Regulation of Floral Organ Identity in Arabidopsis by Ectopic Expression of OsMADS58
2012-03-01WangYanmeiYanDaweiZhangYingyingLiJingandCangJing
Wang Yan-mei,Yan Da-wei,Zhang Ying-ying,Li Jing,and Cang Jing*
1 College of Life Sciences,Northeast Agricultural University,Harbin 150030,China
2 Institute of Plant Physiology and Ecology,Shanghai Institutes for Biology Sciences,Chinese Academy of Sciences,Shanghai 200032,China
Introduction
The principle to explain the mechanism of floral organ specification has developed from ABC model to ABCDE model (Coen and Meyerowitz,1991;Theissen,2001).It is shown that the expressions of five classes of genes are required for the formation of different floral organs,that is,class-A for the sepals,class-A,B and E for the petals,class-B,C and E for the stamens,class-C and E for the carpels and class-D and E for the ovules.Most of the elements in ABCDE model are MADS-box genes,and such genes have been found in numerous eukaryotes (Shore and Sharrocks,1995;Ng and Yanofsky,2001).As the nomenclature indicated that all the MADS-box proteins contained a highly conserved MADS-box domain with DNA binding function.
AGAMOUS (AG) is the class-C MADS-box gene in Arabidopsis which participates in the formation of stamens and carpels as described in the ABCDE models.Except the MADS-box domain,it also contains a moderately conserved K-box domain.In the AG mutant,both the stamens and carpels are replaced by petals in a new flower (Yanofsky et al.,1990;Bowman et al.,1991).Inversely,over-expression of AG in Arabidopsis led to the production of abnormal floral organs,and most of the transformants present the first whorl organs with stigmatic papillae at the tips and ovules on the margins.Some flowers have narrow and short staminoid petals or even lack the second whorl organs (Mizukami and Ma,1992).AG regulates the flower development through interacting with many other genes such as APETALA1,APETALA3,LEAFY and TERMINAL FLOWER1 (Parcy et al.,2002).APETALA2 and miR172 also participate in the AG pathway (Wollmann et al.,2010).
The AG homologues have been found in many organisms including maize (Schmidt et al.,1993),cucumber (Kater et al.,1998),rice (Yamaguchi et al.,2006) and so on.In rice,the AG subfamily members OsMADS3 and OsMADS58 belong to the C-lineage,whereas OsMADS13 and OsMADS21 belong to the D-lineage according to the phylogenetic relationship (Yamaguchi et al.,2006).The OsMADS58-silenced plants form florets which reiterate a set of floral organs consisting of lodicules,stamens/ectopic lodicules,and abnormal carpel organs indicate that OsMADS58 are crucial for the carpel morphogenesis and floral meristem determinacy.Meanwhile,OsMADS58 affects the lodicules and stamens identity.
Other than the RNA-silenced plants,no more researches about the OsMADS58 had been reported so far.In this study,OsMADS58 was ectopicly expressed in Arabidopsis resulting in the alterations of floral organ formation.
Materials and Methods
Plant growth condition
The rice variety Nipponbare (japonica) was grown in the field under natural conditions.The Arabidopsis variety Columbia-0 (Col-0) was used for genetic transformation and therefore the wide type in comparison.The seeds were surface-sterilized and stratified at 4℃ for 3 days on MS medium.After germination,plants were grown in soil at 22℃ in long day conditions (16 h-light/8 h-dark photoperiod).Plant organs were sampled at proper time and stored in low tempreture refrigerator (–80℃).
Sequence analysis
The accession numbers of the protein sequences were NP_567569 (AG) and BAE54300.1 (OsMADS58).The amino acid sequences were aligned by ClustalW and processed by BOXSHADE (http://www.ch.embnet.org/software/BOX_form.html).
