文心兰生物钟基因OnELF3克隆与表达分析
2025-02-09谭玉荣裴小华翟珂吴双李梦桃蒙青长宋灏明杨玉靖潘英谢光明李秀梅刘进平


摘""要:ELF3是重要的生物钟基因,调节光信号的输入与昼夜节奏输出,参与调控植物的重要过程。为了了解其在文心兰花瓣衰老启动中的调控作用,本研究对OnELF3基因进行克隆和表达分析。结果表明:利用转录组文库中的ELF3基因序列,从文心兰花瓣中扩增得到OnELF3基因(GenBank登录号:OL613896);OnELF3基因全长2208"bp,编码735个氨基酸。亚细胞定位表明其在细胞核中表达。同源序列比对与系统发育树分析显示OnELF3与铁皮石斛兰和蝴蝶兰亲缘关系最近。原核表达分析表明,OnELF3基因能原核表达出功能蛋白。该基因在长光照、全光照及全黑暗条件下表达模式与其作为生物钟基因功能一致。实时荧光定量分析表明,OnELF3基因在绽口期和盛开期表达量最高,表明OnELF3基因可能在开花和花衰老启动中均发挥作用。本研究为进一步鉴定OnELF3基因在开花和花衰老启动中的功能奠定基础。
关键词:文心兰;花衰老;ELF3基因;基因克隆;表达分析中图分类号:S31""""""文献标志码:A
Cloning"and"Expression"Analysis"of"the"Biological"Clock"Gene"OnELF3"in"Oncidium
TAN"Yurong1,2,3,"PEI"Xiaohua1,2,"ZHAI"Ke1,2,"WU"Shuang1,2,"LI"Mengtao1,2,"MENG"Qingchang1,2,SONG"Haoming1,2,"YANG"Yujing1,2,"PAN"Yingwen4,"XIE"Guangming1,2,5,"LI"Xiumei5,"LIU"Jinping1,2*
1."School"of"Breeding"and"Multiplication"(Sanya"Institute"of"Breeding"and"Multiplication),"Hainan"University,"Sanya,"Hainan"572024,"China;"2."School"of"Tropical"Agriculture"and"Forestry,"Hainan"University,"Danzhou,"Hainan"571737,"China;"3."Guangxi"Academy"of"Argicultural"Sciences,"Nanning,"Guangxi"530007,"China;"4."Post-entry"Quarantine"Station"for"Tropical"Plant,"Hainan"Entry-exit"Inspection"and"Quarantine"Bureau,"Haikou,"Hainan"570311,"China;"5."Wuzhishan"Wanquan"Horticulture"Co.,"Ltd."/"Hongshan"Farm,"Zhongshan"Town,"Wuzhishan,"Hainan"572216,"China
Abstract:"ELF3"is"an"important"biological"clock"gene"that"regulates"the"input"of"light"signal"and"the"output"of"circadian"rhythm,"participating"in"modulation"of"many"important"processes"of"plants."In"order"to"understand"its"regulation"in"the"initiation"of"petal"senescence"of"Oncidium,"we"conducted"cloning"and"expression"analysis"of"OnELF3"in"Oncidium."OnELF3"(GenBank"accession"number:"OL613896)"was"isolated"from"the"Oncidium"petals."OnELF3"had"a"length"of"2208"bp"and"encoded"a"protein"of"735"amino"acids."Subcellular"localization"showed"its"expression"in"the"nucleus."Homologous"sequence"alignment"and"phylogenetic"tree"analysis"showed"that"OnELF3"was"closest"to"the"counterparts"of"Dendrobium"candidum"and"Phalaenopsis."Prokaryotic"expression"analysis"showed"that"the"cloned"OnELF3"was"capable"of"prokaryotic"expression"of"functional"proteins."Its"expression"patterns"under"long"day,"full"light,"and"full"darkness"conditions"were"consistent"with"its"function"as"a"circadian"clock"gene."Real"time"fluorescence"quantitative"analysis"showed"that"the"expression"of"OnELF3"was"the"highest"in"cracked"buds"and"full-open"flowers,"indicating"that"OnELF3"may"play"a"role"in"both"flowering"and"flower"senescence"initiation."This"study"would"lay"a"foundation"for"further"functional"identification"of"OnELF3"in"the"initiation"of"flower"senescence.
