Plant DNA extraction for PCR: A simple and environment-friendly approach
2010-07-15ZHOUQianLILanzhiZHUHongjianGAOBidaCollegeofBioSafetyScienceandTechnologyHNAUChangsha410128China
ZHOU Qian,LI Lan-zhi,ZHU Hong-jian,GAO Bi-da(College of Bio-Safety Science and Technology,HNAU,Changsha 410128,China)
1 Introduction
PCR has revolutionized the genetic and mole- cular biology studies since its introduction. However,successful amplification with reproducible results in PCR,depends upon the quality and quantity of template DNA. Numerous DNA extraction procedures for isolation genomic DNA from various plant mater- ials have been published,including the conventional CTAB method[1],SDS method[2]and its modifi- cations[3-4]. These protocols use phenol- chloroform extraction to remove protein,which is hazardous both to environment and operator’s health especially when hundreds or even thousands of plant samples need to be analyzed such as in marker- assisted breeding and high-resolution mapping studies. Some hazardous- reagent-free DNA extraction protocols have been developed such as enzymatic methods[5],salt extraction method[6]and so on. However,these methods are tedious and therefore not time efficient when it comes to large-scale DNA extraction. Others high-throughput,single-step DNA extraction methods described for PCR-based markers[7-8]are undoubtedly rapid and avoid the use of hazardous chemicals. But the crude extracts containing many contaminating substances were easily degraded and often interfere with further processing of PCR. Kotchoni and Gachomo[9]have successfully developed an enviroment- friendly DNA extraction procedure to isolate DNA of Arabidopsis thaliana. However,this method is unsuitable for most plant materials containing high quantities of poly- saccharides,polyphenolics,tannins and pigments. Here we developed a new simple DNA extraction method for plant materials without using hazardous regeants,which is safe and time-efficient especially suitable for large-scale DNA extraction and can even be easily performed by non-specialists.
2 Materials and methods
2.1 Plant material
Leaves of rice,cotton,rape,oat,maize,wheat,tobacco,sweet potato,potato,tobacco,peanut and soybean obtained from the experimental field of Hunan Agriculture University.
2.2 Reagents
Extraction Buffer:2%(w/v) CTAB,1% (w/v) SDS,2%(w/v) PVP,100 mmol/L Tris-Cl (pH8.0),20 mmol/L EDTA (pH8.0),1.4 mol/L NaCl (The buffer is emulsion which will be hierarchical on standing,mix well before use).
Ethanol,AR grade .
2.3 Primer
The primers for plant species are ITS4,5′-TCC TCCGCTTATTGATATGC-3′ and ITS5,5′-G GAAG TAAAAGTCGTAACAAGG -3′.
2.4 DNA extraction protocol
Put 100 mg leaf tissue in an eppendorf tube and grind the tissue with a homogenizing pestle in liquid nitrogen. Add 600 μL of the boiling preheated extraction buffer and vortex for 5~10 s to mix thoroughly,and then incubate at a boiling water bath for 15 min and invert tubes every 5 min to allow mixing. Centrifuge at 15 kg for 10 min at room temperature. Carefully transfer the middle layer of aqueous solution to a new tube. Add double volume alcohol and mix gently by inversion the tube several times,place at room temperature for 2 min. Centrifuge tubes at 15 kg for 30 s at room temperature. Discard the supernatant and resuspend the pellet in 500 μL 70% alcohol. Centrifuge eppendorf tubes at15 kg for 1 min at room temperature. Discard the supernatant and air-dry the pellets at room temperature. Dissolve the pellets in 100 μL 0.1 TE for 1 hr at room temperature or 4 ℃ over night.
