Genome-wide identification of the CONSTANS-LIKE (COL) family and mechanism of fruit senescence regulation by PpCOL8 in sand pear (Pyrus pyrifolia)
2024-05-13YueXuShuruiSongHuiyingWangXilongCaoXinranZhaoWenliWangLiyueHuoYaweiLiMisganawWassieBinLuLiangChenHaiyanShi
Yue Xu ,Shurui Song ,Huiying Wang ,Xilong Cao ,Xinran Zhao ,Wenli Wang ,Liyue Huo,Yawei Li,Misganaw Wassie,Bin Lu,Liang Chen,Haiyan Shi#
1 College of Horticulture,Hebei Agricultural University,Baoding 071001,China
2 Wuhan Botanical Garden,Chinese Academy of Sciences,Wuhan 430074,China
Abstract Pyrus pyrifolia Nakai ‘Whangkeumbae’ is a sand pear fruit with excellent nutritional quality and taste.However,the industrial development of pear fruit is significantly limited by its short shelf life.Salicylic acid (SA),a well-known phytohormone,can delay fruit senescence and improve shelf life.However,the mechanism by which SA regulates CONSTANS-LIKE genes (COLs) during fruit senescence and the role of COL genes in mediating fruit senescence in sand pear are poorly understood.In this study,22 COL genes were identified in sand pear,including four COLs (PpCOL8,PpCOL9a,PpCOL9b,and PpCOL14) identified via transcriptome analysis and 18 COLs through genome-wide analysis.These COL genes were divided into three subgroups according to the structural domains of the COL protein.PpCOL8,with two B-box motifs and one CCT domain,belonged to the first subgroup.In contrast,the other three PpCOLs,PpCOL9a,PpCOL9b,and PpCOL14,with similar conserved protein domains and gene structures,were assigned to the third subgroup.The four COLs showed different expression patterns in pear tissues and were preferentially expressed at the early stage of fruit development.Moreover,the expression of PpCOL8 was inhibited by exogenous SA treatment,while SA up-regulated the expression of PpCOL9a and PpCOL9b.Interestingly,PpCOL8 interacts with PpMADS,a MADS-box protein preferentially expressed in fruit,and SA up-regulated its expression.While the production of ethylene and the content of malondialdehyde (MDA) were increased in PpCOL8-overexpression sand pear fruit,the antioxidant enzyme (POD and SOD) activity and the expression of PpPOD1 and PpSOD1 in the sand pear fruits were down-regulated,which showed that PpCOL8 promoted sand pear fruit senescence.In contrast,the corresponding changes were the opposite in PpMADS-overexpression sand pear fruits,suggesting that PpMADS delayed sand pear fruit senescence.The co-transformation of PpCOL8 and PpMADS also delayed sand pear fruit senescence.The results of this studyrevealed that PpCOL8 can play a key role in pear fruit senescence by interacting with PpMADS through the SA signaling pathway.
Keywords: Pyrus pyrifolia,CONSTANS-LIKE gene,salicylic acid,fruit senescence,MADS
1.lntroduction
Pyrus pyrifoliaNakai ‘Whangkeumbae’ is an essential sand pear fruit in China (Meng and Song 2003).The fresh pear fruit can be stored at room temperature for only two weeks or two months in cold storage,indicating a limited shelf life of the fruit (Wanget al.2009).Hence,the industrial development of sand pear is limited due to its short shelf life and fast aging after harvest.Therefore,delaying fruit senescence and prolonging the shelf life of sand pear is a critical challenge in the ‘Whangkeumbae’ pear industry.Understanding the molecular mechanisms underlying early fruit senescence and the identification of candidate genes that can delay fruit senescence in sand pear could be crucial for increasing the shelf life of the fruit.Phytohormones,such as salicylic acid (SA),are critical in delaying fruit ripening and senescence (Shiet al.2021).For instance,exogenous application of SA before harvest reduced the anthocyanin content,increased the chlorophyll content,and delayed fruit ripening (Hassanet al.2007).Additionally,treatment with SA after harvest delayed fruit senescence in sand pear (Hassanet al.2007).
Ethylene (ET) is an important hormone regulating climacteric fruit ripening and senescence (Barry and Giovannoni 2006).SA and its derivative acetylsalicylic acid (ASA) could inhibit the synthesis of ET (Leslie and Romani 1988).For instance,SA treatment inhibited ethylene synthesis in apple pulp slices and the reduction in ethylene was more pronounced with an increasing SA concentration (Fan and He 1998).Our previous studies found that SA could regulate the expression of two ethylene synthesis genes,the 1-aminocyclopropane-1-carboxylate (ACC) oxidase and ACC synthetase genes,in ‘Whangkeumbae’ pear fruit (Shi and Zhang 2012;Shiet al.2013).SA also regulated the expression of the ethylene receptor gene (PpERS) and the transcription factor gene (PpEIN3b),which are involved in the ethylene signaling pathway during fruit ripening and senescence in ‘Whangkeumbae’ pear (Zhanget al.2013;Shiet al.2019).Transcriptome analysis showed that postharvest treatment of pear fruits with SA regulated genes related to plant hormone biosynthesis and metabolism,cell wall metabolism and modification,antioxidant systems,and senescence-associated transcription factors (Shiet al.2021).
CONSTANS/CONSTANS-LIKE (COs/COLs) are zinc finger transcription factors containing a B-boxtype zinc finger and a CCT domain.The B-box-type zinc finger is found in the N-terminal and is believed to regulate protein-protein interactions,while the CCT domain is in the C-terminal region and is crucial for the nuclear localization of the proteins (Robsonet al.2001;Griffithset al.2003).InArabidopsis,there are 17 CO/COL members that can be divided into three subgroups according to the variations in the B-box domains (Robsonet al.2001;Hassidimet al.2009).Group I members,including CO and COL1-COL5,contain two B-box motifs.Group II contains only one B-box motif,such as in COL6-COL8 and COL16.Group III includes COL9-COL15 with one B-box motif and a second divergent B-box motif (Griffithset al.2003).In recent years theCO/COLgenes belonging to Group I have been identified in many plant species (Gangappa and Botto 2014;Zhanget al.2015;Chaurasiaet al.2016).