Keywords:"Oncidium;"flower"senescence;"ELF3"gene;"gene"cloning;"expression"analysis
DOI:"10.3969/j.issn.1000-2561.2025.02.005
文心兰(Oncidium)花型酷似婆娑起舞的女郎,又名跳舞兰、舞女兰;又似飞翔的蝴蝶,因此又名金蝶兰。文心兰因其别致优美的花型、亮黄鲜艳的花色,为极具观赏价值的重要切花类型[1]。花衰老不仅是重要的植物生理过程,也是影响文心兰这类切花商品价值的一个十分重要的指标。研究文心兰花衰老的分子机制,不仅有助于了解植物器官的衰老与调控,也有助于从分子水平上控制花衰老,从根本上解决切花保鲜问题[2-11]。
EARLY"FLOWERING3(ELF3)是生物钟(circadian"clock)基因,而生物钟是生物为适应外界变化的内在昼夜节奏,参与调控几乎所有的植物的重要过程[12-16]。ELF3作为生物钟核心振荡器中晚间复合体(evening"complex,"EC)即ELF3-"ELF4-LUX"ARRHYTHMO(LUX)的重要组成部分,调节光信号的输入与生物钟的输出,不仅控制下胚轴伸长生长与植物开花时间[17-20],还在植物耐盐性[21]和叶片衰老[22]方面发挥重要作用。文心兰花发育和衰老过程的转录组研究表明,ELF3基因还可能参与文心兰花瓣衰老的启动调控[23]。
本研究从文心兰中克隆到生物钟基因OnELF3的全长序列,并对其进行蛋白质分析、亚细胞定位分析,以及其在不同花发育时期和昼夜中的表达分析,研究结果为研究OnELF3基因功能鉴定及其在文心兰花瓣衰老启动的调控中的作用奠定基础。
1""材料与方法
1.1""材料
供试材料为南茜黄金3代文心兰鲜切花,购自海南省出入境检疫局热带植物隔离检疫中心。亚细胞定位表达载体pCAMBIA1300、原核表达载体pET-30a、农杆菌菌株GV3101、大肠杆菌DH5α、大肠杆菌BL21(DE3)及本生烟草均由本实验室保存。
1.2""方法
1.2.1""OnELF3基因的克隆与生物信息学分析""参照CTAB法提取文心兰花瓣总RNA[24]。利用1%浓度凝胶电泳检测RNA条带的完整性,并利用微量分光光度计测量RNA浓度。按照TaKaRa反转录试剂盒(PrimeScript"RT"regent"Kit"with"gDNA"Eraser)合成cDNA第1条链。根据转录组测序获得的序列设计引物OnELF3-F/R(表1),以获得的文心兰cDNA为模板扩增OnELF3基因CDS区(去掉终止密码子),将获得的目的片段纯化后连接T载体,菌落检测正确后测序,便于后续实验。
以扩增得到的正确的序列在ORFfinder(https://www.ncbi.nlm.nih.gov/orffinder/)在线软件上预测氨基酸序列;通过ExPASy服务器的ProtParam"tool(https://web.expasy.org/protparam/)在线软件对预测的氨基酸序列进行理化性质分析;利用SignalP"4.1(http://www.cbs.dtu.dk/services/"SignalP/)在线软件对氨基酸序列的信号肽进行预测;用Plant-mPLoc(http://www.csbio.sjtu.edu.cn/"cgi-bin/PlantmPLoc.cgi)在线软件预测OnELF3蛋白亚细胞定位。以预测到的OnELF3蛋白序列在NCBI数据库中作Blast检索,并使用MEGA"6软件构建系统进化树。
