基于转录组测序筛选新疆野苹果种子休眠解除过程中植物激素相关基因
2024-12-31房震李静马娟张凯叶春秀



摘" " 要: 【目的】研究新疆野苹果(Malus sieversii)种子不同贮藏阶段转录组差异和激素含量变化,筛选新疆野苹果种子休眠解除过程中植物激素相关基因,为后续新疆野苹果种子休眠解除激素调控机制研究提供依据。【方法】以新疆野苹果种子为材料,对照未层积种子和经4 ℃低温沙藏层积处理(30、60、90、120 d)后种子进行转录组测序,采用酶联免疫吸附法测定种子中的脱落酸(abscisic acid,ABA)、赤霉素(gibberellin,GA)、生长素(auxin,IAA)、细胞分裂素(cytokinin,CTK)含量以及乙烯氨基环丙烷羧酸氧化酶(1-aminocyclopropane-1-carboxylic acid oxidase,ACO)和氨基环丙烷羧酸合成酶(1-aminocyclopropane-1- carboxylic acid oxidase synthase,ACS)的活性。【结果】新疆野苹果种子GA、IAA、CTK含量随贮藏时间的增加呈上升趋势,而ABA含量呈下降趋势。乙烯(ethylene,ETH)合成途径ACO和ACS活性随贮藏时间的增加呈增强趋势。GO富集分析筛选出85个差异基因调控种子萌发(GO:0010029),114个GA相关差异基因,313个ABA相关差异基因和156个ETH相关差异基因。KEGG通路富集分析显示,主要富集通路有植物激素信号转导、MAPK信号通路-植物、内质网中的蛋白加工、淀粉和蔗糖代谢、糖酵解/糖原生成等。ABA信号传导通路中有3个PYR/PYL基因下调表达,2个蛋白磷酸酶2C(PP2C-type protein phosphatases,PP2C)、3个蔗糖非发酵相关的蛋白激酶(sucrose non-fermenting-1-related protein kinase 2,SnRK2)上调表达和2个ABA分解代谢8'-羟化酶(8'-hydroxylases)基因表达显著上调。GA信号转导途径中2个受体GID1(gibberellin insensitive dwarf 1)和6个负调控因子DELLA蛋白上调表达;ETH受体ETR(Ethylene receptor)的直接前体1-氨基环丙烷-1-羧酸(1-aminocyclopropane-1-carboxylic acid,ACC)和ACO基因上调表达。【结论】新疆野苹果种子经低温沙藏层积处理,ERF2-like表达量与ACO、ACS活性变化呈相反趋势,PYR1-like、WRKY33基因表达量与ABA含量变化均下降,说明以上基因可能参与ABA信号通路调控新疆野苹果种子休眠解除过程。
关键词:新疆野苹果;转录组;内源激素;差异表达基因;种子休眠
中图分类号:S661.1 文献标志码:A 文章编号:1009-9980(2024)10-1961-18
Screening of plant hormone-associated genes during seed dormancy release in Malus sieversii based on transcriptome sequencing
FANG Zhen, LI Jing, MA Juan, ZHANG Kai, YE Chunxiu*
(College of Forestry and Landscape Architecture, Xinjiang Agricultural University, Urumqi 830052, Xinjiang, China)
Abstract: 【Objective】 The study aimed to study the transcriptome differences and hormone content changes of the seeds of Malus sieversii at different stages of stratification in order to screen the plant hormones genes related to dormancy release and provide a basis for subsequent studies on the hormonal regulation mechanism of seed dormancy release in M. sieversii. 【Methods】 The seeds of M. sieversii were used as materials, transcriptome sequencing was performed on the seeds at different stages of the stratification at 4 ℃ (0, 30, 60, 90 and 120 d). The content of abscisic acid (ABA), gibberellins (GA), auxin (IAA), cytokinin (CTK), the activities of ethylene 1-aminocyclopropane-1-carboxylate oxidase (ACO) and 1-aminocyclopropane-1-carboxylate oxidase synthase (ACS) were measured using an enzyme-linked immunosorbent assay. 【Results】 The ABA content of the seeds showed a decreasing trend with the increase of stratification time, the maximum content was 80.22 ng·g-1 on 0 d of the storage, the content on 60 d of the storage was significantly lower than that at the three periods of 0, 30 and 90 d (p<0.05), the content reached a minimum of 43.67 ng·g-1 on 120 d. The GA content showed an increasing trend with the increase of the storage time, the content was the lowest at 78.42 pmol·g-1 on 0 d, and the content on 120 d significantly higher than those of the other periods (p<0.05), and reached a maximum of 170.67 pmol·g-1. The IAA content showed an increasing trend with the storage time, and the content on 90 d was significantly higher than those of the other periods, reaching a maximum value of 41.36 nmol·g-1. The CTK content on 90 d was significantly higher than those of the other periods, reaching a minimum value of 43.67 ng·g-1, it showed a decreasing trend in the storage time of 0-30 d, the content on 30 d was significantly lower than those of the other periods (p<0.05), reaching a minimum value of 31.34 ng·g-1, and showed an increasing trend in the storage time of 30-60 d, suggesting that CTK would promote the accumulation of seed assimilates in this period. The activities of ACC oxidase and ACC synthase in the ethylene biosynthesis pathway showed inconsistent trends during the storage. The activity of ACO was 262.52 ng·g-1 on 90 d of the storage, which was significantly higher than that of other periods (p<0.05), and the activity of ACO was the lowest on 30 d of the storage, which was 157.38 ng·g-1. The activity of ACS reached the maximum value of 418.92 ng·g-1 on 120 d of the storage, which was significantly higher than that of the three periods of 0, 30 and 60 d (p<0.05). It indicated that the stratification promoted the synthesis of GA, IAA, CTK, ACC oxidase and ACC synthase, and two enzymes of the ethylene synthesis pathway might be more sensitive to low temperature. There were more significant DEGs on M120d_CK120d compared with M0d_CK0d, M30d_CK30d, M60d_CK60d and M90d_CK90d, suggesting that there were more DEGs on M120d for the regulation of seed germination and physiological changes There were a total of 7384, 4875 and 3236 significantly and differentially expressed genes, significantly and differentially up-regulated genes and significantly and differentially down-regulated genes on the M30d_CK30d and M60d_CK60d periods, respectively. The gene ontology enrichment of DEGs was performed, and the biological processes were mainly involved in the response to osmotic stress and water deprivation, abscisic acid response, response to salt stress, transcriptional regulation, regulation of seed germination, and gibberellin response. The cellular components were mainly chloroplast stroma, chloroplast envelope and thylakoid. The molecular functions were mainly related to DNA-binding transcription factor activity, phosphatase activity and protein homodimer activation. The multiple plant hormone biological processes remained active and changed during the course of stratification of the seeds, suggesting that they would play a role in seed dormancy release activities. The KEGG pathway enrichment analysis showed that the main pathways enriched in seeds at different periods of the stratification were plant hormone signaling, MAPK signaling pathway-plant, protein processing in endoplasmic reticulum, starch and sucrose metabolism, and glycolysis/glycogenesis. Among them, plant hormones would play a key role in regulating seed dormancy and germination in M. sieversii, and hormone signal transduction pathway-related genes such as ABA, GA, and ETH might be involved in the processes such as seed dormancy release. The starch and sucrose metabolic pathways were involved in the process of carbon metabolism in the seed embryo, providing carbon source for the seed embryo. The glycogen production metabolic pathway was involved in the synthesis and metabolism of cellular proteins, providing nitrogen source and energy for seed embryo germination. The ABA receptor PYR/PYL had 12 genes up-regulated and 3 genes down-regulated. The ABA signal transducer protein phosphatases 2C (PP2C) and positive regulator GA receptor gibberellin insensitive dwarf 1 (GID1) genes were up-regulated Three genes were up-regulated and one gene was down-regulated for the sucrose non-fermenting-1-related protein kinase 2 (SnRK2), and two genes were up-regulated for the ABA catabolic hydroxylases 8'-hydroxylases. The GA signaling the negatively regulated growth inhibitor DELLA protein was up-regulated by 6 genes and down-regulated by 1 gene, indicating that low-temperature stratification treatment enhanced GA signaling. The ETH and IAA were significantly and differentially expressed at different stratification stages of M. sieversii embryos. The ETH receptor ETR, ETHYLENE INSENSITIVE 3/EIN3-LIKE, 1-aminocyclopropane-1-carboxylic acid, a direct precursor of ETH, and ACO were up-regulated expression. The 5 IAA-related genes were up-regulated and one was down-regulated, and small auxin up-regulated RNA was up-regulated, indicating that the stratification would promot IAA synthesis. The cell cycle protein genes were up-regulated to meet the nutritional growth of the seed dormancy release. In addition, the genes related to sucrose metabolism were screened, sucrose synthase and endoglucanase were up-regulated during the stratification. The sucrose transport protein STP13 and STP14 were up-regulated, while STP5 and STP10 were down-regulated. 【Conclusion】 The expression of the ERF2-like showed an opposite trend to the changes of ACO and ACS activities, and the expression of the PYR1-like and WRKY33 genes decreased in relation to the changes of ABA content, suggesting that the above genes might be involved in the ABA signaling pathway to regulate the process of dormancy release process of M. sieversii seeds.