Functional studies showed thatCOs/COLscan play crucial roles in plant development,flowering,fruit ripening,and stress responses.For instance,overexpression ofCOL9delayed flowering inArabidopsisby reducing the expression of theCOandFTgenes (Cheng and Wang 2005).In rice,overexpression ofOsCOL9reduced the flowering time by repressing theEhd1pathway (Liuet al.2016b).In banana,the transcription level ofMaCOL1was increased during fruit ripening and stress treatments,suggesting thatMaCOL1may participate in fruit ripening and stress responses (Chenet al.2012).Meanwhile,few studies have investigated theCOLgenes and their role in fruit ripening in sand pear.Identifying critical genes involved in fruit ripening and senescence could be crucial for increasing the shelf life of sand pear fruit.In this study,22COLgenes were identified from sand pear (P.pyrifoliaNakai ‘Whangkeumbae’),including four previously identifiedCOLs viaRNA-sequencing (Shiet al.2021):PpCOL8,PpCOL9a,PpCOL9b,andPpCOL14.ThePpCOLgenes were divided into three subgroups according to their nuclear localization signal (NLS) motifs and CCT domains.
The accumulations of the fourCOLtranscripts identifiedviatranscriptome analysis were determined in all pear tissues,and all the transcripts were preferentially expressed at the early stage of fruit development.Moreover,PpCOL8andPpCOL9swere responsive to SA during pear fruit senescence.Interestingly,PpCOL8 interacted with PpMADS,a protein belonging to the MADS-box family,whose gene was predominantly expressed in fruit and up-regulated by exogenous SA.Furthermore,PpCOL8andPpCOL9were characterized during fruit senescence.These results suggest that the COL proteins,especially PpCOL8,could play a key role in fruit senescence by interacting with PpMADS through the SA signaling pathway.
2.Materials and methods
2.1.Plant materials
The plant materials were prepared according to our previous study (Shiet al.2019).Briefly,sand pear (P.pyrifoliaNakai ‘Whangkeumbae’) fruits were collected at 30,60,90,120,130,140,145,and 150 d after full bloom from the experimental farm of Hebei Agricultural University,Baoding,China.‘Whangkeumbae’ fruit growing at 150 d after full bloom is considered a fully mature fruit.After harvest,separate samples of the pear fruits were placed at room temperature for 5,10,15,20,25,and 30 d.The mesocarp of the pears was collected for further study.Young tissues,including shoots,stems,leaves,petals,and anthers,were collected from the same pear trees.Diseased fruits were chosen from pear fruit stored at room temperature for 30 d after harvest.The above samples were ground into a powder with liquid nitrogen for RNA isolation,respectively.
2.2.Fruit treatments
Pear fruits at 160 d after full bloom or 10 d after natural harvest,were used to prepare mesocarp discs using a hole punch.Subsequently,SA (Solarbio,Beijing,China) treatment was performed by immersing the mesocarp discs into solutions with various concentrations (0.002,0.02,0.2,and 2.0 mmol L-1) for 12 h,and control samples were treated with ddH2O for 12 h (Shiet al.2019).A total of 20 mesocarp discs were prepared from 20 fruits of the same pear tree for this experiment.Total RNA was extracted from SA-treated and control mesocarp discs with three replicates.
2.3.lsolation of PpCOLs
In a previous study,fourCOLgenes (PpCOL8,PpCOL9a,PpCOL9b,andPpCOL14) were identified from pear fruit through RNA sequencing (Shiet al.2021).Total RNA was isolated from pear fruit at 150 d after full bloom with a 0.2 mmol L-1SA treatment.Poly(A)+mRNA was purified with an mRNA purification kit (TIANGEN,Beijing,China).DEGs were screened,and fourCOLclones with complete sequences were identified.
For the genome-wide analysis ofCOLgenes fromP.pyrifolia,the genome file and genome annotation file (assembly number: GWHBAOS00000000) of the cultivar ‘Cuiguan’ were obtained from the National Genomics Date Center (NGDC) (Gaoet al.2021).The COL protein sequences of theArabidopsisCOL family (Robsonet al.2001;Griffithset al.2003) were downloaded from the NCBI database (https://www.ncbi.nlm.nih.gov/).We used two approaches to identify theCOLfamily members in theP.pyrifoliagenome.First,two conserved domains of COL transcription factors,CCT (PF06203) and zinc finger B-box (PF00643),were downloaded from the Pfam database (http://pfam.sanger.ac.uk/;http://pfam.janelia.org/) by a Hidden Markov Model (HMM) (Eddy 2011).The SPDE (Xuet al.2021) and TBtools (Chenet al.2020) software were used to perform the HMM search.Second,to obtain candidateCOLfamily members,an extensive local BLASTP alignment (E-value 1e-5) against all protein sequences ofP.pyrifoliawas performed,usingArabidopsis thalianaCOL proteins as a query.After removing the redundant sequences,the presence of CCT and B-box conserved domains were verified in the Pfam and NCBI-CDD databases (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi).
2.4.RNA extraction and real-time quantitative PCR (RT-qPCR) analysis
To quantify the expression levels of the fourPpCOLs,total RNA was extracted from young tissues,including shoots,stems,leaves,petals,anthers,and developing pulp,as described previously (Shiet al.2019).Total RNA was extracted from pear fruit samples to detect the expression ofPpCOL8,PpMADS,PpPOD1andPpSOD1.SYBR Green fluorescent dye (CoWin Biosciences.,Beijing,China) was used for RT-qPCR.PpUBI(GenBank accession number: AF195224) was used as a standard control.The gene-specific primers used for this experiment are shown in Appendix A.The RT-qPCR procedure was carried out according to our previous study (Shiet al.2019).Data were analyzed using SPSS software.
2.5.DNA sequencing and protein analysis
The NCBI Conserved Domain Search was carried out to identify the conserved domains of the PpCOL proteins (http://blast.ncbi.nlm.nih.gov/Blast.cgi).The physicochemical properties of the PpCOL proteins were investigated using the ProtParam online tool (https://web.expasy.org/protparam/).Furthermore,the Plant-mPLoc (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/) search was performed to predict the subcellular localization of the PpCOL proteins.Protein sequence homology analysis was carried out with the Clustal Omega tool (http://www.ebi.ac.uk/Tools/msa/clustalo/).We also identified protein motifs using the motif scan online tool (http://myhits.isb-sib.ch/cgi-bin/motif_scan),and proteinprotein interaction was predicted using the string (https://string-db.org/).The evolutionary relationships of the pear COLs were determined using MEGA7 software based on the Neighbor-Joining method (Kumaret al.2016).The genomic DNA sequences of the fourPpCOLswere analyzed according to genome information of sand pear (Gaoet al.2021) using the GDR database (https://www.rosaceae.org/).TheP.pyrifoliacultivar ‘Cuiguan’ genome annotation file was acquired from NGDC (https://ngdc.cncb.ac.cn/),and TBtools (Chenet al.2020) was used to mapPpCOL8,PpCOL9a,PpCOL9b,andPpCOL14to the sand pear chromosomes.
