below we discuss molecular methods that can identify the agents to species level.
Over the last few years,PCR-based molecular methods have become increasingly popular among epidemiologists interested in onychomycosis[10-14].These methods have primarily been based on sequence differences among themain causative agents at the ribosomal RNA gene cluster.Specific fragments including portions of the large ribosomal RNA subunit(or 28S rRNA),the small ribosomal RNA subunit(or 18S rRNA),and the internal transcribed spacer(ITS)region have been investigated.For several reasons,this gene cluster is highly effective for fungal identification,including the diagnosis of fungi causing onychomycosis[15].First,multiple copies of the gene cluster exist in individual cells,making direct PCR identification using a small number of cells from the host tissue possible (i.e.without going through culturing of themicrobes).Second,this gene cluster is overall highly conserved in both function and DNA sequence.As a result,both universal and group-specific PCR primers can be(and have been)designed and used.Some of these primers can be used to obtain DNA fragments from multiple divergent species,making it easy for labs to do the diagnosis.Third,a large GenBank database for this gene cluster already exists,making direct comparisons between sample sequences and reference sequences possible for species identification.Indeed,because of the high variability between many closely related species and low variability among strains within the same species,the ITS region has been recently suggested as a DNA barcode for fungi,making standardization and database construction and maintenance much easier and more convenient to use[16].Below we briefly describe a few representative studieswhich use a portion of the rRNA gene cluster for identification purposes.
In one study,Monod et al.[8]directly extracted DNA using a commercial kit after dissolving nail fragments in a Na2S solution.They then amplified part of the 28S rRNA gene by PCR with universal primers.The amplified fragments were then purified,digested by restriction endonucleases for preliminary screening,and sequenced.The obtained sequences were compared with those in GenBank database.The obtained sequence results were highly sensitive and specific to the individual species.In addition,mixed infections could be directly demonstrated in 10%of cases by RFLP analysis of PCR products,without sequencing.This type of analysis could be routinely conducted inmost research and clinical labs and the infectious agents could be identified in as few as 2 days.In contrast,identification to species level by othermethods(i.e.the culture-based methods)takes from at least a week (for fast growing fungi)to three weeks(for slow growing fungi).The PCR-RFLP and sequencing approach[8]wasmodified in a recent follow-up study that incorporated fluorescence-labeling and terminal restriction fragment length polymorphism analysis into PCR[13].The method,called PCR-TRFLP(Polymerase Chain Reaction–Terminal Restriction Fragment Length Polymorphism),is similar to amethod developed for the rapid identification of bacteria using polymorphisms at the 16S rRNA gene.However,to effectively use this approach,known standards of reference strains from most or all of the common fungal pathogens need to be established in the database and run together with patient samples.In the study by Verrier et al.[13],a total of two dermatophytes(T.rubrum and T.interdigitale)and 11 non-dermatophyte fungi(Fusarium oxysporum,F.solani,Aspergillus versicolor,A.flavus,Acremonium alternatum,A.strictum,Candida parapsilosis,C.albicans,Alternaria spp.,Penicillium citrinum,and Scopulariopsis brevicaulis)were used as references to establish the fluorescent banding pattern of the restriction enzyme digested PCR fragments.The PCR fragments were amplified using forward primer LSU1 (5’-GATAGCGMACAAGTA-
enced study.Themethods refer to:(i)KOH,potassium hydroxide treatment and microscopy;(ii)Culture,growth on medium conducive for fungal propagation;(iii)DTM,growth on dermatophyte testmedium;(iv)CW,Calcofluor white stain and microscopy;(v)Bx/PAS,nail plate biopsy using periodic acid-Schiff andmicroscopy;and (vi)Molecular,any tests that involve analyzing DNA from the sample,either indirectly by culturing first or directly by analyzing the nail DNA.GAGTG-3’)and reverse primer LSU2 (5’-GTCCGTGTTTCAAGACGGG-3’).LSU1 primerwas fluorescently labeled at the 5’-terminus with either red-ATTO565 or yellow-ATTO550.The PCR products were digested by several restriction endonucleases (AvaI,AvaII and StuI)before capillary electrophoresis and analysis by laser detection using a 3730 DNA Analyzer(Applied BioSystems).Using these reference species profiles,PCR-TRFLP allowed the identification of infectious agents in 74%(162/219)of cases in which the culture results were negative.Though the equipment,chemical and operational requirements are complex,the PCR-TRFLP method is conceptually simple and has the potential to be automated so that it could be routinely applied to the rapid diagnosis of a large number of clinical specimens in dermatology laboratories.