1.2.2""OnELF3的亚细胞定位分析""利用Primer"5.0软件设计OnELF3基因带有p1300载体多克隆酶切位点的引物OnELF3-1300-F/R(表1)。以含有目的片段的T载体质粒为模板克隆目的条带,采用酶切酶连法构建亚细胞定位载体35S::OnELF3:GFP,将构建好的载体质粒转化到农杆菌GV3101中。采用瞬时表达的方法,将含有亚细胞定位载体35S::OnELF3:GFP的农杆菌注射到生长期为1个月的烟草叶片背部,28"℃弱光培养3"d后剪取叶片,利用激光共聚焦荧光显微镜分析基因的定位情况。
1.2.3""OnELF3基因的原核表达分析""利用Primer"5.0软件设计OnELF3基因带有pET30a载体多克隆酶切位点的引物OnELF3-30a-F/R(表1)。以含有目的片段的T载体质粒为模板克隆目的条带,构建亚细胞定位载体T7"promoter::"OnELF3:His-Tag,将构建好的载体质粒转化到大肠杆菌BL21(DE3)中。0.8"mol/L"IPTG,37"℃诱导pET30a和OnELF3-pET30a表达0、1、2、3"h后收集菌液进行SDS-PAGE电泳分析;0.8"mol/L"IPTG"pET30a和OnELF3-pET30a表达3"h进行Western"Blotting分析。
1.2.4""OnELF3基因的表达模式分析""生物钟节律表达分析取样:将150枝文心兰花枝平均分到3个量杯中,对文心兰切花进行2~3"d的暗处理、全光照和长光照处理后,从早上6:00开始时每隔2"h采1次样,采样时根据转录组数据选择采集盛开期花瓣(盛开前期、盛开期及衰老初期3个时期中在盛开期表达量最高),连续采样72"h,液氮速冻后保存于‒80"℃冰箱。文心兰切花不同发育时期表达分析取样:根据文心兰南茜品种花的形态变化特征,将其花发育分为花苞期(A)、绽口期(B)、半开放期(C)、盛开前期(D)、盛开期(E)、衰老初期(F)、衰老期(G)等8个不同的时期[25],在同一时间点采集不同时期的花瓣,装于无RNA酶的离心管中,于‒80"℃冰箱保存备用。取样结束后提取花瓣RNA,反转录为cDNA。以文心兰肌动蛋白基因为内参基因,设计OnELF3基因荧光定量PCR的引物OnELF3Q-F/R(表1)。
2""结果与分析
2.1""OnELF3基因CDS区序列克隆与生物信息学分析
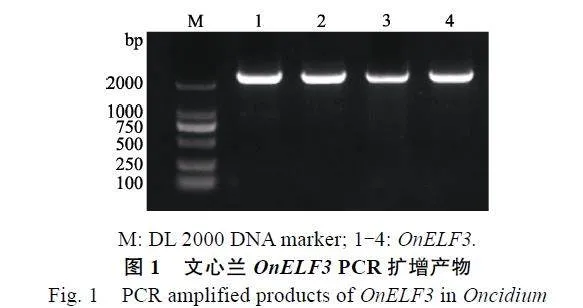
据转录组测序结果得到OnELF3基因的mRNA序列,设计特异性引物扩增出2432"bp序列,根据NCBI数据库Blast分析得到OnELF3基因的开放阅读框全长为2208"bp(GenBank登录号:OL613896)。根据该开放阅读框设计特异性引物扩增得到OnELF3的CDS,长度为2240"bp(图1)。