2.6.Yeast two-hybrid assays
Yeast two-hybrid assay (Y2H) was conducted according to the Matchmaker Gold Y2H Library Screening System.The ORF sequences ofPpCOL8,PpCOL9aandPpCOL9bwere amplified and cloned into the MCS of pGBKT7 to generate the bait vector.The ORF sequence of the STRING predicted-interacting protein,PpMADS,was amplified and inserted into the pGADT7 vector to generate the prey vector (pGADT7-PpMADS).All primers used in this experiment are listed in Appendix B.The plasmid constructs were co-transformed into the competent yeast strain AH109.Subsequently,the yeast cells were spread-plated on SD/-Trp/-Leu agar plates.After three days incubation at 30°C,colonies were selected and streaked onto SD/-Trp/-Leu/-His and SD/-Trp/-Leu/ His/-Ade agar plates supplemented with 0,10,and 25 mmol L-13-AT.Protein-protein interactions were observed after 3 d of incubation at 30°C.
2.7.Bimolecular fluorescence complementation (BiFC) assays
BecausePpCOL8showed interaction withPpMADS,we conducted a BiFC assay to verify the Y2H results.The CDS ofPpCOL8andPpMADSwithout the termination codons were fused with the N-terminal and C-terminal of the yellow fluorescence protein (YFP) to generatePpCOL8-N-YFPandPpMADS-C-YFP,respectively.All primers used are presented in Appendix B.The transformedAgrobacterium tumefaciensstrain GV3101 was co-infected intoNicotiana benthamianaleaves.The infiltrated tobacco was cultivated for 24 h in the dark and then transferred to a 14 h:10 h/light:dark photoperiod at 24°C for another 24 h.Subsequently,protein-protein interaction was verified by the appearance of yellow fluorescence using laser scanning confocal microscopy (Leica,Wetzlar,Germany).
2.8.Transient overexpression of the PpCOL8 and PpMADS genes in sand pear fruit
The transient overexpression genetic transformation experiment in sand pear followed Voinnetet al.(2003) with slight modifications.PpCOL8andPpMADSwere transferred into the overexpression vector.The primer sequences are presented in Appendix A.The integrated vector was transferred into theAgrobacteriumstrain GV3101,and the bacteria were suspended to an OD of 1.0 with an infiltrating buffer,which was then held at 4°C for 2-4 h.A mixture of about 300 μLAgrobacteriumwas injected into mature sand pear fruits and co-cultured for 2 d.Empty vector was used as a negative control.All samples were frozen in liquid nitrogen and stored at -80°C.
2.9.Measurement of ethylene production in sand pear fruits
The ethylene production determination followed Liet al.(2015) with slight modifications.For each sample,the sand pear fruits were packed into an airtight container with a rubber stopper and kept at room temperature for 2 h,then 1 mL of gas was collected with a syringe to measure ethylene production.Ethylene was detected by using a gas chromatograph (Agilent,7820A,USA).Ethylene production was measured for at least three sand pear fruits at each sampling site.
2.10.Assays of malondialdehyde (MDA) content and the peroxidase (POD) and superoxide dismutase (SOD) activities
The determination of MDA content and the POD and SOD enzyme activities followed Chenet al.(2019) with slight modifications.Sand pear fruit samples (1.5 g) were homogenized in 9 mL of 10% TCA.The homogenate was centrifuged at 10,000×g for 20 min.A 2.0 mL supernatant sample (2.0 mL of 10% TCA as control) was absorbed and 2 mL of 2.067% TBA was added to it,and then the solution was cultured in boiling water for 20 min.The reaction was terminated by cooling on ice.The MDA absorption was measured using a spectrophotometer at 450,532,and 600 nm.
Sand pear fruit samples (1 g) were ground in a chilled mortar with 10 mL of 50 mmol L-1potassium phosphate buffer (pH 7.8).Each homogenate was centrifuged at 12,000×g for 15 min at 4°C,and the supernatant was used for the following assays.The 3.0 mL reaction system contained 100 mmol L-1potassium phosphate buffer (pH 7.0),1 mL of 45 mmol L-1H2O2and 0.5 mL enzyme extraction,and POD was determined at 470 nm.For SOD activity determination,the reaction mixture (4.5 mL) contained 50 mmol L-1potassium phosphate buffer (pH 7.8),104 mmol L-1methionine,0.3 mmol L-1nitroblue tetrazolium (NBT),0.32 mmol L-1riboflavin,0.8 mol L-1ethylene diamine tetraacetic acid (EDTA),and 500 μL enzyme extract.The reaction mixtures were illuminated at 4,000 lux for 20 min.The SOD activity was assayed by determining the activity of the enzyme extracts at 560 nm.
3.Results
3.1.ldentification of COL genes in pear
According to the conserved domains,the fourCOLswere designated asPpCOL8(GenBank accession number: MT879750),PpCOL9a(GenBank accession number: MT879753),PpCOL9b(GenBank accession number: MT879752),andPpCOL14(GenBank accession number: MT879751).Moreover,comparative genomics identified 19 COL transcription factors containing B-box and CCT conserved domains from theP.pyrifoliagenome.Among the 19COLs,oneCOLgene (EVM0031482.1) shared 100% identity withPpCOL9b.Hence,a total of 22COLgenes were identified from sand pear.
As shown in Table 1,the lengths of the amino acid sequences of the PpCOL proteins vary from 340 aa (Pp0011588,Pp0033434) to 488 aa (PpCOL14).ThePpCOLgenes show distinct physical and chemical properties,with molecular weights ranging from 37 to 53 kDa and isoelectric points (pI) from 5.11 to 7.46.The pI values of 21 PpCOLs are weakly acidic,indicating that most of the COL proteins of sand pear are rich in acidic amino acids.Among the 22 PpCOL members,PpCOL14 and Pp0034404 are stable,while the other 20 PpCOL proteins are unstable.According to the predicted subcellular localization,allPpCOLgene family members are located in the nucleus,so this family of genes may play an important role in the nucleus.

Table 1 The characteristics of COLs in sand pear1)
3.2.Phylogenetic and conserved motif analyses of PpCOLs
A phylogenetic tree was constructed using the full-length PpCOL protein sequences to explore their evolutionary relationships.As shown in Fig.1-A,the 22 PpCOL proteins were divided into three groups.Group I contained seven PpCOL proteins,including PpCOL8 which contains one CCT domain and two B-box motifs (Fig.2).Group II included six PpCOL members (Fig.1-A).The remaining nine PpCOL members,including PpCOL9a,PpCOL9b,and PpCOL14,each with one CCT,one B-box,and a more divergent zinc finger domain belonged to Group III.