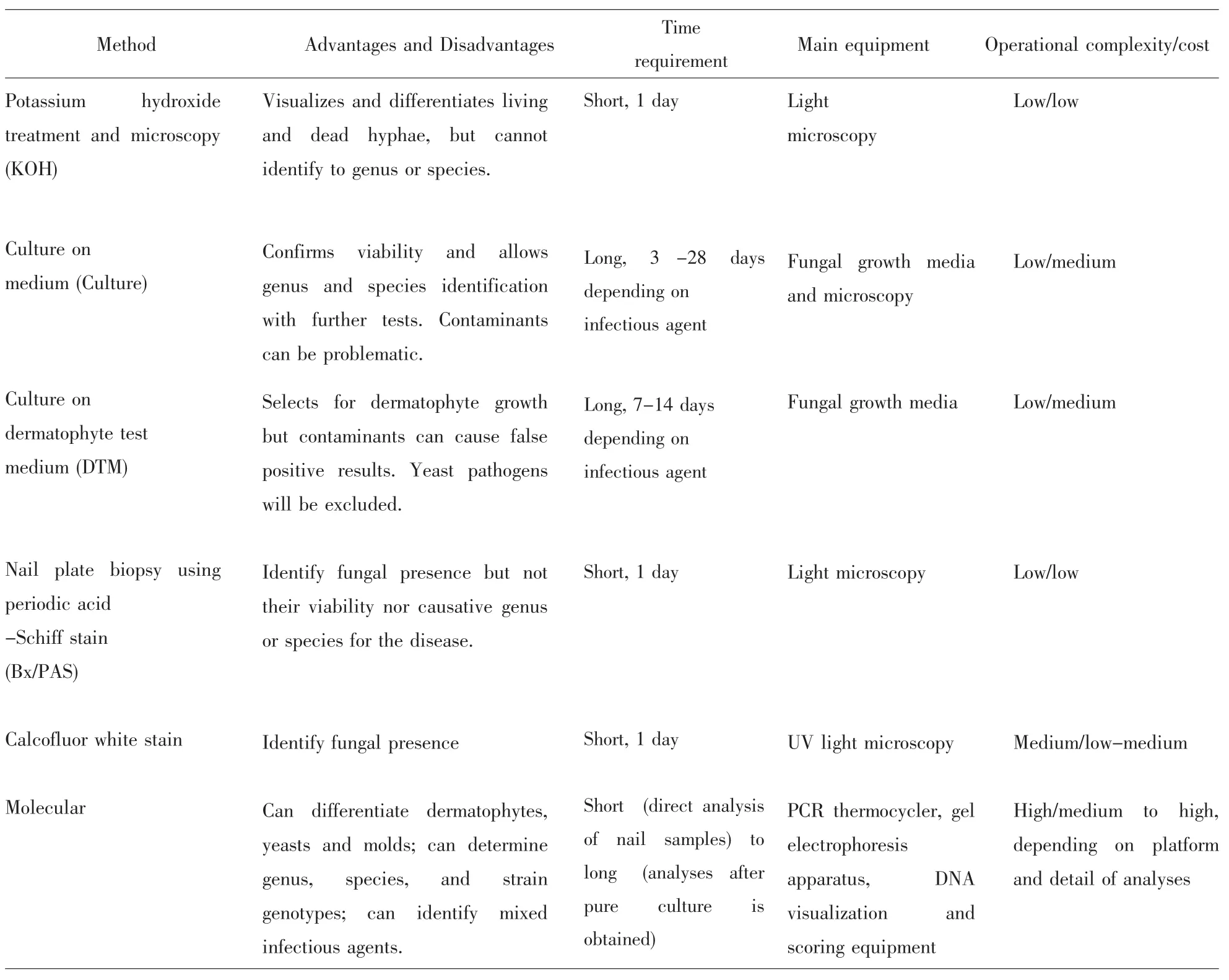
Table2 Methods for diagnosing onychomycosis and identifying infectious causal agent(s)
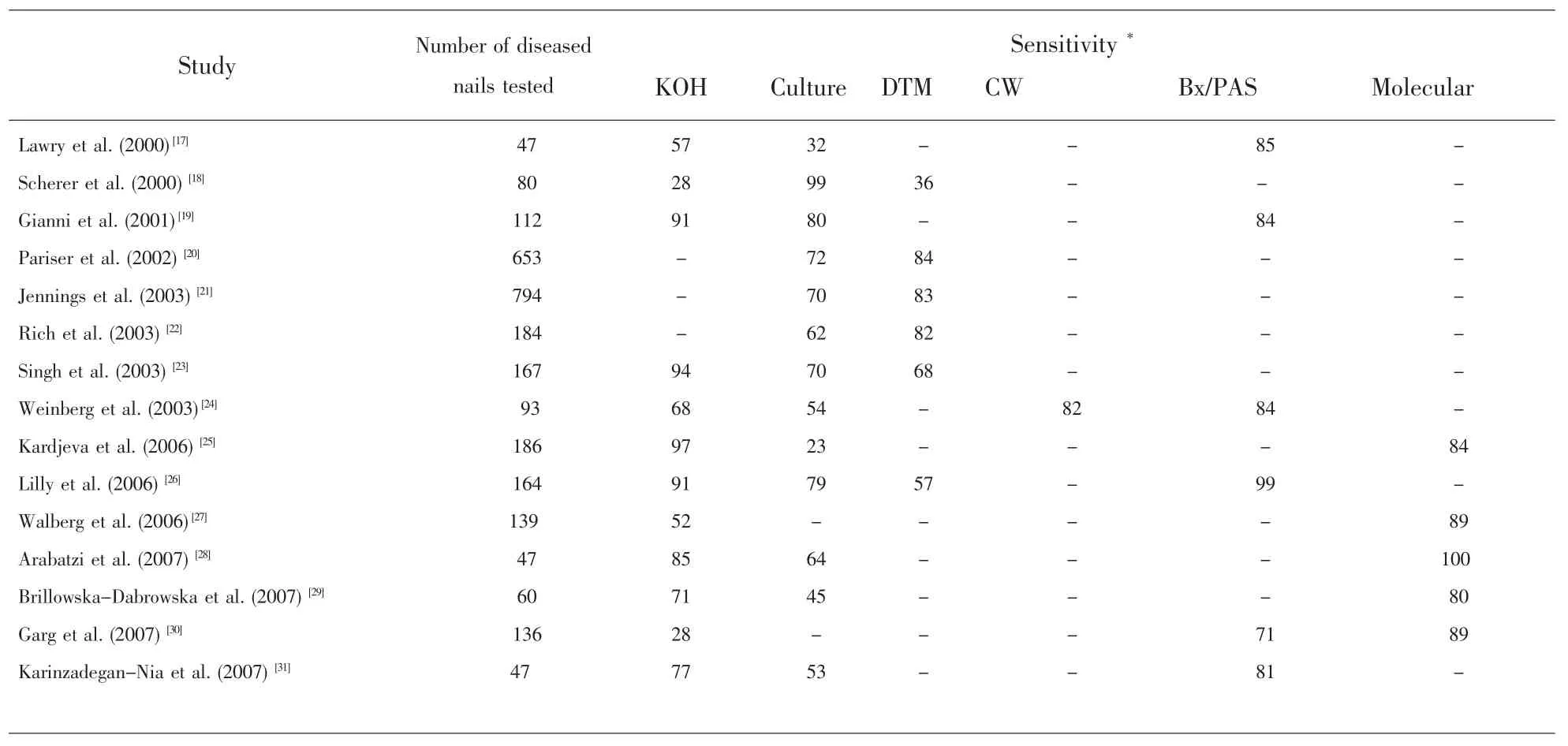
Table3 Sensitivity comparisons among sixmethods for diagnosing onychomycosis(Only studieswith more than 40 samples and which employed at least two differentmethods are included here)
Aside from DNA fragments in the rRNA gene cluster,other species-specific gene fragments have also been developed and could be used for targeted identification.For example,Brasch et al.[10]reported recently the successful application of a T.rubrum/T.violaceum specific PCR primer pair to directly amplify DNA from nail clippings and skin scalings.They used the following primers to amplify a fragment about 280 base pairs long that contains a GT-microsatellite repeat specific for strains of the closely related T.rubrum/Trichophyton violaceum clade:forward 5’-TGGTCTGGCCTTGACTGACC-3’,reversed 5’ –GTAAGGATGGCTAGTTAGGGGG-3’.Strains of other common fungi that cause onychomycosis,including the closely related T.mentagrophytes,failed to produce any PCR product by this method.
Nail infections are often long-term and correspondingly entail long-term therapy.Because longterm treatments of microbial infections are likely to lead to the emergence of drug resistance,targeted treatments are essential for successful outcomes.Sincemany fungal species have intrinsically different susceptibilities to different antifungal drugs,efficient and accurate identification of the causative agents is especially important to ensure appropriate treatment to eradicate infection.With decreasing cost and broad availability,molecular methods will likely become increasingly common in clinical settings.