Key words: Malus sieversii; Transcriptome; Endogenous hormone; Differentially expressed gene; Seed dormancy
新疆野苹果[Malus sieversii (Ledeb.) Rome.]主要分布在中亚天山山脉[1],是现代栽培苹果的祖先和珍贵的种质资源,其种群遗传多样性丰富。作为世界苹果基因库的组成和中国栽培苹果的砧木之一[2],具有保护价值和研究意义[3]。新疆野苹果种子休眠受到种皮结构及生理生化指标双重影响[4],进而影响新疆野苹果育种工作的推进和种质资源的维护。
目前转录组学已应用在NaCl胁迫下筛选新疆野苹果叶和根糖酵解途径相关基因,发现磷酸丙糖异构酶(triosephosphate isomerase,TPI)、果糖-1,6二磷酸酶(fructose-1,6-bisphosphatase,FBPase)、磷酸烯醇式丙酮酸羧激酶[phosphoenolpyruvate carboxy kinase (ATP),pckA]、丙酮酸磷酸双激酶(pyruvate,phosphate dikinase,PPDK)等基因的表达量显著变化[5]。马红喜等[6]筛选获得-3 ℃冻害新疆野苹果组培苗光合调控相关基因PsbQ与PsbY。苏永峰等[7]筛选新疆野苹果组培苗应答冻害谷胱甘肽代谢相关基因,冻害胁迫响应与谷胱甘肽过氧化物酶(glutathione peroxidase,GPX)和谷胱甘肽转硫酶(glutathione S-transferase,GST)表达上调有关。因此,研究新疆野苹果种子休眠解除中的差异基因,对苹果的遗传育种及保护种质资源提供基因资源。
植物激素是植物产生的少量有机化合物,可以促进或抑制多种生理过程[8]。脱落酸(abscisic acid,ABA)和赤霉素(gibberellin,GA)在调控种子休眠和萌发中发挥主要作用,种子中的ABA浓度除了控制ABA生物合成外,还通过将ABA转化为相酸(phasic acid)导致ABA失活来决定,负责转化是ABA-8'-羟化酶(ABA-8'-hydroxylases),它是由CYP707A1和CYP707A2编码的2个细胞色素P450(cytochrome P450,CYP450)[9]。Urbanova等[10]研究发现3种拟南芥(Arabidopsis)GA受体GID1(gibberellin insensitive dwarf 1)与GAI(GA-insensitive)、RGA(repressor-of-ga1-3)、RGL1(RGA-like1)、RGL2、RGL3等5种DELLA蛋白结合,作为GA信号转导的阻遏物。其中RGL1在种子萌发中比GAI和RGA具有更重要的作用,但RGL2是拟南芥中响应GA种子萌发最重要的调节因子。乙烯(ethylene,ETH)是另一种发芽促进剂,减少休眠3(reduced dormancy 3)是ETH受体反应因子1(ethylene response factor 1,ETR1)的功能丧失突变体,ETR1通过延迟休眠1(delay of germination 1,DOG1)乙烯部分途径控制种子休眠。ETR1抑制ERF12的表达,ERF12募集TOPLESS形成阻遏复合物并与DOG1启动子结合,从而抑制DOG1的表达解除种子休眠[11]。马娟等[12]发现新疆野苹果种子低温层积处理生长素(auxin,IAA)、GA3、细胞分裂素(cytokinin,CTK)含量增加,ABA含量降低,IAA、GA3和CTK促进种子解除休眠。转录组筛选分析花生(Arachis hypogaea)种子休眠过程中的ABA、GA、ETH、IAA相关差异表达基因,ABA合成和代谢基因调控花生种子休眠解除[13]。紫荆(Cercis chinensis Bunge)种子休眠解除主要与植物激素信号转导和能量代谢通路差异基因的显著变化相关[14]。对低温处理豆梨(Pyrus calleryana Dence)[15]和强弱休眠花生种子[16]进行转录组测序,表明植物激素(ABA、GA、ETH、IAA)的信号转导和生物合成在调控种子休眠维持和解除中起关键作用。前期的生理生化指标表明内源激素对种子休眠解除具有显著影响[12]。笔者在本研究中以新疆野苹果种子为材料,选取未层积和低温沙藏层积处理(30、60、90、120 d)的种子进行转录组测序,同时测定种子中植物激素ABA、GA、IAA、CTK含量、乙烯氨基环丙烷羧酸氧化酶(1-aminocyclopropane-1-carboxylic acid oxidase,ACO)和氨基环丙烷羧酸合成酶(1-aminocyclopropane-1- carboxylic acid oxidase synthase,ACS)的活性,分析在低温层积过程中的激素变化,旨在揭示新疆野苹果种子休眠解除植物激素相关的关键基因,为解析种子休眠解除的激素调控机制提供参考。