Isolation of OsMADS58 and construction of vectors
Total RNA was isolated from Nipponbare flowers with Trizol reagent (Invitrogen).1 μg total RNA was used for the cDNA synthesis by the SuperScript reverse transcriptase (Invitrogen).The primers that amplified the OsMADS58 coding sequence (accession number:AB232157) were M58-F(GGATCCATGCACATAT ACAAAGAGCAG) and M58-R (GGTACCTTATCT TTCAT CTGACATAAAGG) with restriction sites Bam HⅠand KpnⅠ,respectively.PCR was performed with a 3 min 95℃ denaturation step,followed by 35 cycles of 30 s denaturation at 95℃,30 s annealing at 60℃ and 30 s extension at 72℃,and a final extension period of 10 min using Trans-StartFastPfu DNA Polymerase (TransgGen Biotech,China).The amplified cDNA was cloned into pGEM-Teasy vector (Promega) and sequenced.The sequencing-checked plasmid was then digested by Bam HⅠ and KpnⅠ and cloned into the downstream of the UBIQUITIN (UBI) promoter in the binary vector pUN1301 to form plasmid UBI::OsMADS58.
Arabidopsis transformation and selection
The UBI::OsMADS58 recombinant plasmid was introduced into Agrobacterium tumefaciens GV3101 to transform Arabidopsis thaliana (Columbia-0) by the floral dip method.The transgenic plants were screened on the MS agar plates containing hygromycin (25 μg • mL-1).Resistant seedlings were transferred to soil for growth.Transformed plants were identified by the GUS staining and the RT-PCR.T1,T2and T3progenies were selected according to their hygromycin resistance and used for further experiments.
Scanning electron microscopy
Various floral organs were fixed overnight at 4℃ in FAA and dehydrated in a graded ethanol series.The samples were then critical-point dried,mounted,coated with gold and visualized by the scanning electron microscope (JSM6360LV).
RT-PCR and real-time PCR assays
Total RNA of Arabidopsis leaves and flowers were isolated and reversely transcribed as described above.The real-time PCR was performed using Eppendorf realplex2system.Reaction system was prepared in 20 μL solution,containing 2 μL cDNA,0.4 μmol • L-1of each primer,and 1× SYBR Green RCR Master Mix (TaKaRa).Relative mRNA level was calculated as 2-ΔΔCT,and the expression of S18 and ACTIN2 were used as controls.The primers used for RT-PCR and real-time PCR were as the followings:


Results
Sequence alignment of AGAMOUS and OsMADS58
AGAMOUS encodes a transcription factor with 252 amino acids containing a MADS-box and a K-box domain.The MADS-box contains DNA binding site,putative phosphorylation site and dimerization interface.The K-box is a possible coiled-coil structure.The overall 1181-nucleotide cDNA of OSMADS58 contains a complete 819-nucleotide coding sequence(CDS),which encodes a protein with 271 amino acids.Amino acid sequence alignment of AG and OsMADS58 is displayed in Fig.1.
It is shown that the MADS-box domain was more conserved than the K-box domain.There were 17 more amino acids at the N-terminal of the MADS domain in OsMADS58,but only one non-conserved substitution in other parts of this domain.These indicated the conserved function for DNA binding between two genes,while the function of the K-box was not explicit.

Fig.1 Amino acid sequences alignment of AGAMOUS and OsMADS58
Alterations of floral organs in UBI::OsMADS58 transgenic Arabidopsis
A binary vector carrying UBI::OsMADS58 was constructed and transformed into Arabidopsis (Col-0) by Agrobacterium.The phenotypes of flowers were analyzed in the T1and T2generations.
A total of 49 T1lines were obtained by kanamycin selection and the GUS staining.The ectopic expression of OsMADS58 in transgenic plants was confirmed by RT-PCR using seedlings,leaves and flowers,which indicated that the UBIQUITIN promoter worked successfully.The expression of five transformed lines are shown in Fig.4A,and no expression of OsMADS58 was found in wild type.Different kinds of abnormal flowers were found in the T1plants.Some plants had incomplete petals which were shorter and narrower than those of the wild type (Fig.2A,B and C),while the numbers of the floral organs of each whorl were normal in these plants.Some plants had flowers without any petals but more stamens (Fig.2B).Scanning electron micrograph showed that the petals were replaced by stamens more or less (Fig.2D).Other plants produced ball-like flowers without petals and less stamens,which could not open.Such plants bear the most significant floral defects including bended sepals with stigmatic papillae at the apex and ovule at the margin (Fig.2B and E),and most of the seeds produced by these plants were sterile.