2.5 PCR amplification
The total PCR reaction volume was 25 μL containing 2.5 μL 10× Qiagen PCR Buffer,1 μL DNA template,1.5 mmol/L MgCl2,0.1 mmol/L each dATP,dCTP,dGTP and dTTP,0.1 μmol/L each forward and reverse primer,and 0.5 U Taq DNA polymerase. Amplifications were carried out with thermal cycles (MJ Research,waltham,MA,USA). The initial step of 95 ℃ for 5 min was followed by 35 cycles of 95 ℃ for 30 s,55 ℃ for 30 s,and 72 ℃ for 45 s,and 1 cycle of 10 min at 72 ℃.
3 Results and discussion
The quantity and quality of DNA extracted by this method was highly variable among the 12 plants species (Table 1). The A260/A280ratio of DNA was 2.0~2.2,all higher than 1.8,which indicated insigni- ficant levels of contaminating proteins but much RNA content without RNase treatment. However,the presence of RNA did not hinder any later PCR amplification and was not considered a problem. The A260/A230ration of most DNA samples was lower than 2.0 especially those DNA prepared from mature leaves,which illuminated there are some contami- nating such as carbohydrate or salt. But the tiny contamination did not interfere with further pro- cessing of PCR,using these DNA as template we successfully amplified ribosomal DNA with the primer ITS4 and ITS5 from all 12 plants examined. All the products gave a clear,sharp and reproducible band on agarose gel,only difference in the intensity of the band was observed (Figure 1),which may be due to the different template concentration used for the PCR reaction.
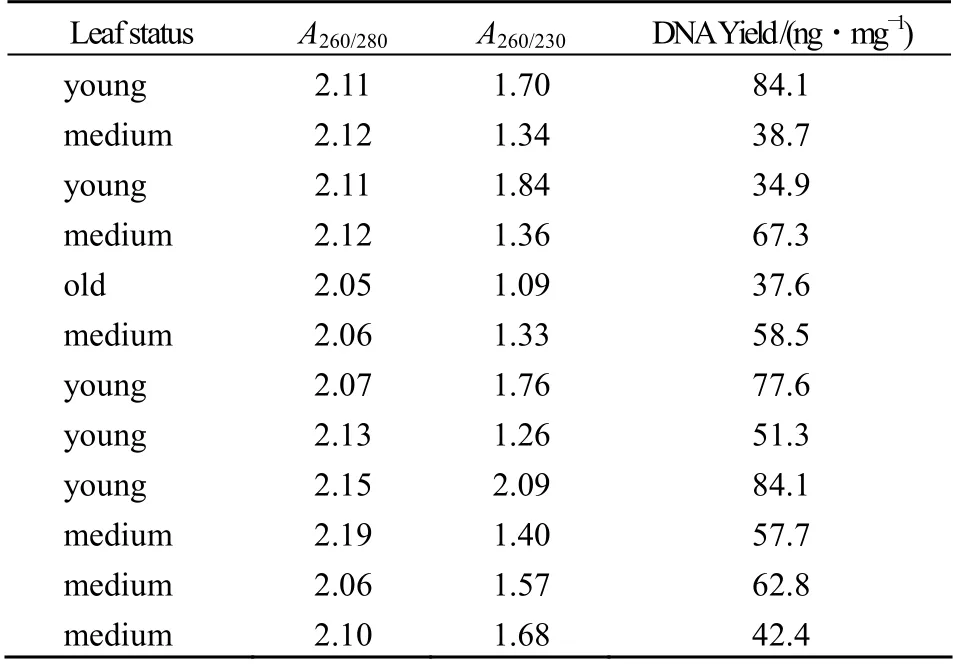
Table 1 Quantity and quality of DNA extracted by our method表1 DNA 的产量与质量
We are interested in plant breeding for disease resistance,which always involves marker-assisted breeding and high-resolution mapping studies. Therefore,an amount of DNA samples suitable for PCR analysis need to be prepared in a short time. Using this method,more than 1 700 DNA samples of rice leaves were successfully extracted used for map-based cloning of a lesion mimic gene (data not show). One person can extract more than 100 samples per day with minimal average cost of consumables per sample. Some DNA extraction methods based on 96-well microtitre extract more DNA samples per day. However,using 96-well microtitre increase the risk of cross contamination during the extracting procedure. Cross contamination is unacceptable if the DNA extracted is to be used for PCR-based marker analysis,such as SSR and RAPD. Compared with other DNA extraction protocols,the main advantage of our method is extracting high-quality DNA without use of hazardous chemicals. When hundreds and thousands DNA samples need to be prepared with phenol- chloroform extraction,the used hazardous chemicals without treatment will put hard pressure upon the environment.