1 材料和方法
1.1 材料
试验材料为新疆塔城额敏县野果林新疆野苹果[M. sieversii(Ledeb.)Rome.]种子,对照组为室温保存5个时期(0、30、60、90、120 d)的种子样品,处理组M0d是CK0d清水浸泡24 h,4 ℃低温沙藏层积4个时期(30、60、90、120 d)的种子样品,标记为M30d、M60d、M90d和M120d。样品迅速于液氮中冷冻,之后在-80 ℃超低温冰箱中保存备用。各时期种子3次生物学重复。种子萌发率测定,将层积不同时期的种子均匀摆放于培养皿脱脂棉表面,盖上皿盖置于自然光下,定期换水并观察种子发芽情况,各时期处理30粒种子,3次重复。
1.2 植物激素和乙烯合成途径酶浓度检测
采用双抗体夹心酶联免疫吸附法(ELISA)测定植物激素ABA、GA、IAA、CTK含量,ETH合成途径ACC氧化酶和ACC合成酶活性,试剂盒购自睿信生物科技有限公司。
1.3 RNA提取与转录组测序分析
所有新疆野苹果种子样本转录组测序分析委托新疆康普森有限公司。使用Trizol法提取5个时期种胚的总RNA,检测RNA样品的浓度和完整性。完成转录组测序文库构建,使用Illumina平台进行PE150测序,得到150 bp的双端测序reads。利用Trimmomatic[17]进行数据质量过滤,去除read中包含的接头序列和质量低于20的碱基,过滤后的数据按照长度进行筛选,去掉长度小于50 bp或者只有一端的reads。过滤之后得到的高质量reads称为Clean reads。金冠苹果参考基因组和基因模型注释文件直接从基因组网站下载,使用Hisat2[18] v2.0.5建立参考基因组索引,将成对的Clean reads与苹果参考基因组进行比对,获得Mapped Data。根据基因长度和Htseq-count统计映射到该基因上的读数,计算出每个基因的每百万碱基对测序的转录本序列片段数(fragments per kilobase of transcript sequence per millions base pairs sequenced,FPKM)进行基因表达量的量化。
1.4 差异表达分析、DEGs功能注释和富集分析
差异表达分析使用DESeq2[19],采用皮尔逊(Pearson)相关系数作为生物重复之间的相关性指标[20]。通过DESeq2发现Padj<0.05的基因被归为普通差异表达基因(differentially expressed genes,DEGs)。研究各组合的显著差异表达基因,选取错误发现率(1 discovery rate,FDR)<0.05且差异倍数|log2(Fold Change)|≥2作为筛选标准。采用层次聚类分析DEGs的表达模式,并将DEGs与基因本体(gene ontology,GO)[21]和(kyoto encyclopedia of genes and genomes,KEGG)[22]数据库比较,使用topGO R和clusterProfiler R[23]软件包来测试GO和KEGG通路中DEGs的统计富集,通过KEGG通路显著性富集来确定植物激素相关差异表达基因。
1.5 差异表达转录因子分析
通过植物转录因子数据库(plant transcription factor database,PTFDB)与DEGs进行比对,筛选差异表达转录因子(transcription factor,TFs)。采用皮尔逊相关系数作为转录因子与植物激素有关DEGs的相关性指标,利用TBtools[24]绘制热图分析差异转录因子的表达模式。
1.6 qRT-PCR验证
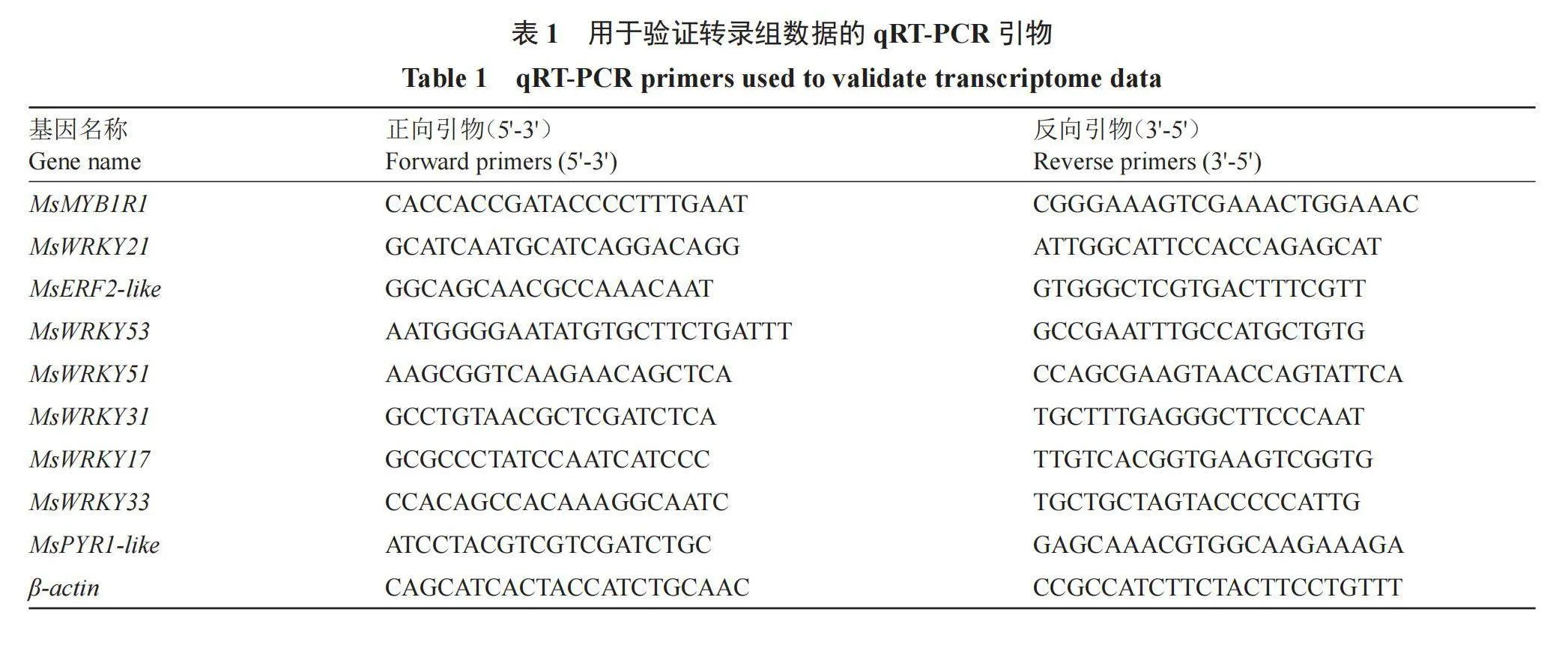
随机选择与种子休眠和激素相关的9个DEGs进行实时荧光定量(qRT-PCR),使用Primer 5设计引物(表1)。以不同时期的新疆野苹果种子RNA为模板,使用SYBR Green I染料法qPCR预混液(Enzy Artisan,Q204)进行qRT-PCR:反应条件为预变性95 ℃ 30 s;三步扩增95 ℃ 10 s;60 ℃ 15 s;72 ℃ 30 s;40个循环;溶解95 ℃ 10 s;60 ℃ 60 s;95 ℃ 15 s;冷却37 ℃ 30 s。以β-actin为内参基因,根据2-ΔΔCt算法[25]计算相对表达量,每个处理3次重复。