6 Conclusions and Perspectives
Onychomycosis is among themost common fungal infections and impacts the overall quality of life for an increasingly large number of people,especially those of old age who are unable to care for themselves.A large diversity of fungi can cause onychomycosis.The causative agents can differ depending on the subtype of nail infection and geographic location of the patient.While current diagnostic methods rely on microscopic examination and fungal culturing,directmolecularmethods are being developed and thesemethods have shown high sensitivity and specificity.With decreasing cost and increasing DNA sequence information in databases,it is likely that DNA sequence-based methods will become the gold standard for diagnosis of nail infections in the foreseeable future.
[1]Scher RK.Onychomycosis:A significantmedical disorder[J].JAm Acad Dermatol,1996,35(3):S2-S5.
[2]Gupta AK,Jain HC,Lynde CW et al.Prevalence and epidemiology of onychomycosis in patients visiting physicians’ offices:a multicenter Canadian survey of 15,000 patients[J].J Am Acad Dermatol,2000,43(2):244–248.
[3]Seebacher C,Bouchara JP,Mignon B.Updates on the epidemiology of Dermatophyte infections[J].Mycopathologia,2008,166(5-6):335–352.
[4]Elewski BE. Onychomycosis: Pathogenesis, Diagnosis, and Management[J].Clin Microbiol Rev,1998,11(3):415–429.
[5]Wu SX,Liu WD,Guo NR,et al.Epidemiological survey of pathogenic fungi of onychomycosis in China[J].JClin Dermatol,2005,9(1):51-57.
[6]Chen LN,Wen H,Liao WQ,et al.The mycological analysis of
1036 cases of clinically suspected onychomycosis[J].Chinese JDermatovenereol,2004,4(1):10-16.
[7]Summerbell RC,Cooper E,Bunn U,et al.Onychomycosis:a critical study of techniques and criteria for confirming the etiologic significance of nondermatophytes[J].Med Mycol,2005,43(1):39-59.
[8]Monod M,Bontems O,Zaugg C,et al.Fast and reliable PCR/sequencing / RFLP assay for identification of fungi in onychomycoses[J].JMed Microbiol,2006,55(9):1211-1216.
[9]Baudraz-Rosselet F,Ruffieux C,Lurati M,et al.Onychomycosis insensitive to systemic terbinafine and azole treatments reveals non-dermatophytemoulds as infectious agents[J].Dermatol,2010,220(2):164-168.
[10]Brasch J,Beck-Jendroschek V,Glaser R.Fast and sensitive detection of 436 Trichophyton rubrum in superficial tinea and onychomycosis by use of a direct polymerase chain reaction assay[J].Mycoses,2011,54(5):e313-317.
[11]Ebihara M,Makimura K,Sato K,et al.Molecular detection of dermatophytes and nondermatophytes in onychomycosis by nested polymerase chain reaction based on 28S ribosomal RNA gene sequences[J].Br JDermatol,2009,161(5):1038-1044.
[12]Yang G,Zhang M,LiW.Direct species identification of common pathogenic dermatophyte fungi in clinical specimens by seminested PCR and restriction fragment length polymorphism[J].Mycopathologia,2008,166(4):203-208.
[13]Verrier J,Pronina M,Peter C,et al.Identification of infectious agents in onychomycoses by Polymerase Chain Reaction-Terminal Restriction Fragment Length Polymorphism[J].J Clin Microbiol,2012[published online ahead of print on 14 December 2011].doi:10.1128/JCM.05164-11
[14]Gupta AK,Zaman M,Singh J.Fast and sensitive detection of Trichophyton rubrum DNA from the nail samples of patients with onychomycosis by a double-round polymerase chain reactionbased assay[J].Br JDermatol,2007,157(4):698-703.
[15]Xu J.Fundamentals of fungalmolecular population genetic analyses.In Evolutionary Genetics of Fungi(ed.J.Xu),2005,Horizon Scientific Press,England.Pp.87-116.
[16]Schoch CL,Seifert KA,Huhndorf S,and the Fungal Barcoding Consortium.The nuclear ribosomal internal transcribed spacer(ITS)region as a universal DNA barcodemarker for Fungi[J].Proc Natl Acad Sci(USA),2012,published ahead of print March 27,2012,doi:10.1073/pnas.1117018109
[17]Lawry MA,Haneke E,Strobeck K,et al.Methods for diagnosing onychomycosis:a comparative study and review of the literature[J].Arch Dermatol,2000,136(9):1112-1116.
[18]Scherer WP,Kinmon K.Dermatophyte testmedium culture versus mycology laboratory analysis for suspected onychomycosis:a study of 100 cases in a geriatric population[J].JAm Podiatr Med Assoc.2000,90(9):450-459.