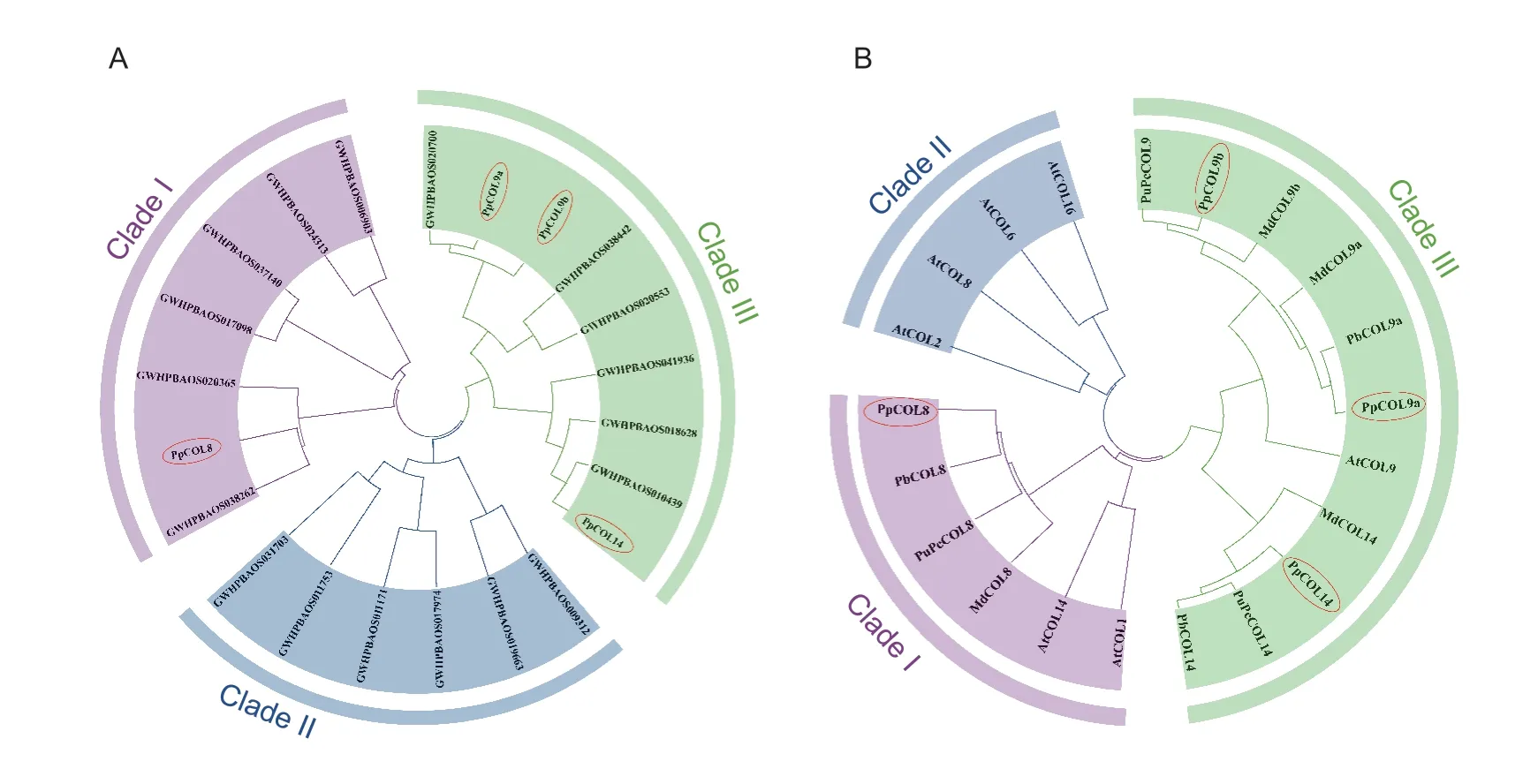
Fig.1 Phylogenetic trees of the COL proteins in the Pyrus pyrifolia genome (A) and in pear,apple and Arabidopsis (B).The phylogenetic trees were constructed based on the bootstrap analysis of 1,000 replicates using the Neighbor-Joining (NJ) method in MEGA7.0,and are divided into three clades.GenBank accession numbers are MT879752 (PpCOL9b,Pyrus pyrifolia),KAB2634449 (PuPcCOL9,Pyrus ussuriensis×Pyrus communis),ARO70339 (MdCOL9b,Malus domestica),RXH67859 (MdCOL9a),MT879753 (PpCOL9a),XP_009361520 (PbCOL9a,Pyrus×bretschneideri),NP_001280792 (MdCOL14),KAB2599070 (PuPcCOL14),MT879751 (PpCOL14),XP_009336468 (PbCOL14),KAB2634042 (PuPcCOL8),XP_009378312 (PbCOL8),MT879750 (PpCOL8),XP_008379481(MdCOL8),RXH67638 (MdCOL13),KAB2610965 (PuPcCOL13),and XP_009378335 (PbCOL13).The Arabidopsis COL accession numbers are NM_121590 (AtCOL1),NM_111105 (AtCOL2),AY081541 (AtCOL6),NM_103803 (AtCOL8),NM_111644 (AtCOL9),NM_128910 (AtCOL14),and NM_102355 (AtCOL16).