利用PtptParam软件预测分析OnELF3基因的编码蛋白,表明OnELF3蛋白分子量大小为80.70"kDa,等电点为8.22,不稳定系数为54.07,属于不稳定蛋白,易降解。Signal"P4.1"Server"软件预测OnELF3蛋白无信号肽,不是分泌蛋白。Plant-mPLoc软件预测OnELF3蛋白定位在细胞核上。利用NCBI分析OnELF3保守结构域,结果显示,未找到与OnELF3基因编码的蛋白相对应的已知结构域,但与其他物种的ELF3蛋白的结构域分析结果一致,因此,认为克隆到的基因的确是ELF3基因。利用DNAMAN软件对OnELF3氨基酸与其他9种植物的ELF3氨基酸序列进行同源序列比对发现,这10个植物的ELF3存在保守的部分序列,OnELF3蛋白与铁皮石斛、蝴蝶兰等兰科植物的ELF3蛋白同源性在63%~"81%之间,与其他物种的ELF3蛋白同源性在40%~50%之间(图2)。
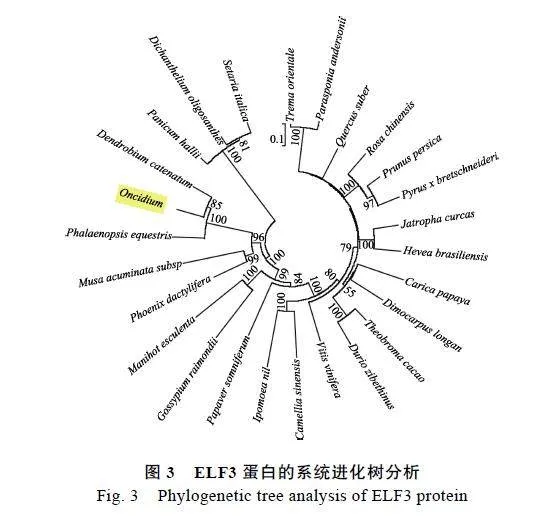
利用MEGA"6软件对OnELF3与NCBI数据库Blast检索到的蛋白序列构建系统进化树。结果显示,文心兰OnELF3蛋白与铁皮石斛兰和蝴蝶兰ELF3亲缘关系最近(图3)。
2.2""OnELF3在烟草表皮细胞中的亚细胞定位
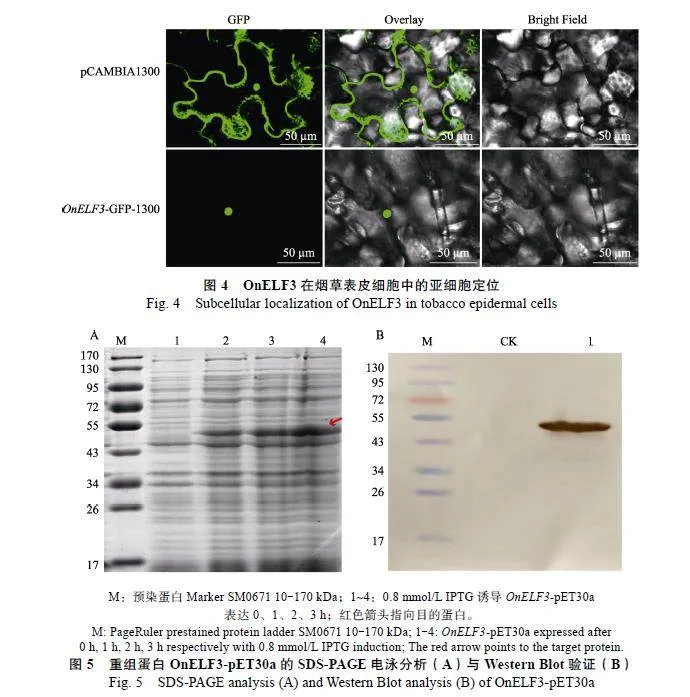
将含有OnELF3基因的重组质粒农杆菌悬浮液注射到烟草叶片中,使其在烟草表皮细胞中表相似性水平≥100%为黑色;相似性水平≥75%为粉红色;相似性水平≥50%为绿色;相似性水平lt;50%则为白色。The"similarity"was"indicated"by"different"colors:"blue-black=100%,"pink≥75%;"green≥50%,"and"whitelt;50%.达,利用激光共聚焦显微镜观察OnELF3蛋白的定位情况。当注射的是空载pCAMBIA"1300时,在细胞膜、细胞质和细胞核均能看到GFP荧光,而当GFP与OnELF3蛋白融合表达时,荧光信号主要在细胞核上(图4)。OnELF3蛋白的核定位使其具有作为转录因子的基本特征。
2.3""OnELF3基因的原核表达分析
OnELF3基因原核表达经SDS-PAGE电泳分析,结果显示,OnELF3-pET30a与空载pET30a相比,条带较深,大小与预测的蛋白大小一致(图5A)。Western"Blot验证结果表明,重组蛋白OnELF3-pET30a诱导成功,重组蛋白OnELF3-"pET30a可被特异性抗体识别,得到与预测时大小一致的单一蛋白条带(图5B)。上述结果说明本研究成功克隆了OnELF3基因并能原核表达出功能蛋白。
2.4""OnELF3基因的生物钟节律表达分析
为确定OnELF3基因的表达是否受生物钟的影响,利用实时荧光定量表达分析对OnELF3基因在长光照(LD)、全光照(LL)及全黑暗(DD)条件下的表达进行定量分析。结果表明,OnELF3基因只在长光照条件下呈节律性振荡表达,在22:00和6:00分别有1个高峰值,在全光照与全黑暗条件下表现出节律不齐(图6)。
2.5""OnELF3基因在文心兰不同花期花瓣中的表达分析
对OnELF3在文心兰切花发育各个时期的花瓣实时荧光定量分析表明,OnELF3基因在绽口期表达量达到最高值,之后表达量下降,在盛开期上升到第二高峰值,之后表达量下降,在衰老期达到最低值(图7)。
3""讨论