[19]Gianni C,Morelli V,Cerri A,et al.Usefulness of histological examination for the diagnosis of onychomycosis[J].Dermatol,2001,202(4):283-288.
[20]Pariser D,Opper C.An in-office diagnostic procedure to detect dermatophytes in a nationwide study of onychomycosis patients[J].Manag Care,2002,11(1):43-48,50.
[21]Jennings MB,Rinaldi MG.Confirmation of dermatophytes in nail specimens using in-office dermatophyte testmedium cultures[J].J Am Podiatr Med Assoc,2003,93(3):195-202.
[22]Rich P,Harkless LB,Atillasoy ES.Dermatophyte test medium culture for evaluating toenail infections in patientswith diabetes[J].Diabetes Care,2003,26(5):1480-1484.
[23]Singh S,Beena PM.Comparative study of different microscopic techniques and culturemedia for the isolation of dermatophytes[J].Indian JMed Microbiol,2003,21(1):21-24.
[24]Weinberg JM,Koestenblatt EK,TutroneWD,et al.Comparison of diagnostic methods in the evaluation of onychomycosis[J].J Am Acad Dermatol,2003,49(2):193-197.
[25]Kardjeva V,Summerbell R,Kantardjiev T,et al.Forty-eight-hour diagnosis of onychomycosiswith subtyping of Trichophyton rubrum strains[J].JClin Microbio,2006,44(4):1419-1427.
[26]Lilly KK,Koshnick RL,Grill JP,et al.Cost-effectiveness of diagnostic tests for toenail onychomycosis:a repeated-measure,singleblinded,cross-sectional evaluation of 7 diagnostic tests[J].JAm Acad Dermatol,2006,55(4):620-626.
[27]Walberg M,Mork C,Sandven P,et al.18S rDNA polymerase chain reaction and sequencing in onychomycosis diagnostics[J].Acta Derm Venereol,2006,86(3):223-226.
[28]Arabatzis M,Bruijnesteijn van Coppenraet LES,Kuijper EJ,et al.Diagnosis of common dermatophyte infections by a novelmultiplex real-time polymerase chain reaction detection/identification scheme[J].Br JDermatol,2007,157(4):681-689.
[29]Brillowska-Dabrowska A,Saunte DM,Arendrup MC.Five-hour diagnosis of dermatophyte nail infections with specific detection of Trichophyton rubrum[J].JClin Microbiol.2007,45(4):1200-1204.
[30]Garg J,Tilak R,Garg A,et al.Rapid detection of dermatophytes from skin and hair[J].BMCRes Notes,2009,2:60.
[31]Karimzadegan-Nia M,Mir-Amin-Mohammadi A,Bouzari N,et al.Comparison of direct smear,culture and histology for the diagnosis of onychomycosis[J].Australia JDermatol,2007,48(1):18-21.
Onychomycosis:an overview with a focus on diagnosis
XU Jianping,Heather Yoell
(Departmentof Biology,McMaster University,Hamilton,Ontario,L8S 4K1,Canada)
Onychomycosis refers to nail infections by fungi.It includes infections of both fingernails and toenails by any fungi,including dermatophytes,yeasts,and non-dermatophyte molds.While rarely fatal,onychomycosis is becoming an increasingly importantmedical problem.About 5%of the global population suffers from onychomysis.A large number of fungal species can cause this disease.Because causal agents of onychomycosis differ in their pathogenesis and susceptibility to antifungal drugs,successful treatments require rapid and accurate diagnosis of the infectious agent(s).As a result,there have been significant efforts among researchers to develop accurate,rapid and inexpensive diagnostic techniques,and among pharmaceutical companies to develop targeted antifungal drugs and efficient drug formulations for treatments.In thismini-review,we provide a brief overview of onychomycosis,with a focus on recent advances in the diagnosis of onychomycosis.
Nail infections;Trichophyton;Candida;Diagnosis;Species identification
Name:XU Jianping;Date of birth:November 5,1965;Sex:Male;Ethnic group:Han Chinese;Place of birth:Zhangshu,Jiangxi Province,P.R.China;Current Position:Professor;University attended for the highest degree:University of Toronto(Ph.D.);Major:Mycology;Current area of research:Fungal Genetics,Epidemiology,and Evolutionary Biology
Address for Correspondence:Dr XU Jianping,Department of Biology,McMaster University,1280 Main StW.,Hamilton,Ontario,L8S 4K1,Canada;Phone number:001-905-525-9140 extension 27934;Email:jpxu@mcmaster.ca
R756.4,R379,R519
A
1674-1129(2012)02-0111-08
10.3969/j.issn.1674-1129.2012.02.003