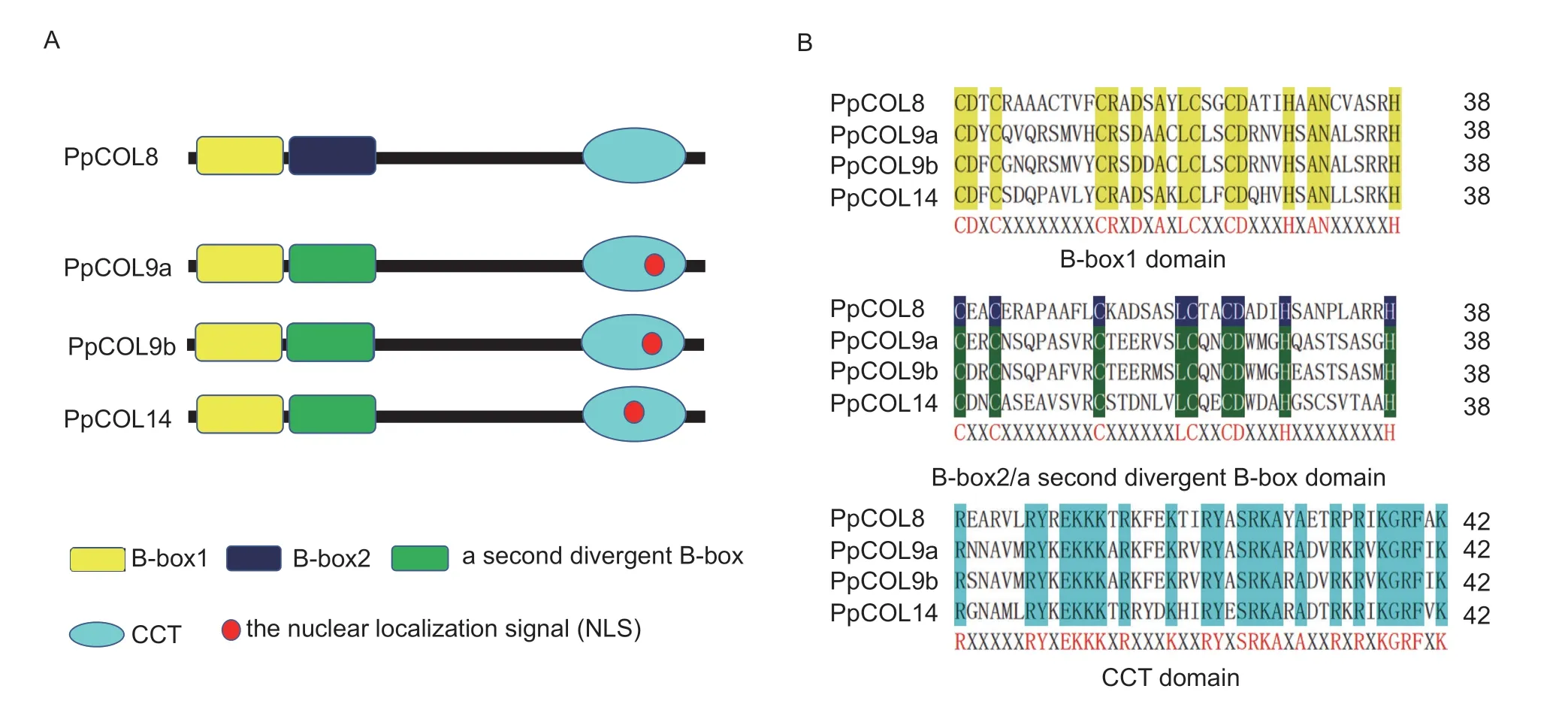
Fig.2 Domain analysis of PpCOLs.A,models of the PpCOL domain.B,conserved domain sequence alignment of PpCOLs.The red letters under the alignments represent the conserved amino acid residues among PpCOL domains.In each domain alignment set,the highlighted letters show the conservation of residues across all domains.
Another phylogenetic tree was constructed using the four PpCOLs identified by Shiet al.(2021) and the COLs from pear,apple,andArabidopsisto understand the relationships of sand pear COL proteins with other orthologous COLs (Fig.1-B).PpCOL8 showed a high evolutionary relationship with the white pear COL protein,PbCOL8 (Pyrus×bretschneideriXP_009378312).PpCOL9a and PpCOL9b were closely related to PbCOL9a (Pyrus×bretschneideriXP_009361520) and PuPcCOL9 (Pyrus ussuriensis×Pyrus communisKAB2634449),respectively.Similarly,PpCOL14 shares a high evolutionary similarity with PuPcCOL14 (P.ussuriensis×P.communisKAB2599070) and PbCOL14 (Pyrus×bretschneideriXP_009336468).
Additionally,the conserved motifs of the four PpCOLs were analyzed.The positions (Fig.2-A) and sequences (Fig.2-B) of the CTT motif and B-box 1/B-box 2/a second divergent B-box domain were conserved.The conserved B-boxes and CCT motifs in the four PpCOLs were aligned and represented by their amino acid sequence logos.As shown in Fig.2-B,the B-box1 and B-box2/a second divergent B-box domains were separated according to their consensus sequences and the spacing between zinc binding residues (Crocco and Botto 2013).According to the amino acid alignment of COL homologs,the conserved sequence of the B-box 1 domain among the PpCOLs was C-X2-C-X8/7-C-X2-D-X-A-X-L-C-X2-C-DX3-H-X-A-N-X5-H,and the homology sequence of the B-box 2/a second divergent B-box domain of the PpCOLs was C-X2-C-X8-C-X6-L-C-X2-C-D-X3-H-X8-H (Fig.2-B).These results indicate that the conserved cysteine (C) and histidine (H) residues are essential for the function of PpCOLs and might be involved in protein-protein zinc ligation (Liet al.2018).The sequences of the CCT domain were much more conserved than those of the two B-box in PpCOLs (Fig.2-B).However,the nuclear localization signals (NLS) of PpCOL9a and PpCOL9b were conserved.The NLS motif of PpCOL14 was in the CTT motif,just like in PpCOL9s,although their locations were different.Nevertheless,PpCOL8 has no NLS motif.
The percent identity matrix of the four PpCOLs was created using Clustal2.1 (Appendix C).The results showed that PpCOL9a shares relatively high homology (87.47% identity) with PpCOL9b at the protein level.PpCOL8 shows the lowest percent identity (27.13) with PpCOL9a at the protein level.
3.3.Chromosomal location and gene structure analyses of the four PpCOL genes
Chromosomal distribution analysis showed that the fourPpCOLswere mapped onto three of the 17 sand pear chromosomes (Appendix D).BothPpCOL8andPpCOL9bwere located on chromosome 9,whilePpCOL9aandPpCOL14were located on chromosomes 17 and 13,respectively.
The genomic DNA sequences of the fourPpCOLswere analyzed according to the sand pear genome database (Gaoet al.2021).Sequence comparison between the cDNA and the corresponding genomic DNA sequence revealed that thePpCOL8gene contains one intron splitting its ORF into two exons.The intron length is 942 bp and it is inserted between the 278th and 279th amino acid sites (Appendix E-a).Interestingly,bothPpCOL9aandPpCOL9bcontain three introns splitting their ORFs into four exons.The positions of all three introns are conserved.PpCOL14also contains three introns splitting its ORF into four exons.With a 488 bp length,the first intron is located between the 297 and 298 aa,while the second intron has a length of 583 bp and is found between 364 and 365 aa.Additionally,the third intron is 80 bp in length within the 454th amino acid site (Appendix E-a).