本研究克隆到1个文心兰生物钟基因OnELF3,保守结构域分析发现其无已知功能的结构域;亚细胞定位分析发现,OnELF3定位于细胞核。与拟南芥[26]的研究结果一致,ELF3编码无已知功能结构域的植物特异性核蛋白。在拟南芥中,ELF3是生物钟晚间阶段转录翻译反馈环的重要成员,呈现24"h节律性振荡,并在傍晚出现表达峰。本研究结果显示,在长日照条件下,OnELF3呈24"h节律性振荡表达,出现2个表达峰,这与拟南芥中ELF3在傍晚出现1个表达峰有所不同,说明不同植物间生物钟基因的节律表坐标轴下方的黑色矩形代表正常条件下的黑暗时间,白色代表光照时间。
达存在一致性与差异性[27]。
ELF3在植物中不仅维持生物钟节律性振荡,还在开花调控过程中扮演重要角色[27]。通过分析OnELF3基因在花不同发育时期的表达量,发现该基因在花的各个发育时期中均有表达,其中在绽口期的表达量最高,在盛开期出现第二个高峰,这一结果与之前的转录组分析相吻合[23],表明OnELF3基因可能参与开花过程及花衰老的启动。小麦属的ELF3突变后,突变系比野生型更早开花[28];水稻ELF3抑制Ghd7的表达,促进水稻开花[29];大豆ELF3与LUX1、LUX2形成复合物,抑制开花抑制因子E1表达,促使大豆提前开花[30]。尽管ELF3在花瓣衰老中的作用尚未见报道,但已有研究表明,ELF3在叶片衰老的调控中起着重要作用[22,"31-34]。拟南芥ELF3通过与ELF4和LUX组成的晚间复合体(EC)直接与MYC2的启动子结合,对茉莉酸盐(JA)诱导的叶片衰老起着负调控作用[32];在自然条件下和黑暗诱导条件下的叶片衰老中,ELF3可在转录水平上调控PIF4和PIF5,不依赖于EC而对叶片衰老发挥负调控作用[33];在高温条件下,ELF3对PIF4抑制作用减弱,从而加速叶片衰老[34]。据此可推测OnELF3基因在文心兰开花和花衰老启动中发挥作用,但由于开花和花衰老过程中的调控机制十分复杂,OnELF3基因功能仍需进一步研究。
参考文献
[1]"刘晓荣,"王碧青,"朱根发."文心兰研究进展[J]."亚热带植物科学,"2007,"36(3):"85-90.LIU"X"R,"WANG"B"Q,"ZHU"G"F."Research"advances"of"Oncidium[J]."Subtropical"Plant"Science,"2007,"36(3):"85-90."(in"Chinese)
[2]"杨光华,"刘进平."文心兰ACC氧化酶基因OnACO1克隆与表达分析[J]."热带作物学报,"2014,"35(4):"693-699.YANG"G"H,"LIU"J"P."Cloning"and"expression"analysis"of"OnACO1"gene"in"Oncidium[J]."Chinese"Journal"of"Tropical"Crops,"2014,"35(4):"693-699."(in"Chinese)
[3]"YANG"G"H,"LIU"J"P."Isolation"of"an"1-aminocycloprop an e-1-carboxylate"synthase"gene"from"Oncidium"Gower"Ramsey[J]."Genetics"and"Molecular"Research,"2014,"13(4):"8480-"8488.