1.7 数据分析
使用Excel进行数据统计,使用SPSS 25 Duncan新复极差法进行差异显著性检验,使用Origin 2022 软件绘图。
2 结果与分析
2.1 新疆野苹果种子不同贮藏时期植物激素含量、酶活性及萌发率比较
新疆野苹果种子ABA含量随贮藏时间的增加呈下降趋势(图1-A),贮藏0 d时ABA含量(w,后同)最高为80.22 ng·g-1,在贮藏60 d时ABA含量显著低于0、30、90 d三个时期(p<0.05),120 d时ABA含量达到最低为43.67 ng·g-1;GA含量随贮藏时间的增加呈上升趋势(图1-B),贮藏0 d时GA含量(b,后同)最低为78.42 pmol·g-1,120 d时GA含量显著高于其他时期(p<0.05),达到最高为170.67 pmol·g-1;IAA含量随贮藏时间的增加呈上升趋势(图1-C),贮藏60 d时IAA含量显著高于其他时期,达到高41.36 nmol·g-1;CTK含量在贮藏时间0~30 d时呈下降趋势(图1-D),贮藏30 d时CTK含量显著低于其他时期(p<0.05),达到最低值为31.34 ng·g-1,在贮藏30~60 d时CTK含量呈显著上升趋势;表明该时期CTK能促进种子同化物的积累。ETH生物合成途径中的ACC氧化酶(ACO)和ACC合成酶(ACS)两个关键酶在贮藏过程中的活性变化趋势不一致(图1-E、F)。在贮藏90 d时,ACO活性为262.52 ng·g-1,显著高于其他时期(p<0.05),贮藏30 d时,ACO活性最低为157.38 ng·g-1。ACS的活性在贮藏120 d时达到最大值418.92 ng·g-1,显著高于0、30、60 d三个时期(p<0.05)。说明低温沙藏层积促进GA、IAA、CTK、ACC氧化酶和ACC合成酶的合成,且ETH合成途径的两个酶可能对低温更加敏感。种子萌发率在低温沙藏层积30、60、90、120 d时萌发率分别为0%、7%、95%、100%。种子萌发率与GA、IAA含量及ACO活性变化趋势一致,与ABA含量变化趋势相反。
2.2 层积期间种子的转录组测序数据
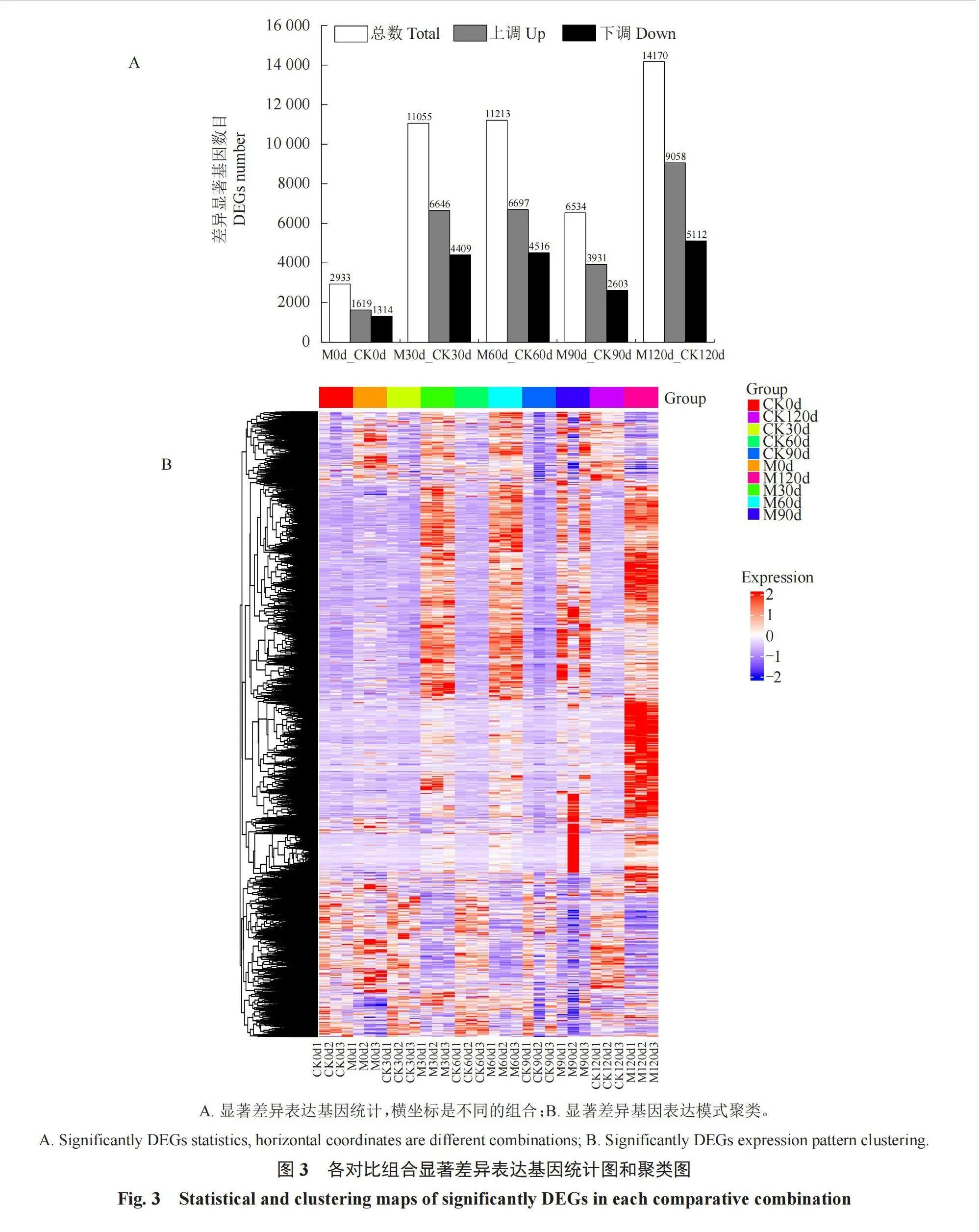
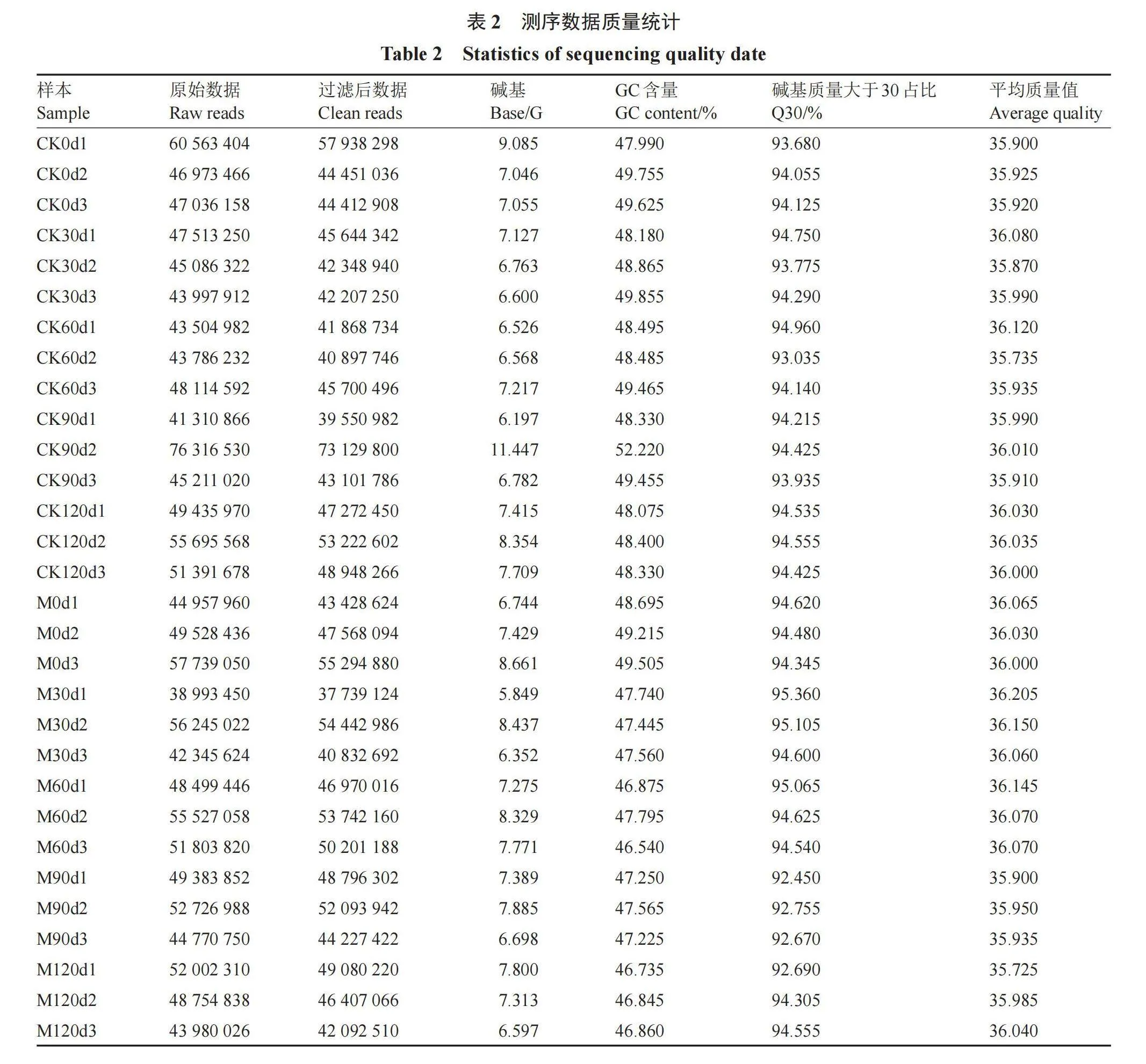
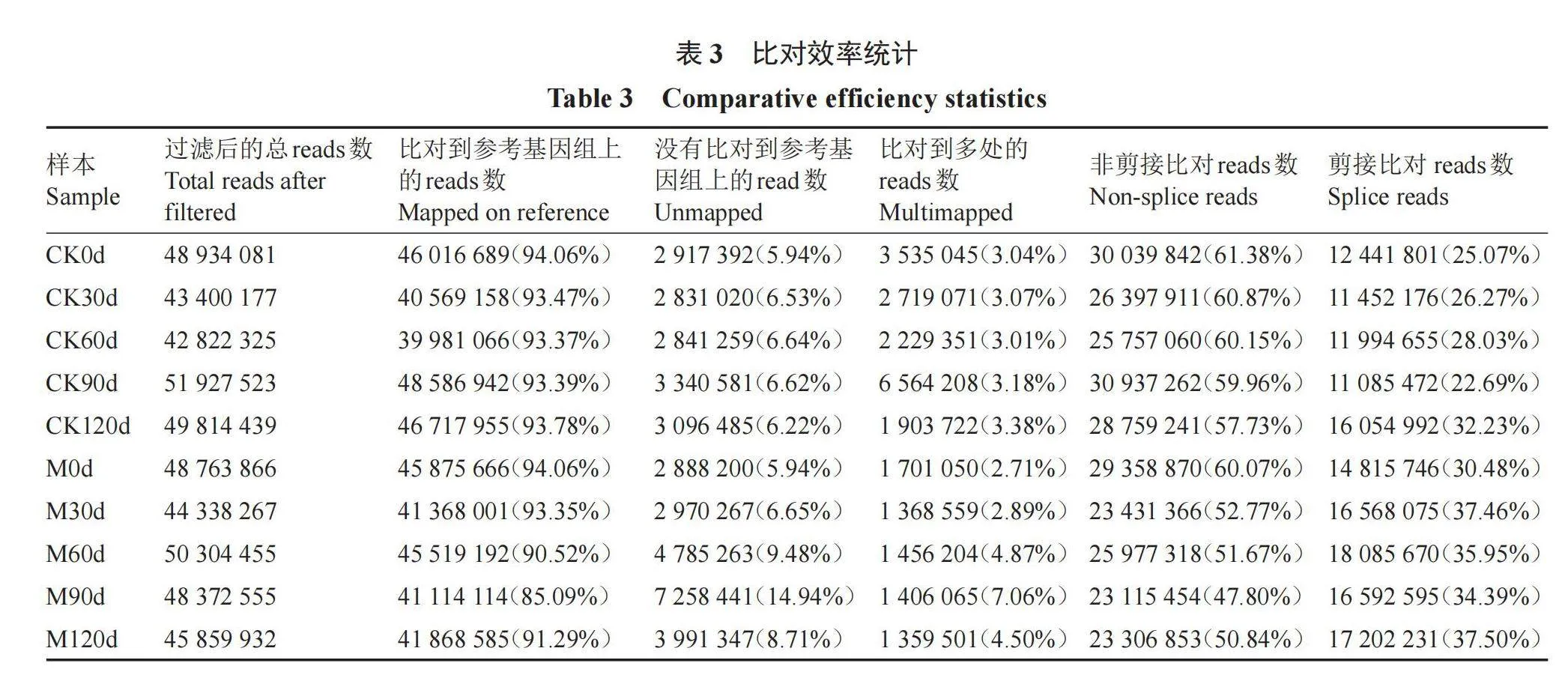
五个休眠解除时期总共得到209.45 G的高质量数据(表2),各样本Q30碱基百分比均高于92.9%,说明样品测序数据以及从头组装后的质量较高。将优化后的高质量数据序列与金冠苹果(M. domestica)的参考基因组进行比对(表3),比对效率为85.09%~94.06%,比对效率经分析后如果高于70%,说明所选参考基因组可满足信息分析需求。reads在参考基因组不同区域的分布情况,每个样品没有比对到基因间区的reads在5.94%~26.81%,可能来源于ncRNA或少许DNA片段污染;reads在基因组外显子区域在66.44%~91.47%,说明物种的参考基因注释较为完善。
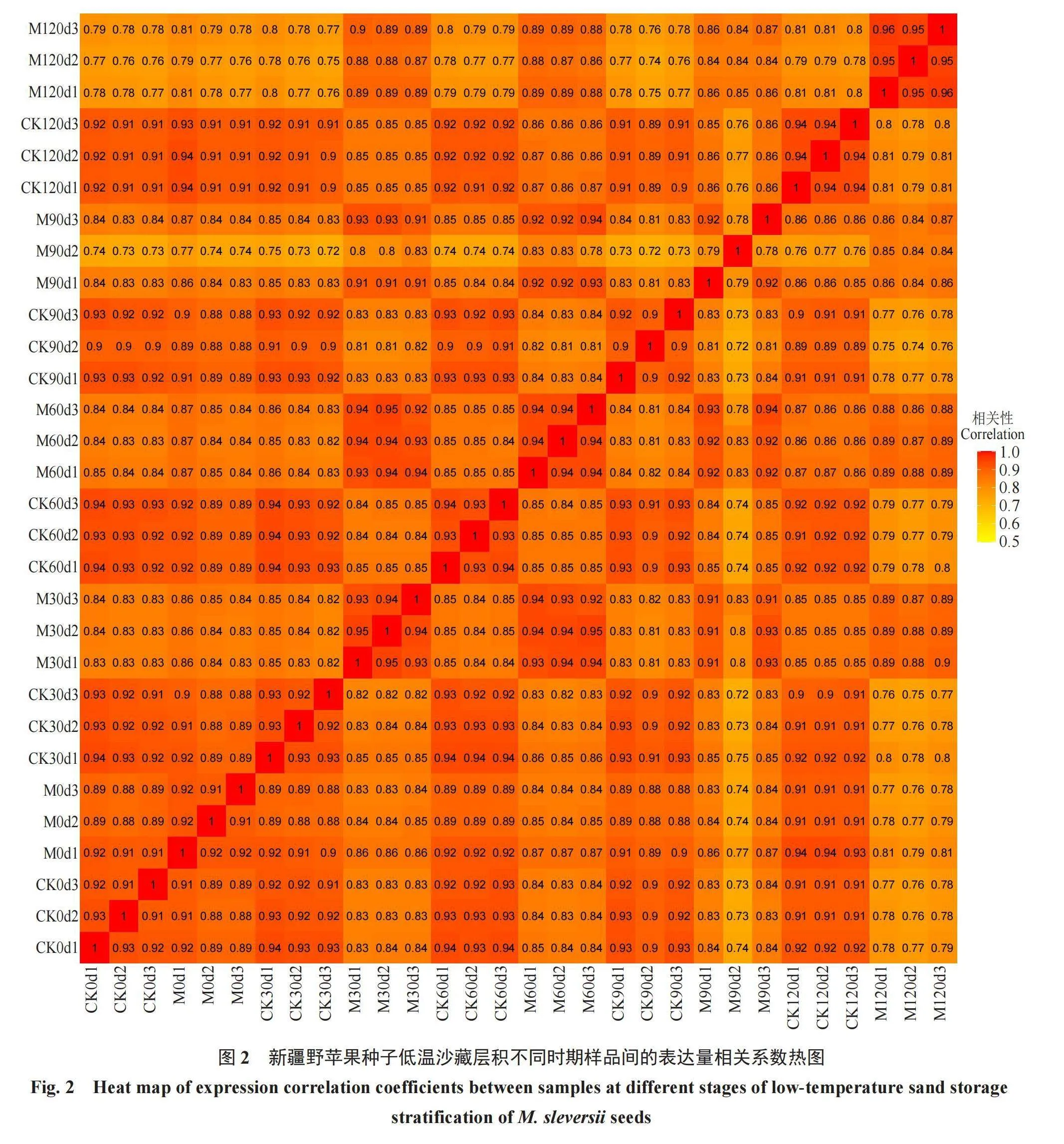
在差异表达分析时使用Pearson相关系数进行计算,样品间基因表达水平相关性是检验试验可靠性和样本选择是否合理的重要指标。根据各样本所有基因的FPKM值计算组内及组间样本的相关性系数,绘制成热图(图2),可直观显示组间样本差异及组内样本重复情况。不同颜色代表皮尔逊相关系数的大小,颜色越偏向红色代表样品间相关系数的绝对值越大。生物学重复样品间R2均大于0.7,相关系数越接近1,表明样品之间表达模式的相似度越高。相关性分析确保后续的差异基因分析得到更可靠的结果。
2.3 显著差异基因分析
显著差异基因分析表明DEGs在低温层积过程中显著富集(图3-A)。M120d_CK120d相对于M0d_CK0d和M30d_CK30d、M60d_CK60d和M90d_CK90d中有更多的显著DEGs,这表明M120d中具有更多的DEGs来调控种子萌发和生理变化。使用R软件包pheatmap进行基因和样品的双向层次聚类分析(图3-B),同处理不同重复间的基因表达模式相似,可能功能相似或参与生物学过程相同,而同时期不同处理间差异基因表达模式存在较大差异。韦恩图展示了层积和对照比较组合共有或独有的差异基因数(图4)。M30d_CK30d与M60d_CK60d时期显著差异表达基因、显著差异上调基因和显著差异下调基因分别共有7384、4875和3236个,表明在低温层积两个时期(M30d_CK30d与M60d_CK60d)之间有更多的显著差异基因参与种子休眠解除。