3.4.Expression of the four PpCOLs in tissues and during fruit development
We also investigated the expression patterns of the fourCOLsin all sand pear tissues.As shown in Fig.3,the four pearCOLswere found to be expressed in all tissues.Moreover,PpCOL8andPpCOL9awere predominantly expressed in petals,whilePpCOL9bexhibited higher expression in anthers than in the other tissues.PpCOL14was highly expressed in shoots (Fig.3).

Fig.3 Expression patterns of the four PpCOLs in pear tissues.Total RNA was isolated from different tissues (shoots,young stems,young leaves,petals,anthers,and mesocarp of fruits).The relative value for the expression of PpCOLs is shown as a percentage of the PpUBI expression.Mean values and standard deviations (SD) (bar) are shown from three independent experiments.
To check whether the expression ofPpCOLsis regulated during fruit development,the expression profiles of the four pearCOLswere investigated during fruit development and after harvest (from 30 d after full bloom to 30 d after harvest).The expression ofPpCOL8was relatively higher at 30 and 60 d after full bloom (Fig.4).PpCOL9aandPpCOL9bhad similar expression patterns with the highest expression levels at 30 d after full bloom (Fig.4).PpCOL14showed a higher expression level during early fruit development (30-90 d after full bloom).

Fig.4 Expression profiles of the four PpCOLs during pear fruit development.Total RNA was isolated from mesocarp at different developmental stages (from 30 d after full bloom to 30 d after harvest).The relative value for the expression of PpCOLs is shown as a percentage of the PpUBI expression.Mean values and standard deviations (SD) (bar) are shown from three independent experiments.
3.5.SA regulates the expression of PpCOLs during fruit senescence
The pear fruit exhibited peak ethylene respiration at 160 d after full bloom or 10 d after natural harvest (Zhanget al.2013).Thus,mesocarps of fruits at 160 d after full bloom were treated with different SA concentrations for expression analysis of thePpCOLs.As shown in Fig.5,treatment with 0.02 and 0.2 mmol L-1SA for 12 h could inhibit the expression ofPpCOL8,while the expression ofPpCOL9swas significantly induced by the 0.02 and 0.2 mmol L-1SA treatments (Fig.5).However,exogenous SA treatments did not affect the expression ofPpCOL14in pear fruit.
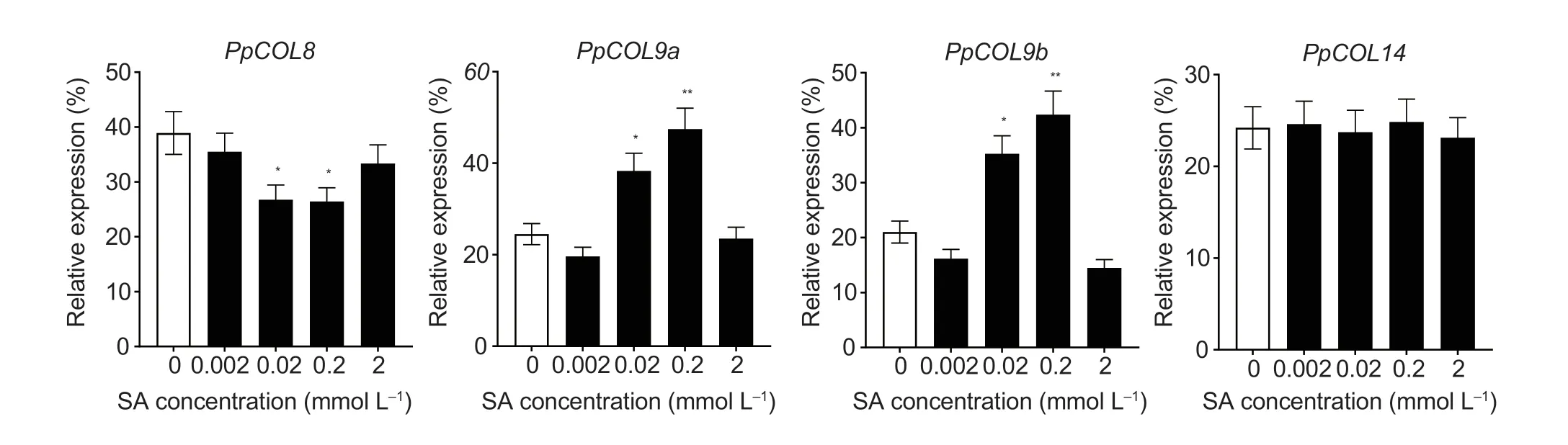
Fig.5 RT-qPCR analysis of PpCOL expression in fruit under salicylic acid (SA) treatment for 12 h.Relative values for PpCOL expression in 10 d after natural harvest fruit treated with 0,0.002,0.02,0.2,and 2 mmol L-1 SA for 12 h are shown as a percentage of the PpUBI expression.Mean values and standard deviations (SD) (bar) are shown from three independent experiments.Independent t-tests for equality of means demonstrated significant differences between control and treated fruits (*,P<0.05;**,P<0.01).
3.6.PpCOL8 interacts with PpMADS in vitro and in vivo
COLgenes have essential roles in plant growth and development (Cuiet al.2022).However,the role ofCOLgenes in regulating fruit senescence is not well understood.Our previous study identifiedPpCOL8,PpCOL9aandPpCOL9bin senescent sand pear fruit.Furthermore,the possible interacting partner of the PpCOL proteins was identified using an online STRING protein interaction tool (https://cn.string-db.org/).As shown in Fig.6-A,PpMADS was identified as an interacting partner ofPpCOL8.Furthermore,a yeast two-hybrid assay was carried out to verify the interactions of PpMADS with the PpCOL8,PpCOL9a,and PpCOL9b proteins.The full-length amino acid sequences of the COLs were fused with GAL4BD,while the sequence of PpMADS was fused with GAL4AD.The transformed yeast cells grew well on SD-Trp/-Leu.However,further growth on SD/-Trp/-Leu/-His/-Ade plates showed that only PpCOL8 could interact with PpMADS (Fig.6-B).Similar results were observed when PpCOL8 and PpMADS were fused with GAL4AD and GAL4BD,respectively (Fig.7-A).These results suggest thein vitrointeraction of PpCOL8 with PpMADS.

Fig.6 Protein-protein interactions between PpCOLs and PpMADS.A,interaction relationship network of pear COL homologs in STRING (https://string-db.org/).B,assay of physical interactions between PpCOL8/9s and PpMADS in yeast.DDO,double dropout supplements;QDO,quadruple dropout supplements.