As the bended sepals had the characteristics of carpels,we speculated that they had become a mosaic organ.To verify this transformation,we observed the morphology of epidermal cells.Scanning electron micrographs showed that the sepals had longer cells with wavy ridges through them which did not exist in the carpel cells (Fig.3A and B).The sepal cells of short-and-narrow-petal flowers were normal (Fig.3C),but most of the surface cells on the carpel-like sepals resembled carpel cells (Fig.3D) suggesting that some sepal cells had developed into carpel cells.

Fig.2 Light and scanning electron micrographs of Arabidopsis flowers
Effects of different OsMADS58 expression levels on floral alterations
To investigate the effect of OsMADS58 expression level on the alteration of floral organ identity,we carried out real-time PCR assays in three types of plants according to the phenotypes described above,and the W,M and S were used for short to denote weak,moderate and strong phenotypes hereafter.As a result,the expression levels of M and S types were approximately twice and five times higher than the mean level of W type separately (Fig.4B).The quantitative results coincided with the separate phenotype as shown by the flower defects,that was,the severer the more defects it had,the higher the expression level was.There were no obvious defects in the low-expressed plants,except for the shorter petals.However,the petals were lacked,when the expression increased,and the sepals,the petals and the stamens were all affected in the highest expression plants.

Fig.3 Scanning electron micrographs of sepal and carpel surface morphology
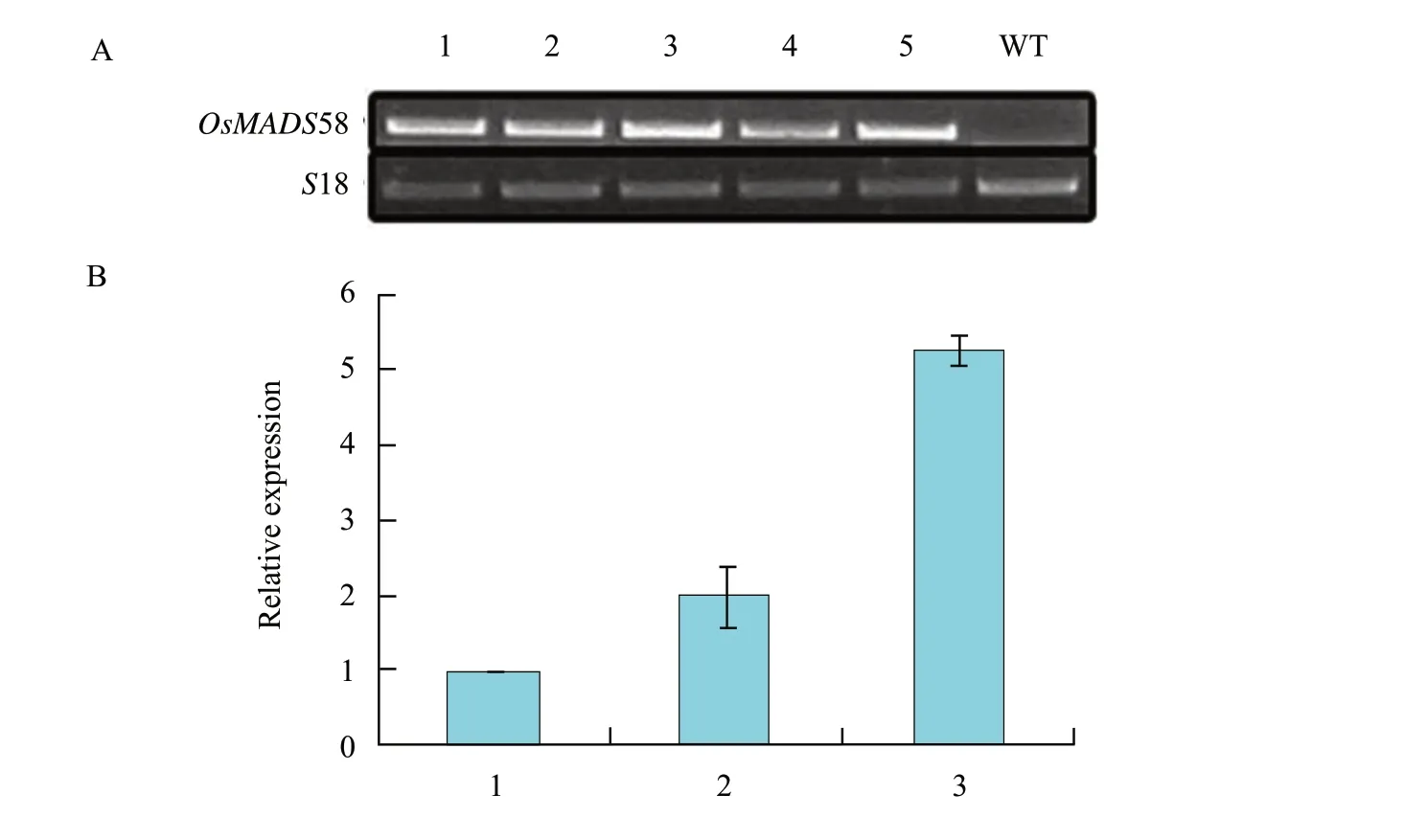
Fig.4 RT-PCR and real-time PCR assay
Other phenotypes of the UBI::OsMADS58 transgenic plants
Besides the alterations in flowers,the OsMADS58 over-expression plants showed different flowering time and leaf morphology.All transgenic plants bolted and flowered earlier than wild type (Fig.5B),especially for the strong phenotype plants.By RT-PCR,we found FLC was down-regulated and SOC1 was upregulated,which were both important regulators in the flowering pathway.This suggested that the redundant OsMADS58 could affect some flowering factors directly or indirectly to promote flowering in Arabidopsis (Fig.5A).The leaves except the cotyledons were curled upwards in varying degrees (Fig.5C).The rosette leaves were smaller and the total number was less than wild type in the whole life.Those suggested that the ectopic expression of OsMADS58 had pleiotropic effects which could both affect the flower formation and leaf development,and it might interact with members of different pathways to regulate the vegetative and reproductive development.

Fig.5 Other phenotypes of the UBI::OsMADS58 transgenic plants
Discussion
In the development of floral organ in Arabidopsis,the expression of the AG is regulated strictly by many factors to fuction properly.It is activated or repressed in different whorls of flowers and different development stages.Therefore,the change of the AG level will cause defects in flower development.In our study,OsMADS58,the rice homologue of the AG,was ectopicly expressed in Arabidopsis.It was shown that the floral organs were altered to different degrees in transgenic plants,according to the elevated levels of OsMADS58 expression.These suggested that OsMADS58 had dosage effect on floral organs development.In the first whorl floral organ development,overexpression of OsMADS58 affected the cell differentiation resulting in the formation of carpel-like cells.In the second whorl,the function of B-class genes may be concealed by OsMADS58,which led to the decrease or loss of petals.
The flowering time of Arabidopsis were regulated by environment conditions and internal factors.The UBI::OsMADS58 transgenic plants flowered earlier than wild type.We speculated that OsMADS58 might repress the FLC in the flowering pathway by unknown ways according to the down-regulation of the FLC.The regulations of leaf morphogenesis in Arabidopsis by auxin (Mattsson et al.,2003;Scarpella et al.,2010),CURLY LEAF (Kim et al.,1998) and microRNA (Rodriguez et al.,2010) had been reported.In the AG over-expression Arabidopsis,the lateral margins of leaves rolled up toward to the midrib (Mizukami and Ma,1992).This phenotype was also found in the UBI::OsMADS58 transgenic plants,which suggested that the redundant transcripts of OsMADS58 disrupt the normal process of leaf development.All of these could be clues for the further functional researches of OsMADS58 in rice.