2.4 DEGs功能注释和富集分析
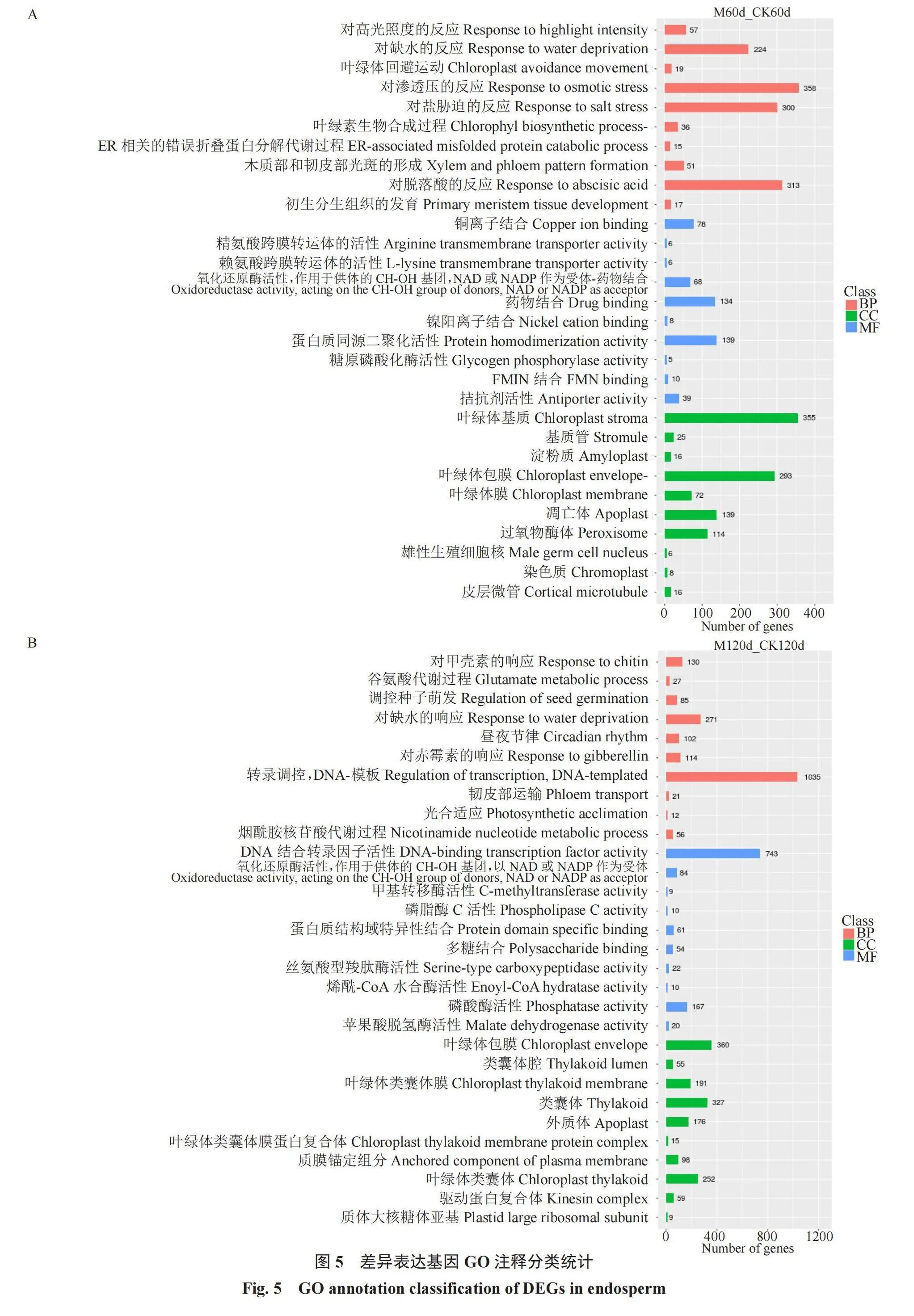
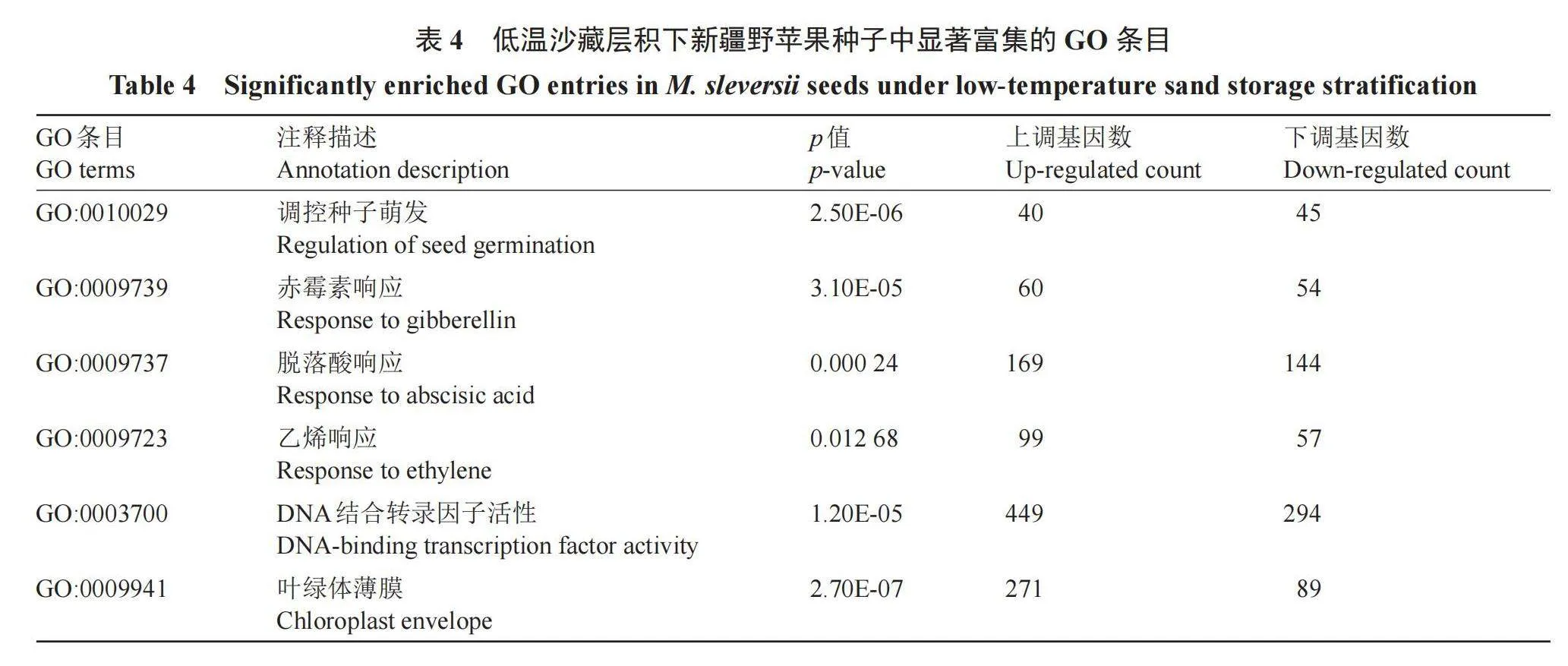
对差异基因进行显著富集GO条目(表4)分析,进一步了解DEGs的生物学功能。在生物过程中,有85个DEGs富集在调控种子萌发(GO:0010029),调控种子萌发中有45个DEGs下调表达,40个DEGs上调表达;在赤霉素响应(GO:0009739)和脱落酸响应(GO:0009737)中分别有114个和313个DEGs,其中赤霉素响应中有60个DEGs上调表达,54个DEGs下调表达;其中脱落酸响应中有169个DEGs上调表达,144个DEGs下调表达;富集在乙烯响应有156个DEGs,99个DEGs上调表达,57个下调表达。分子功能中,富集DEGs最高的是DNA结合转录因子活性(GO:0003700),449个上调表达,294个下调表达。细胞组成中,叶绿体薄膜(GO:0009941)中有271个上调表达,89个下调表达。说明植物激素相关调控基因对种子休眠与萌发的影响较大。
对DEGs进行GO富集(图5),生物过程主要参与渗透压和缺水的反应、脱落酸响应、盐胁迫的反应、转录调控、调控种子萌发和赤霉素响应等。细胞组分主要有叶绿体基质、叶绿体包膜和类囊体。分子功能主要与DNA结合转录因子活性、磷酸酶活性和蛋白质同源二聚体活化有关。新疆野苹果种子低温层积休眠解除过程中,多个植物激素生物学过程保持活跃变化,表明植物激素在种子休眠解除活动中发挥作用。
KEGG通路富集分析显示(图6),种子在不同时期低温沙藏层积过程中,主要富集的通路有植物激素信号转导、MAPK信号通路-植物、内质网中的蛋白加工、淀粉和蔗糖代谢、糖酵解/糖原生成等。ABA、GA、ETH等激素信号转导通路相关基因参与种子休眠解除等过程,淀粉和蔗糖代谢通路参与种胚的碳代谢过程,为种胚提供营养物质。糖原生成代谢通路参与细胞蛋白质的合成和代谢过程,为种胚萌发提供氮源和能量。
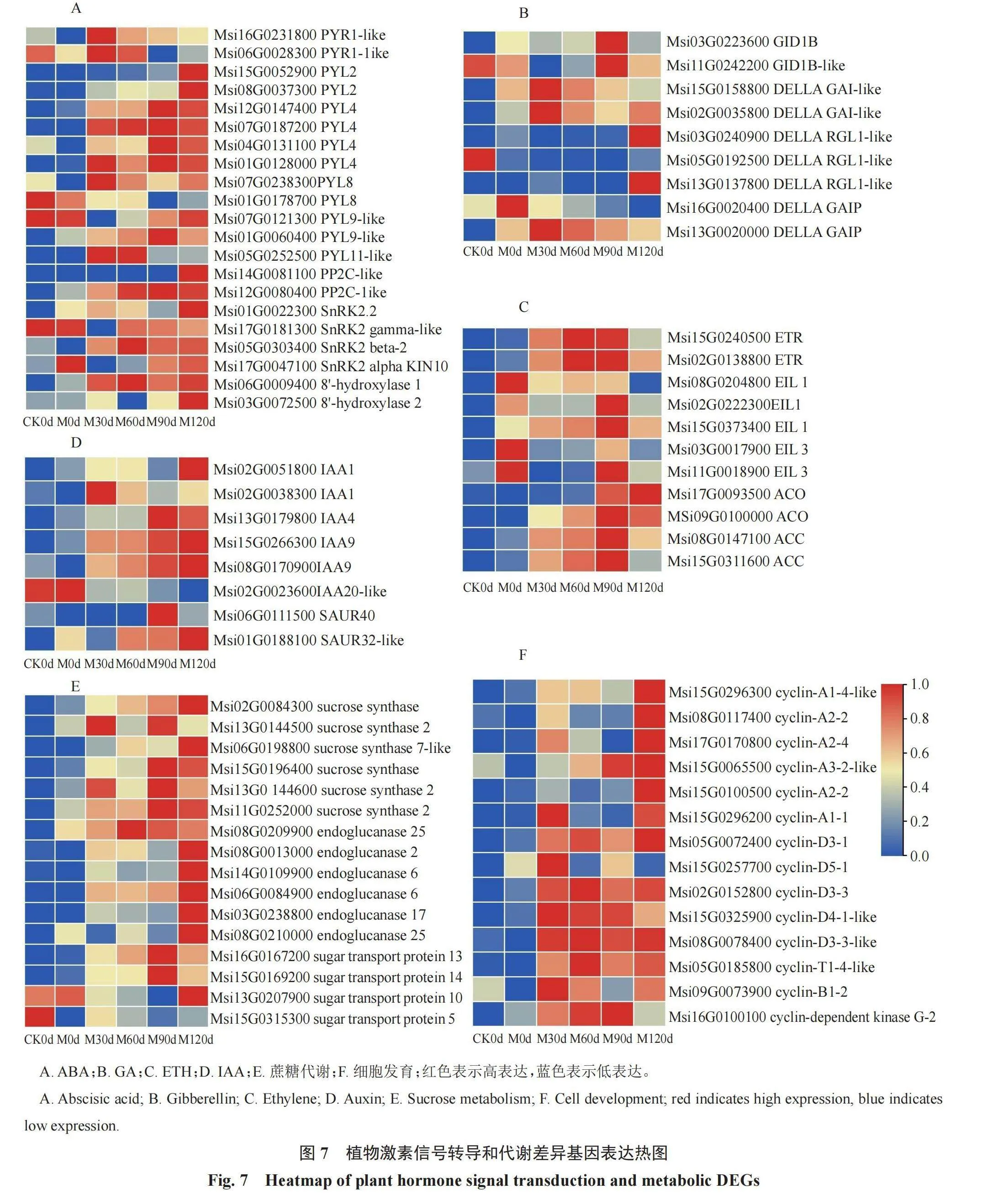
2.5 植物激素信号转导和代谢差异表达基因的筛选
根据KEGG通路分析,筛选在植物激素信号转导和代谢通路上富集的差异表达基因,分析新疆野苹果种子不同低温沙藏层积阶段的激素和代谢相关差异基因表达模式(图7),基于已报道的参与植物激素信号转导相关基因,参考苹果全基因组序列,筛选ABA、GA、ETH、IAA等激素差异表达基因。在贮藏处理下新疆野苹果植物激素相关差异基因均表现为上调基因数多于下调基因数。在新疆野苹果种子低温沙藏层积过程中,ABA受体PYR/PYL有12个基因上调和3个基因下调表达,ABA信号转导因子PP2C和正调控因子GA受体GID1基因呈上调表达趋势,SnRK2有3个基因上调和1个基因下调表达,ABA分解代谢8'-羟化酶2个基因呈上调表达。GA信号转导起负调控作用的生长抑制因子DELLA蛋白有6个基因上调和1个基因下调表达,表明低温沙藏层积处理增强GA信号转导。ETH、IAA在新疆野苹果种子不同层积阶段中呈现出显著差异表达,ETH受体ETR、正调控信号因子EIL(ETHYLENE INSENSITIVE 3/ EIN3-LIKE)、ETH的直接前体ACC和ACC氧化酶(ACO)呈上调表达;IAA相关基因5个上调和1个下调表达,生长素上调小RNA(small auxin up-regulated,SAUR)上调表达,表明低温沙藏层积促进IAA的合成。
细胞发育生理变化在种子休眠到休眠解除过程中发挥作用,筛选发现许多DEGs参与细胞分裂和生长发育。细胞周期蛋白A(cyclin-A)和细胞周期蛋白D(cyclin-D)调控细胞有丝分裂活动,细胞周期蛋白基因上调表达,满足种子休眠解除的营养生长需求。另外,筛选到与蔗糖代谢有关的基因,蔗糖分解基因(sucrose synthase,SUS)和内切葡聚糖酶(endoglucanase,EG)在低温沙藏层积过程中上调表达;蔗糖转化酶13、14和1(sugar transport protein,STP)上调表达,而STP5下调表达,这说明蔗糖代谢提供种子发育所需的能量。
2.6 转录因子分析
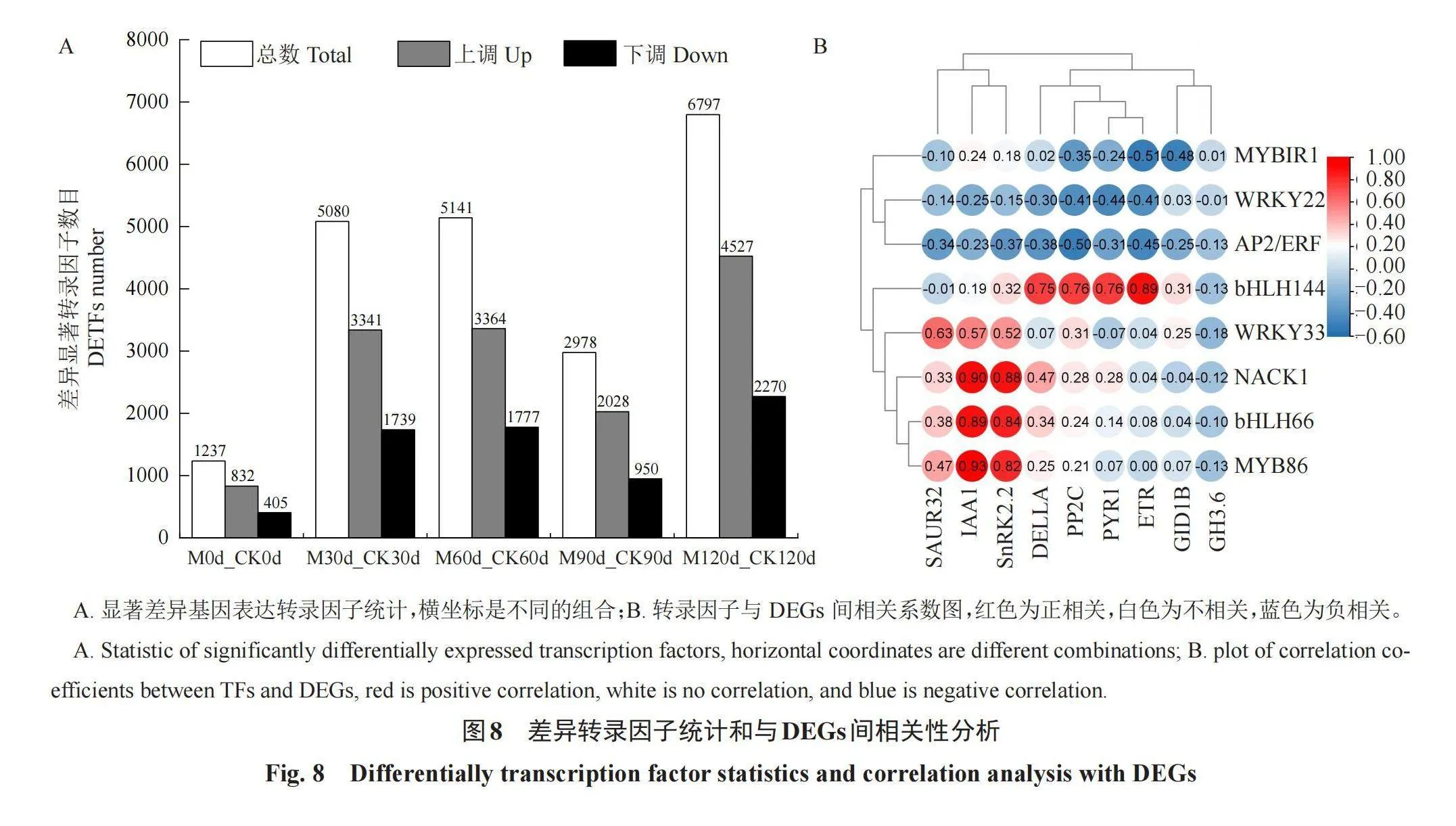