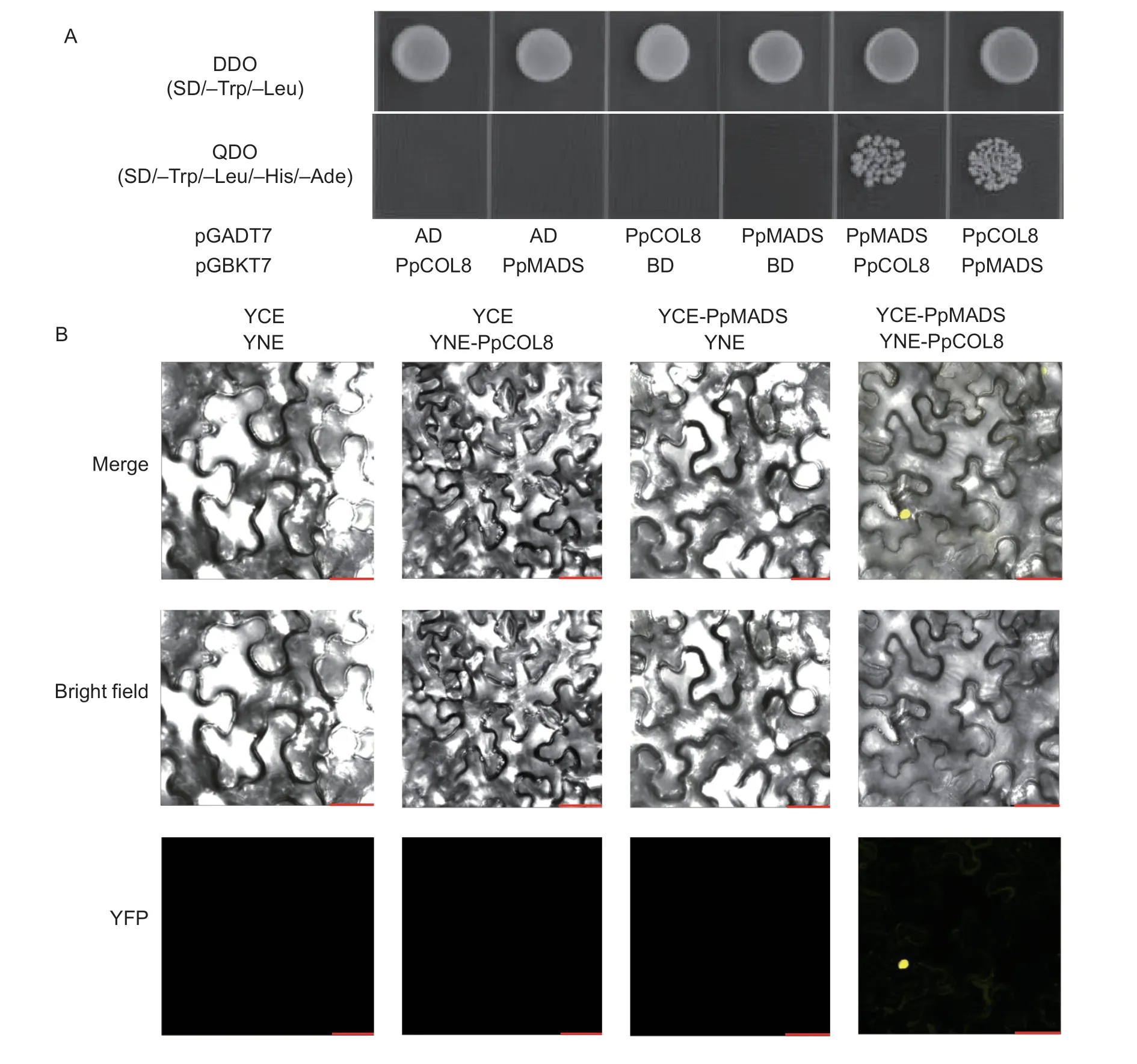
Fig.7 PpCOL8 interacts with PpMADS.A,Y2H assays of PpCOL8 and PpMADS.B,the interaction of PpCOL8 with PpMADS in the BiFC assay in Nicotiana benthamiana.Scale bar=25 μm.
Moreover,a BiFC assay was performed to verify the Y2H results.For the BiFC assay,PpMADSwas fused with the C-terminal of YFP,whilePpCOL8was fused with the N-terminal of YFP.As shown in Fig.7-B,the YFP signal was observed only in the nuclei of cells co-expressingPpCOL8andPpMADS,indicating that PpCOL8 could interact with PpMADS.The Y2H and BiFC assays revealed that PpCOL8 could interact with PpMADSin vitroandin vivo.
3.7.PpMADS is preferentially expressed in fruit and up-regulated by SA
PpMADS(GenBank accession number: ON216316) was isolated from the pear fruit cDNA library (Shi and Zhang 2012).The theoretical pI and instability index (II) values of PpMADS are 8.72 and 55.76,respectively,indicating that PpMADS is an unstable protein.PpMADScontains six introns (Appendix E-b) and is located on chromosome 16.Subcellular localization prediction revealed that PpMADS is a nucleus localized protein.As shown in Fig.8-A,PpMADSwas predominantly expressed in the mesocarp.With weak expression in anthers,petals and shoots,PpMADSwas not expressed in stems or leaves.Additionally,PpMADSwas highly expressed during fruit development,especially during fruit senescence (Fig.8-B).Interestingly,the expression ofPpMADSwas significantly up-regulated by the 0.2 mmol L-1SA treatment (Fig.8-C).

Fig.8 Expression analyses of PpMADS in pear tissues (A),during fruit development (B),and under salicylic acid (SA) treatments (C).Relative values of PpMADS expression in tissues (shoots,young stems,young leaves,petals,anthers and mesocarp of fruits),during fruit development (from 30 d after full bloom to 30 d after harvest),and in 10 d after natural harvest fruit treated with 0,0.002,0.02,0.2,and 2.0 mmol L-1 SA for 12 h are shown as a percentage of the PpUBI expression.Mean values and standard deviations (SD) (bar) are shown from three independent experiments.Independent t-test for equality of means demonstrated signiffcant differences between control and overexpression fruits (**,P<0.01).