[4]"田晓岩,"石乐松,"潘英文,"刘进平."文心兰OnACO2基因的克隆及表达分析[J]."分子植物育种,"2015,"13(7):"1602-1610.TIAN"X"Y,"SHI"L"S,"PAN"Y"W,"LIU"J"P."Cloning"and"expression"analysis"of"OnACO2"gene"from"Oncidium"Gower"Ramsey[J]."Molecular"Plant"Breeding,"2015,"13(7):"1602-"1610."(in"Chinese)
[5]"田晓岩,"杨翠萍,"胡进,"闫冰玉,"巩笑笑,"庄玉粉,"潘英文,"刘进平."文心兰OnERS1全长基因克隆及表达分析[J]."分子植物育种,"2017,"15(4):"1265-1272.TIAN"X"Y,"YANG"C"P,"HU"J,"YAN"B"Y,"GONG"X"X,"ZHUANG"Y"F,"PAN"Y"W,"LIU"J"P."Cloning"and"expression"analysis"of"OnERS1"gene"from"Oncidium"Gower"Ramsey[J]."Molecular"Plant"Breeding,"2017,"15(4):"1265-1272."(in"Chinese)
[6]"SHI"L"S,"LIU"J"P."Molecular"cloning"and"expression"analysis"of"an"1-aminocyclopropane-1-carboxylate"synthase"gene"from"Oncidium"Gower"Ramsey[J]."Biochemical"and"Biophysical"Research"Communications,"2016,"469:"203-209.
[7]"杨翠萍,"胡进,"闫冰玉,"巩笑笑,"谭玉荣,"高璇,"王丹,"张恒,"刘进平."文心兰生物钟相关基因PRR7克隆与表达分析[J]."热带作物学报,"2018,"39(8):"1670-1579.YANG"C"P,"HU"J,"YAN"B"Y,"GONG"X"X,"TAN"Y"R,"GAO"X,"WANG"D,"ZHANG"H,"LIU"J"P."Cloning"and"expression"analysis"of"circadian-associated"gene"PRR7"(pseudo-res po nse"regulator7)"from"Oncidium[J]."Chinese"Journal"of"Tropical"Crops,"2018,"39(8):"1570-1579."(in"Chinese)
[8]"闫冰玉,"巩笑笑,"谭玉荣,"王鹏,"李双江,"潘英文,"刘进平."文心兰OnRR9基因克隆及表达[J]."热带生物学报,"2018,"9(4):"393-400.YAN"B"Y,"GONG"X"X,"TAN"Y"R,"WANG"P,"LI"S"J,"PAN"Y"W,"LIU"J"P."Cloning"and"expression"analysis"of"OnRR9"gene"in"Oncidium[J]."Journal"of"Tropical"Biology,"2018,"9(4):"393-400."(in"Chinese)
[9]"闫冰玉,"巩笑笑,"谭玉荣,"王丹,"高璇,"张恒,"李双江,"王鹏,"潘英文,"刘进平."文心兰OnRR10基因的克隆及表达分析[J]."热带生物学报,"2020,"11(3):"287-295.YAN"B"Y,"GONG"X"X,"TAN"Y"R,"WANG"D,"GAO"X,"ZHANG"H,"LI"S"J,"WANG"P,"PAN"Y"W,"LIU"J"P."Cloning"and"expression"analysis"of"OnRR10"gene"in"Oncidium[J]."Journal"of"Tropical"Biology,"2020,"11(3):"287-295."(in"Chinese)
[10]"谭玉荣,"高璇,"王丹,"张恒,"李双江,"王鹏,"李晨,"徐丹,"李梦桃,"潘英文,"刘进平."文心兰生物钟基因OnELF3启动子克隆与表达分析[J]."分子植物育种,"2020,"18(15):"4898-4907.TAN"Y"R,"GAO"X,"WANG"D,"ZHANG"H,"LI"S"J,"WANG"P,"LI"C,"XU"D,"LI"M"T,"PAN"YW,"LIU"J"P."Cloning"and"expression"analysis"of"Oncidium"circadian-clock"gene"OnELF3"promoter[J]."Molecular"Plant"Breeding,"2020,"18(15):"4898-"4907."(in"Chinese)
[11]"YANG"C"P,"TAN"Y"R,"YAN"B"Y,"GONG"X"X,"WANG"D,"GAO"X,"ZHANG"H,"WANG"P,"LI"S"J,"WANG"Y,"ZHOU"LY,"PAN"Y"W,"LIU"J"P."Molecular"characterization"of"clock-associated"PSEUDO-RESPONSE"REGULATOR"9"gene"from"Oncidium"‘Gower"Ramsey’[J]."Plant"Growth"Regulation,"2020,"91(3):"371-381.