The procedure described here is simple,safe and reliable,which works well for extracting high quality DNA suitable for PCR from all plant leaves tested and should be widely applicable for analysis of large population from virtually all plant species.
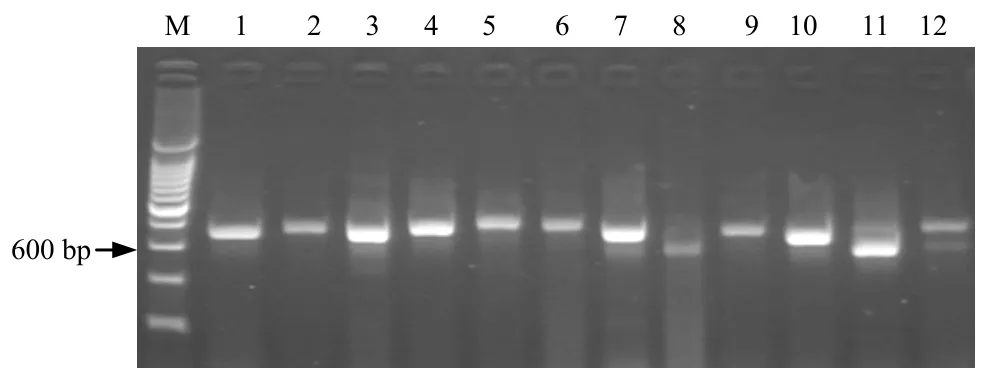
Fig.1 Agarose gel (1.5%) of PCR amplification products of 12 plants图 1 12 种植物DNA PCR 扩增产物电泳结果
[1]Murray M G,Thompson W F. Rapid isolation of high molecular weight plant DNA[J]. Nucleic Acids Research,1980,8(19):4321-4325.
[2]Dellaporta S L,Wood J, Hicks J B. A plant DNA minipreparation:Version II[J]. Plant Molecular Biology Reporter,1983,1(4):19-21.
[3]Allen G C, Flores-Vergara M A, Krasnyanski S, et al. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide[J]. Nature Protocols,2007,1:2320-2325.
[4]Ahmed I,Islam M,Arshad W,et al. High- quality plant DNA extraction for PCR:An easy approach[J]. Journal of Applied Genetics,2009,50(2):105-107.
[5]Manen J-F,Sinitsyna O,Aeschbach L,et al. A fully automatable enzymatic method for DNA extraction from plant tissues[J]. BMC Plant Biology,2005,5:23-32.
[6]Cheung W Y,Hubert N,Landry B S. A simple and rapid DNA microextraction method for plant,animal,and insect suitable for RAPD and other PCR analyses[J]. Genome Research,1993,3:69-70.
[7]Hill-Ambroz K L,Brown-Guedira G L,Fellers J P. Modified rapid DNA extraction protocol for high throughput microsatellite analysis in wheat[J]. Crop Science,2002,42:2088-2091.
[8]Xin Z-G,Velten J P, Oliver M J, et al. High-throughput DNA extraction methord suitable for PCR[J]. Biotechni- ques,2003,34:820-826.
[9]Kotchoni S O,Gachomo E W. A rapid and hazardous reagent free protocol for genomic DNA extraction suitable for genetic studies in plants[J]. Molecular Biology Reporter,2009,36:1633-1636.