M0d_CK0d、M30d_CK30d、M60d_CK60d、M90d_CK90d、M120d_CK120d五个比较组中差异显著TFs分别为1237、5080、5141、2978和6797个(图8-A)。在M0d_CK0d中,832个TFs上调和405个TFs下调;在M30d_CK30d中,3341个TFs上调和1739个TFs下调;在M60d_CK60d中,3364个TFs上调和1777个TFs下调;在M90d_CK90d中,2028个TFs上调和950个TFs下调;在M120d_CK120d中,4527个TFs上调和2270个TFs下调。
采用皮尔逊相关系数分析8个TFs和9个DEGs间的相关性(图8-B),bHLH144与ETR、DELLA、PP2C和PYR1呈显著正相关(p<0.01);WRKY33、MYB86与SAUR32、IAA1和SnRK2.2呈显著正相关(p<0.01);bHLH66与IAA1、SnRK2.2呈显著正相关(p<0.01),MYB家族NACK1与IAA1、SnRK2.2和DELLA呈显著正相关(p<0.01);MYB1R1与ETR和GID1B呈显著负相关(p<0.01),WRKY22与ETR、PP2C和PYR1呈负相关(p<0.05),AP2/ERF与PP2C呈显著负相关(p<0.01),与ETR、DELLA和SnRK2.2呈负相关(p<0.05)。
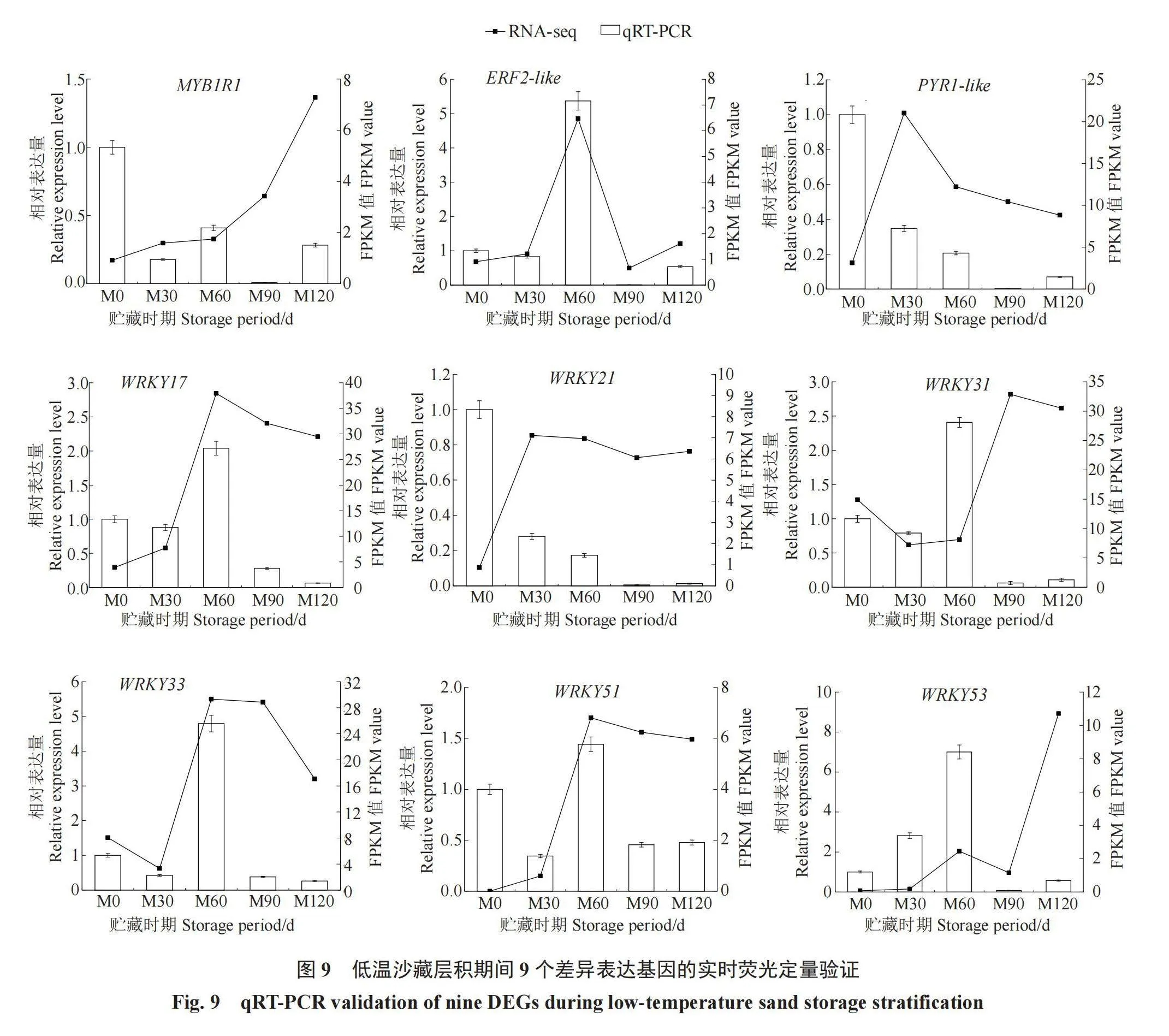
2.7 新疆野苹果种子休眠和激素差异基因的qRT-PCR验证
为验证转录组数据结果,挑选9个与种子休眠和激素相关的DEGs进行qRT-PCR实验(图9),测定其在种子低温层积期间的表达模式,结果显示所选的9个DEGs的qRT-PCR表达模式大多与转录组测序结果基本一致。结合生物学重复的要求,说明转录组数据可靠。
3 讨 论
转录组技术是分析种子休眠和萌发相关问题的常用方法之一。笔者在本研究中采用Illumina平台对新疆野苹果种子未层积和低温沙藏层积过程样品进行转录组测序,总共获得209.45 G的高质量数据,与金冠苹果的参考基因组进行比对,比对效率为85.09%~94.06%。陈静等[13]利用RNA-seq技术对花生种子休眠和萌发四个时期的样本进行转录组测序,四个时期共表达1206个差异unigenes。发现ABA 8'-hydroxylases、GA20ox、ACO、EREBP-like和热激蛋白等调控花生种子休眠解除及萌发,植物激素(GA、ABA、ETH、IAA)相关unigenes在花生种子休眠解除过程中呈显著差异。独行菜(Lepidium apetalum)低温萌发停滞前后的种子,经Illumina HiseqTM 2000高通量测序平台转录组测序,获得2108个差异基因,主要富集在转录调控和植物激素信号转导等相关过程中[26]。鲁强[27]基于比较转录组学研究15 ℃暖温层积处理三桠苦(Melicope pteleifoli)种子休眠解除的分子机制,主要与植物激素信号传导通路中ABA受体、GA受体、PP2C和DELLA蛋白等差异基因的显著变化相关。
笔者在本研究中对未层积和低温沙藏层积处理新疆野苹果种子样品进行对比,筛选出85个显著DEGs调控种子萌发,114个GA相关的显著DEGs,313个ABA相关的显著DEGs和156个ETH相关的显著DEGs。生物过程主要参与转录调控,分子功能主要与DNA结合转录因子活性、磷酸酶活性和蛋白质同源二聚体活化有关。龙佳丽等[28]研究低温处理前后甜菜DEGs功能富集,发现激素信号IAA和ABA信号通路与低温胁迫密切相关。何鑫鑫等[29]使用RNA-Seq测序,分析了Wus2和IPT转基因AC(Ailsa Craig)番茄类愈伤组织与普通下胚轴之间的基因表达差异,在激素信号转导通路中富集到60个差异表达基因,其中上调表达基因34个,下调表达基因26个。KEGG通路富集分析显示,新疆野苹果种子在不同时期低温沙藏层积过程中,主要富集的通路有植物激素信号转导、MAPK信号通路-植物、内质网中的蛋白加工、淀粉和蔗糖代谢、糖酵解/糖原生成等。表明以上途径在新疆野苹果种子低温沙藏层积过程中起积极作用。笔者在本研究中发现,细胞周期蛋白基因、蔗糖分解基因和内切葡聚糖酶在低温沙藏层积过程中上调表达,促使种胚代谢活动增加,将贮藏物质转化为种子萌发营养物质。