[12]"徐小冬,"谢启光."植物生物钟研究的历史回顾与最新进展[J]."自然杂志,"2013,"35(2):"110-126.XU"X"D,"XIE"Q"G."The"circadian"clock"in"plants[J]."Chinese"Journal"of"Nature,"2013,"35(2):"110-126."(in"Chinese)
[13]"GREENHAM"K,"MCCLUNG"C"R."Integrating"circadian"dynamics"with"physiological"processes"in"plants[J]."Nature"Reviews"Genetics,"2015,"16(10):"598-610
[14]"SABRINA"E"S,"RUGNONE"M"L,"KAY"S"A."Light"perception:"a"matter"of"time[J]."Molecular"Plant,"2020,"13(3),"363-"385.
[15]"SU"C,"WANG"Y,"YU"Y,"HE"Y,"WANG"L."Coordinative"regulation"of"plants"growth"and"development"by"light"and"circadian"clock[J]."aBIOTECH,"2021,"2:"176-189.
[16]"ZHU"X,"WANG"H."Revisiting"the"role"and"mechanism"of"ELF3"in"circadian"clock"modulation[J]."Gene,"2024,"913:"148378.
[17]"ZAGOTTA"M"T,"HICKS"K"A,nbsp;JACOBS"C"I,"YOUNG"J"C,"HANGARTER"R"P,"MEEKS-WAGNER"D"R."The"Arabidopsis"ELF3"gene"regulates"vegetative"photomorphogenesis"and"the"photoperiodic"induction"of"flowering[J].nbsp;Plant"Journal,"1996,"10(4):"691-702.
[18]"ZAHO"H,"XU"D,"TIAN"T,"KONG"F,"LIN"K,"GAN"S,"ZHANG"H,"LI"G."Molecular"and"functional"dissection"of"EARLY-FLOWERING"3"(ELF3)"and"ELF4"in"Arabidopsis[J]."Plant"Science,"2021,"303:"110786.
[19]"BODERN"S"A,"WEISS"D,"ROSS"J"J,"DAVIES"N"W,"TREVASKIS"B,"CHANDLER"P"M,"SWAIN"S"M."EARLY"FLOWERING3"regulates"flowering"in"spring"barley"by"mediating"gibberellin"production"and"FLOWERING"LOCUS"T"expression[J]."Plant"Cell,"2014,"26(4):"1557-1569.
[20]"PRESS"M"O,"LANCTOT"A,"QUEITSCH"C."PIF4"and"ELF3"act"independently"in"Arabidopsis"thaliana"thermoresponsive"flowering[J]."PLoS"One,"2016,"11(8):"e0161791.
[21]"SAKURABA"Y,"BULBUL"S,"PIAO"W,"CHOI"G,"PAEK"N"C."Arabidopsis"EARLY"FLOWERING3"increases"salt"tolerance"by"suppressing"salt"stress"response"pathways[J]."Plant"Journal,"2017,"92(6):"1106-1120.
[22]"SAKURABA"Y,"HAN"S"H,"YANG"H"J,"PIAO"W,"PAEK"N"C."Mutation"of"rice"Early"Flowering3.1"(OsELF3.1)"delays"leaf"senescence"in"rice[J]."Plant"Molecular"Biology,"2016,"92(1/2):"223-234.
[23]"YANG"C"P,"XIA"Z"Q,"HU"J,"ZHUANG"Y"F,"PAN"Y"W,"LIU"J"P."Transcriptome"analysis"ofnbsp;Oncidium"petals"provides"new"insights"into"the"initiation"of"petal"senescence[J]."Journal"of"Horticultural"Science"and"Biotechnology,"2019,"94(1):"12-23.