Conclusions
The MADS-box sequences of OsMADS58 and the AG were highly conserved.OsMADS58 was ectopicly expressed in Arabidopsis in our study.The phenotypes of flowers in transgenic plants were very similar with the AG over-expression plants.The results verified the importance of the MADS domain to the function of the MADS-box proteins again and indicated that OsMADS58 could function like the AG in Arabidopsis,which would help us to investigate its role in rice flower development and understand the evolutionary relationship between monocots and dicots.
Bowman J L,Smyth D R,Meyerowitz E M.1991.Genetic interactions among floral homeotic genes of Arabidopsis.Development,112(1): 1-20.
Coen E S,Meyerowitz E M.1991.The war of the whorls: genetic interactions controlling flower development.Nature,353(6339): 31-37.
Dennis E,Bowman J L.1993.Flower development: manipulating floral organ identity.Curr Biol,3(2): 90-93.
Kater M M,Colombo L,Franken J,et al.1998.Multiple AGAMOUS homologs from cucumber and petunia differ in their ability to induce reproductive organ fate.Plant Cell,10(2): 171-182.
Kim G T,Tsukaya H,Uchimiya H.1998.The CURLY LEAF gene controls both division and elongation of cells during the expansion of the leaf blade in Arabidopsis thaliana.Planta,206(2): 175-183.
Mattsson J,Ckurshumova W,Berleth T.2003.Auxin signaling in Arabidopsis leaf vascular development.Plant Physiol,131(3): 1327-1339.
Mizukami Y,Ma H.1992.Ectopic expression of the floral homeotic gene AGAMOUS in transgenic Arabidopsis plants alters floral organ identity.Cell,71(1): 119-131.
Ng M,Yanofsky M F.2001.Function and evolution of the plant MADS-box gene family.Nat Rev Genet,2(3): 186-195.
Parcy F,Bomblies K,Weigel D.2002.Interaction of LEAFY,AGAMOUS and TERMINAL FLOWER1 in maintaining floral meristem identity in Arabidopsis.Development,129(10): 2519-2527.
Rodriguez R E,Mecchia M A,Debernardi J M,et al.2010.Control of cell proliferation in Arabidopsis thaliana by microRNA miR396.Development,137(1): 103-112.
Scarpella E,Barkoulas M,Tsiantis M.2010.Control of leaf and vein development by auxin.Cold Spring Harb Perspect Biol,2(1): a001511.
Schmidt R J,Veit B,Mandel M A,et al.1993.Identification and molecular characterization of ZAG1,the maize homolog of the Arabidopsis floral homeotic gene AGAMOUS.Plant Cell,5(7): 729-737.
Shore P,Sharrocks A D.1995.The MADS-box family of transcription factors.Eur J Biochem,229(1): 1-13.
Theissen G.2001.Development of floral organ identity: stories from the MADS house.Curr Opin Plant Biol,4(1): 75-85.
Wollmann H,Mica E,Todesco M,et al.2010.On reconciling the interactions between APETALA2,miR172 and AGAMOUS with the ABC model of flower development.Development,137(21): 3633-3642.
Yamaguchi T,Lee D Y,Miyao A,et al.2006.Functional diversification of the two C-class MADS box genes OSMADS3 and OSMADS58 in Oryza sativa.Plant Cell,18(1): 15-28.
Yanofsky M F,Ma H,Bowman J L,et al.1990.The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors.Nature,346(6279): 35-39.
杂志排行
Journal of Northeast Agricultural University(English Edition)的其它文章
- Regulatory Network of Transcription Factors in Response to Drought in Arabidopsis and Crops
- Comparison of Net Photosynthetic Rate in Leaves of Soybean with Different Yield Levels
- Multiplex PCR System Optimization with Potato SSR Markers
- Analysis on Combining Ability for Characters of Male Sterile Lines in Rapeseed (Brassica napus L.)
- Study on Mutant Induction of Gladiolus by in vitro Culture of Petals
- Research on Tobacco Transformation of Vacuolar H+-ATPase Subunit c Gene from Iris lacteal