植物激素在种子休眠和萌发过程中,调节激素含量和信号转导的平衡。在静息状态下ABA受体PYR/PYLs/RCARs以二聚体形式存在,与ABA结合时以单体形式与PP2Cs结合,通过抑制PP2Cs,解除PP2Cs对蛋白激酶SnRK2s的抑制,从而产生转录因子ABI5和RAV1被磷酸化,以激活下游ABA响应基因[30]。ABA信号传导通路中PYR/PYL有3个基因下调表达,2个PP2C、3个SnRK2和2个ABA分解代谢8'-hydroxylases基因表达显著上调。相关研究表明,种子特异性磷酸酶PP2C控制在种子发育过程中起作用的高活性ABA信号通路[31]。在水稻低温处理后,受体PYR/PYL蛋白有1个基因显著下调,PP2C有5个差异基因显著上调,SnRK2有2个差异表达基因显著上调[32]。以上说明新疆野苹果ABA信号通路基因表达情况与水稻的结果相似。GA信号转导中GA受体GID与负调控因子DELLA蛋白上调表达,在GA含量低时,DELLA蛋白会结合下游调控因子来抑制GA信号转导;而GA含量高时,DELLA蛋白与受体GID1感知结合,可促进GID1/GA/DELLA复合体的形成,解除DELLA对下游调控因子的抑制,调控植物内激素变化[33],说明低温沙藏层积能促进GA合成。IAA相关基因5个上调表达,SAUR上调表达。ETH受体ETR、ETHYLENE INSENSITIVE 3、ACC氧化酶上调表达。这与新疆野苹果种子低温沙藏层积过程中ABA含量下降、IAA和GA含量增加结果一致[12],低温沙藏层积促进了ACC氧化酶和ACC合成酶的合成。笔者在本研究中进一步证实植物激素信号转导途径在新疆野苹果种子低温沙藏层积中起到重要的调控作用,探究植物激素相关基因的表达,还需进一步研究新疆野苹果种子休眠解除的分子机制。
转录因子的主要作用是激活或阻遏基因的转录调控,其在植物发育、植物激素调节和逆境信号转导等过程中发挥作用[34]。笔者在本研究中发现bHLH144与ETR、DELLA、PP2C和PYR1呈显著正相关(p<0.01);WRKY33、MYB86与SAUR32、IAA1和SnRK2.2呈显著正相关(p<0.01);bHLH66与IAA1、SnRK2.2呈显著正相关(p<0.01),表明bHLH、WRKY和MYB等转录因子可能作为正调节因子参与激素相关基因的转录调控,在植物生长发育等过程中发挥作用。
4 结 论
综合分析了未层积和低温沙藏层积处理新疆野苹果种子的转录组测序结果,挖掘新疆野苹果种子休眠解除的相关基因,筛选出85个DEGs调控种子萌发、114个GA相关DEGs、313个ABA相关DEGs和156个ETH相关DEGs。ERF2-like表达量与ACO、ACS活性变化呈相反趋势、PYR1-like、WRKY33基因表达量与ABA含量变化均下降,说明以上基因可能参与ABA信号通路调控新疆野苹果种子休眠解除过程。
参考文献References:
[1] 阎国荣,许正. 中国新疆野生果树研究[M]. 北京:中国林业出版社,2010:108-109.
YAN Guorong,XU Zheng. Study on the wild fruit trees in Xinjiang,China[M]. Beijing:China Forestry Publishing House,2010:108-109.
[2] 于立洋,李政,韩佩尧,张静,汪敏骅,田晓晓,张军. 8个新疆野苹果优良无性系抗寒性比较[J]. 核农学报,2017,31(9):1827-1835.
YU Liyang,LI Zheng,HAN Peiyao,ZHANG Jing,WANG Minhua,TIAN Xiaoxiao,ZHANG Jun. Comparison of cold hardiness of 8 Malus sieversii clones[J]. Journal of Nuclear Agricultural Sciences,2017,31(9):1827-1835.
[3] 魏景利,冯涛,张春雨,张艳敏,陈学森. 新疆野苹果种质资源的研究与应用[J]. 落叶果树,2009,41(4):16-18.
WEI Jingli,FENG Tao,ZHANG Chunyu,ZHANG Yanmin,CHEN Xuesen. Research and application of germplasm resources of Malus sieversii[J]. Deciduous Fruits,2009,41(4):16-18.
[4] 杨磊,廖康,佟乐,许正,刁永强,陈云华,皮里东. 影响新疆野苹果种子萌发相关因素研究初报[J]. 新疆农业科学,2008,45(2):231-235.
YANG Lei,LIAO Kang,TONG Le,XU Zheng,DIAO Yong-
qiang,CHEN Yunhua,PI Lidong. Primary report on effect of related factor on germination of Malus sieversii (Ledeb.) Rome. seed[J]. Xinjiang Agricultural Sciences,2008,45(2):231-235.