[24]"石乐松,"刘进平."文心兰RNA不同提取方法比较[J]."生物技术,"2012,"22(6):"42-45.SHI"L"S,"LIU"J"P."Comparison"of"different"RNA"extraction"methods"of"Oncidium"orchid[J]."Biotechnology,"2012,"22(6):"42-45."(in"Chinese)
[25]"石乐松,"孙慧,"田晓岩,"吴繁花,"蔡秀清,"庄玉粉,"刘进平."文心兰南茜种花发育进程与分期[J]."热带生物学报,"2016,"7(4):"440-443,"449.SHI"L"S,"SUN"H,"TIAN"X"Y,"WU"F"H,"CAI"X"Q,"ZHUANG"Y"F,"LIU"J"P."Developmental"stages"of"Oncidium"Gower"Ramsey"flower[J]."Journal"of"Tropical"Organisms,"2016,"7(4):"440-443,"449."(in"Chinese)
[26]"LIU"X"L,"COVINGTON"M"F,"FAMLJAISER"C,"CHORY"J,"WAGNER"D"R."ELF3"encodes"a"circadian"clock-regulated"nuclear"protein"that"functions"in"an"Arabidopsis"PHYB"signal"transduction"pathway[J]."Plant"Cell,"2001,"13(6):"1293-1304.
[27]"HICKS"K"A,"ALBERTSON"T"M,"WAGNER"D"R."EARLY"FLOWERING3"encodes"a"novel"protein"that"regulates"circadian"clock"function"and"flowering"in"Arabidopsis[J]."Plant"Cell,"2001,"13(6):"1281-1292.
[28]"ALVAREZ"M"A,"TRANQUILLI"G,"LEWIS"S,"KIPPES"N,"DUBCOVSKY"J."Genetic"and"physical"mapping"of"the"earliness"per"se"locus"Eps-A"(m)"1"in"Triticum"monococcum"identifies"EARLY"FLOWERING"3"(ELF3)"as"a"candidate"gene[J]."Funct"Integr"Genomics,"2016,"16:"365-382.
[29]"SAITO"H,"OGISO-TANAKA"E,"OKUMOTO"Y,"YOSHI TA KE"Y,"IZUMI"H,"YOKOO"T,"MATSUBARA"K,"HORI"K,"YANO"M,"INOUE"H,"TANISAKA"T."Ef7"encodes"an"ELF3-like"protein"and"promotes"rice"flowering"by"negatively"regulating"the"floral"repressor"gene"Ghd7"under"both"short-"and"long-day"conditions[J]."Plant"Cell"Physiol,"2012,"53:"717-728.
[30]"BU"T"T,"LU"S"J,"WANG"K,"DONG"L"D,"LI"S"L,"XIE"Q"G,"XU"X"D,"CHENG"Q,"CHEN"L"Y,"FANG"C,"LI"H"Y,"LIU"B"H,"WELLER"J"L,"KONG"F"J."A"critical"role"of"the"soybean"evening"complex"in"the"control"of"photoperiod"sensitivity"and"adaptation[J]."Proceedings"of"the"National"Academy"of"Sciences"of"the"United"States"of"America,"2021,"118:"e2010241118.
[31]"LEE"J,nbsp;KANG"M"H,"KIM"J"Y,"LIM"P"O."The"role"of"light"and"circadian"clock"in"regulation"of"leaf"senescence[J]."Front"Plant"Science,"2021,"12:"669170.
[32]"ZHANG"Y,"WANG"Y,"WEI"H,"LI"N,"TIAN"W,"CHONG"K,"WANG"L."Circadian"evening"complex"represses"jasmonate-"induced"leaf"senescence"in"Arabidopsis[J]."Molecular"Plant,"2018,"11(2):"326-337.
[33]"SAKURABA"Y,"JEONG"J,"KANG"M"Y,"KIM"J,"PAEK"N"C,"CHOI"G."Phytochrome-interacting"transcription"factors"PIF4"and"PIF5"induce"leaf"senescence"in"Arabidopsis[J]."Nature"Communications,"2014,"5:"4636.
[34]"KIM"C,"KIM"S"J,"JEONG"J,"PARK"E,"OH"E,"PARK"Y"I,"LIM"P"O,"CHOI"G."High"ambient"temperature"accelerates"leaf"senescence"via"PHYTOCHROME-INTERACTING"FACTOR"4"and"5"in"Arabidopsis[J]."Molecular"Cells,"2020,"43(7):"645-661.