[5] 何晨晨,刘俐君,鲁晓燕. 基于转录组测序分析NaCl胁迫下新疆野苹果叶和根糖酵解相关基因的表达[J]. 果树学报,2020,37(7):951-961.
HE Chenchen,LIU Lijun,LU Xiaoyan. Analysis of genes related to glycolysis in the leaves and roots of Malus sieversii under NaCl stress based on transcriptome sequencing[J]. Journal of Fruit Science,2020,37(7):951-961.
[6] 马红喜,刘俐君,苏永峰,张德恩,袁引燕,鲁晓燕. 基于转录组测序筛选新疆野苹果组培苗应答冻害光合特性相关基因[J]. 果树学报,2022,39(9):1529-1539.
MA Hongxi,LIU Lijun,SU Yongfeng,ZHANG De’en,YUAN Yinyan,LU Xiaoyan. Screening of freezing stress-responsive genes related to photosynthesis in in vitro seedlings of Malus sieversii via RNA-seq[J]. Journal of Fruit Science,2022,39(9):1529-1539.
[7] 苏永峰,刘俐君,马红喜,袁引燕,张德恩,鲁晓燕. 基于转录组测序筛选新疆野苹果组培苗应答冻害谷胱甘肽代谢相关的基因[J]. 果树学报,2023,40(5):829-840.
SU Yongfeng,LIU Lijun,MA Hongxi,YUAN Yinyan,ZHANG De’en,LU Xiaoyan. Screening of genes related to glutathione metabolism responding to freezing stress in Malus sieversii seedlings in vitro based on transcriptome sequencing[J]. Journal of Fruit Science,2023,40(5):829-840.
[8] FENG S Y,LIU Z,CHEN H L,LI N,YU T,ZHOU R,NIE F L,GUO D,MA X,SONG X M. PHGD:An integrative and user-friendly database for plant hormone-related genes[J]. Imeta,2024,3(1):e164.
[9] SAJEEV N,KOORNNEEF M,BENTSINK L. A commitment for life:Decades of unraveling the molecular mechanisms behind seed dormancy and germination[J]. The Plant Cell,2024,36(5):1358-1376.
[10] URBANOVA T,LEUBNER-METZGER G. Gibberellins and seed germination[J]. Annual plant reviews online,2016,49:253-284.
[11] LI X Y,CHEN T T,LI Y,WANG Z,CAO H,CHEN F Y,LI Y,SOPPE W J J,LI W L,LIU Y X. ETR1/RDO3 regulates seed dormancy by relieving the inhibitory effect of the ERF12-TPL complex on DELAY OF GERMINATION1 expression[J]. The Plant Cell,2019,31(4):832-847.
[12] 马娟,杨巧玲,徐昕洁,苏布哈尼·买买提江,尼杂木丁·吐逊,叶春秀. 新疆野苹果种子低温层积过程中生理生化指标变化趋势[J/OL]. 分子植物育种,2023:1-17(2023-05-30). https://kns.cnki.net/kcms/detail/46.1068.S.20230529.1534.034.html.
MA Juan,YANG Qiaoling,XU Xinjie,Subhani Maimaitijiang,Nizhamudin Tusun,YE Chunxiu. Variation trends of physiological and biochemical indices during low temperature stratification of seeds of Malus sieversii (Ledeb.) M. Roem[J/OL]. Molecular Plant Breeding,2023:1-17(2023-05-30). https://kns.cnki.net/kcms/detail/46.1068.S.20230529.1534.034.html.
[13] 陈静,江玲,王春明,胡晓辉,翟虎渠,万建民. 花生种子休眠解除过程中相关基因的表达分析[J]. 作物学报,2015,41(6):845-860.
CHEN Jing,JIANG Ling,WANG Chunming,HU Xiaohui,ZHAI Huqu,WAN Jianmin. Expression analysis of genes involved in peanut seed dormancy release (Arachis hypogaea L.)[J]. Acta Agronomica Sinica,2015,41(6):845-860.
[14] 高云鹏. 紫荆种子休眠解除过程中生理生化变化及分子机理研究[D]. 南京:南京林业大学,2020.
GAO Yunpeng. Study on the physiological and biochemical changes and molecular mechanisms in Cercis chinensis seeds during dormancy releasing[D]. Nanjing:Nanjing Forestry University,2020.
[15] ZHANG J,QIAN J Y,BIAN Y H,LIU X,WANG C L. Transcriptome and metabolite conjoint analysis reveals the seed dormancy release process in callery pear[J]. International Journal of Molecular Sciences,2022,23(4):2186.
[16] 王菲菲,张胜忠,胡晓辉,CHU Ye,崔凤高,钟文,赵立波,张天雨,郭进涛,于豪谅,苗华荣,陈静. 比较转录组分析花生种子休眠调控网络[J]. 作物学报,2023,49(9):2446-2461.
WANG Feifei,ZHANG Shengzhong,HU Xiaohui,CHU Ye,CUI Fenggao,ZHONG Wen,ZHAO Libo,ZHANG Tianyu,GUO Jintao,YU Haoliang,MIAO Huarong,CHEN Jing. Comparative transcriptome profiling of dormancy regulatory network in peanut[J]. Acta Agronomica Sinica,2023,49(9):2446-2461.
[17] BOLGER A M,LOHSE M,USADEL B. Trimmomatic:A flexible trimmer for Illumina sequence data[J]. Bioinformatics,2014,30(15):2114-2120.
[18] KIM D,LANGMEAD B,SALZBERG S L. HISAT:A fast spliced aligner with low memory requirements[J]. Nature Methods,2015,12(4):357-360.
[19] LOVE M I,HUBER W,ANDERS S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2[J]. Genome Biology,2014,15(12):550.
[20] SCHULZE S K,KANWAR R,GÖLZENLEUCHTER M,THERNEAU T M,BEUTLER A S. SERE:Single-parameter quality control and sample comparison for RNA-Seq[J]. BMC Genomics,2012,13:524.
[21] ASHBURNER M,BALL C A,BLAKE J A,BOTSTEIN D,BUTLER H,CHERRY J M,DAVIS A P,DOLINSKI K,DWIGHT S S,EPPIG J T,HARRIS M A,HILL D P,ISSEL-TARVER L,KASARSKIS A,LEWIS S,MATESE J C,RICHARDSON J E,RINGWALD M,RUBIN G M,SHERLOCK G. Gene ontology:Tool for the unification of biology. The Gene Ontology Consortium[J]. Nature Genetics,2000,25(1):25-29.
[22] KANEHISA M,GOTO S. KEGG:Kyoto encyclopedia of genes and genomes[J]. Nucleic Acids Research,2000,28(1):27-30.
[23] YU G C,WANG L G,HAN Y Y,HE Q Y. clusterProfiler:An R package for comparing biological themes among gene clusters[J]. OMICS,2012,16(5):284-287.
[24] CHEN C J,WU Y,LI J W,WANG X,ZENG Z H,XU J,LIU Y L,FENG J T,CHEN H,HE Y H,XIA R. TBtools-II:A “one for all,all for one” bioinformatics platform for biological big-data mining[J]. Molecular Plant,2023,16(11):1733-1742.
[25] LIVAK K J,SCHMITTGEN T D. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C(T)) Method[J]. Methods,2001,25(4):402-408.
[26] 周茜. 独行菜种子转录组及低温萌发表达谱分析[D]. 乌鲁木齐:新疆师范大学,2016.
ZHOU Qian. Transcriptome of seed and DGE method research of seed low temperature germination period in Lepidium apetalum Willd[D]. Urumqi:Xinjiang Normal University,2016.
[27] 鲁强. 三桠苦种子休眠的解除方法和初步机理研究[D]. 广州:广州中医药大学,2019.
LU Qiang. Study on the methods and preliminary mechanism of seed dormancy release of Melicope pteleifol[D]. Guangzhou:Guangzhou University of Chinese Medicine,2019.
[28] 龙佳丽,邹奕,邳植,吴则东. 转录组测序揭示激素介导的信号通路参与甜菜低温应答[J]. 中国农学通报,2021,37(31):5-14.
LONG Jiali,ZOU Yi,PI Zhi,WU Zedong. Transcriptome analysis reveals hormone-mediated signaling pathways involved in low temperature response in sugar beet[J]. Chinese Agricultural Science Bulletin,2021,37(31):5-14.
[29] 何鑫鑫,黄家权. Wus2和IPT转基因番茄类愈伤组织的转录组分析[J]. 中国瓜菜,2024,37(6):27-36.
HE Xinxin,HUANG Jiaquan. Transcriptome analysis of transgenic tomato callus tissues from Wus2 and IPT[J]. China Cucurbits and Vegetables,2024,37(6):27-36.
[30] ZHAO Y,JIA K H,TIAN Y T,HAN K J,EL-KASSABY Y A,YANG H,SI H Y,SUN Y H,LI Y. Time-course transcriptomics analysis reveals key responses of Populus to salt stress[J]. Industrial Crops and Products,2023,194:116278.
[31] ANTONI R,GONZALEZ-GUZMAN M,RODRIGUEZ L,RODRIGUES A,PIZZIO G A,RODRIGUEZ P L. Selective inhibition of clade A phosphatases type 2C by PYR/PYL/RCAR abscisic acid receptors[J]. Plant Physiology,2012,158(2):970-980.
[32] 高红秀,朱琳,刘天奇,张忠臣. 水稻植物激素响应低温胁迫反应的转录组分析[J]. 分子植物育种,2021,19(13):4188-4197.
GAO Hongxiu,ZHU Lin,LIU Tianqi,ZHANG Zhongchen. Transcriptomic analysis of plant hormone response to low temperature stress in rice[J]. Molecular Plant Breeding,2021,19(13):4188-4197.
[33] 张珏锋,李芳,钟海英,陈建明. 茭白‘浙农7号’不同生长阶段的叶片转录组学及激素水平分析[J]. 农业生物技术学报,2024,32(2):299-310.
ZHANG Juefeng,LI Fang,ZHONG Haiying,CHEN Jianming. Analysis of transcriptomics and hormone levels of Jiaobai ‘Zhenong 7’ in different growth stages[J]. Journal of Agricultural Biotechnology,2024,32(2):299-310.
[34] BAKSHI M,OELMÜLLER R. WRKY transcription factors:Jack of many trades in plants[J]. Plant Signaling amp; Behavior,2014,9(2):e27700.