3.8.PpCOL8 and PpMADS regulating fruit senescence
To further investigate the functions ofPpCOL8andPpMADSduring sand pear fruit senescence,thePpCOL8andPpMADSgenes were overexpressed in mature sand pear fruits.RT-qPCR results showed thatPpCOL8andPpMADSwere overexpressed in sand pear fruits.The production of ethylene was increased in fruit overexpressingPpCOL8,which accelerated the process of fruit senescence.PpMADSinhibited the production of ethylene,which suggested the senescence process of fruits was delayed.The co-transformation ofPpCOL8andPpMADSalso inhibited the production of ethylene.The MDA content was increased in thePpCOL8-overexpression fruits,but it was reduced in thePpMADSoverexpression fruits.WhenPpCOL8andPpMADSwere co-transformed into fruits,the MDA content was higher than that inPpMADS-overexpression fruits,but lower than that inPpCOL8-overexpression fruits (Fig.9).

Fig.9 The expression levels of PpCOL8 (A) and PpMADS (B),ethylene production (C) and MDA content (D) in sand pear fruits overexpressing the PpCOL8 and PpMADS genes.Mean values and standard deviations (SD) (bar) are shown from three independent experiments.Independent t-test for equality of means demonstrated signiffcant differences between control and overexpression fruits (*,P<0.05;**,P<0.01).
In sand pear fruits,overexpression ofPpCOLandPpMADSchanged the content of MDA,which suggestedPpCOLandPpMADSmay regulate fruit senescence by affecting antioxidant enzymes.According to the sequences ofPcPOD(Zhuet al.2018) andPbSOD(Liuet al.2021),PpPOD1(Gene ID: EVM0013215.1) andPpSOD1(Gene ID: EVM0020924.1) were identified from the sand pear genome.After overexpressingPpCOL8in sand pear fruits,the activities of POD and SOD in the fruits were reduced,whereas after overexpressingPpMADSin sand pear fruits,the activities of POD and SOD were increased.After the co-transformation ofPpCOL8andPpMADSin pear fruits,the POD and SOD activities were higher than those inPpCOL8-overexpression fruits,but lower than those ofPpMADSoverexpression fruits.Moreover,the expression levels ofPpPOD1andPpSOD1inPpCOL8-overexpression sand pear fruits were down-regulated.In contrast,the corresponding changes inPpMADS-overexpression fruits were the opposite.The expression levels ofPpPOD1andPpSOD1inPpCOL8andPpMADSco-transformation sand pear fruits were higher than those inPpCOL8-overexpression fruits,but lower than those inPpMADSoverexpression fruits (Fig.10).

Fig.10 Peroxidase (POD) activity (A),superoxide dismutase (SOD) activity (B),PpPOD1 expression (C),and PpSOD1 expression (D) in PpCOL8 and PpMADS overexpression sand pear fruits.Mean values and standard deviations (SD) (bar) are shown from three independent experiments.Independent t-test for equality of means demonstrated signiffcant differences between control and overexpression fruits (*,P<0.05;**,P<0.01).
4.Discussion
Several members of theCOLgene family have been identified from monocot and dicot plants,includingArabidopsis,banana,rice,barley,soybean,Chrysanthemum lavandulifolium,andLilium×formolongi(Griffithset al.2003;Chenet al.2012;Wuet al.2014;Fuet al.2015;Liet al.2018).Here,we identified 22COLgenes from sand pear (Fig.1),including the four previously identifiedCOLs,PpCOL8,PpCOL9a,PpCOL9b,andPpCOL14,viaRNA-sequencing (Shiet al.2021).All PpCOL family members were located in the nucleus (Table 1),suggesting the COL family may play an important role in the nucleus.Lianget al.(2023) also verified that SlCOL1 is a nuclear protein,which was based on the predicted subcellular localization.
In plants,the COL proteins are divided into three groups according to the number and structure of their B-box domains.Group I members contain two B-box domains,while Group II has only one B-box domain,and Group III comprises one B-box domain and a second divergent B-box domain (Gangappa and Botto 2014;Liet al.2018).TwoCOL2genes,MiCOL2AandMiCOL2B,were identified from ‘SiJiMi’ mango.Sequence analysis showed that MiCOL2A and MiCOL2B both contain two B-box domains and one CCT domain,which are highly conserved (Lianget al.2023).In this study,PpCOL8 was found to have two B-box domains and one CCT motif and belonged to Group I,which was similar to MaCOL1 (Chenet al.2012).PpCOL9a,PpCOL9b,and PpCOL14,each with a CCT motif and a second divergent B-box,were assigned to Group III (Fig.1).These four PpCOLs were found to be localized in the nucleus.They all have NLS motifs,except PpCOL8 (Fig.2),which could be crucial for their specific biological functions.
So far,only a few fruitCOLshave been identified,including one bananaCOL,MaCOL1(Chenet al.2012),and the 22PpCOLsidentified from sand pear in this study.The bananaCOL(MaCOL1) was differentially expressed among various banana plant tissues,with higher expression in the flower.The expression ofMaCOL1in peel changed slightly but its expression was remarkably increased in the pulp during fruit ripening,suggesting thatMaCOL1might be involved in banana fruit pulp ripening (Chenet al.2012).In this study,the four pearCOLs,PpCOL8,PpCOL9a,PpCOL9b,andPpCOL14,were expressed in all sand pear tissues (Fig.3).The expression ofPpCOL8was relatively higher at 60 d after full bloom than in other fruit development stages.PpCOL9aandPpCOL9bshowed the highest expression levels at 30 d after full bloom among all the fruit development stages.PpCOL14was strongly expressed at 90 d after full bloom during fruit development (Fig.4).These results suggest that COL proteins could be involved in fruit development,ripening,and senescence in sand pear.
In addition to fruit development,theCOLgenes can play a crucial role in regulating flowering time.For instance,transgenicArabidopsisplants overexpressingCOL8showed a late-flowering phenotype under long-day conditions (Takaseet al.2011).Similarly,overexpression ofCOL9delayed flowering inArabidopsisby reducing the expression ofCOandFT(Cheng and Wang 2005).Additionally,overexpression ofOsCOL9also delayed the flowering time by repressing theEhd1pathway (Liuet al.2016b).Meanwhile,OsCOL9was induced withMagnaporthe oryzaeinfection and was involved in disease resistance in rice (Liuet al.2016a).OsCOL9 interacted with OsRACK1 through its CCT domain and enhanced rice blast resistance through the SA and ET signaling pathways (Liuet al.2016a).
In this study,PpCOL8andPpCOL9awere predominantly expressed in petals,andPpCOL9bwas mainly expressed in anthers among all tissues (Fig.3),suggesting their role in the regulation of flowering development.Interestingly,the expression levels ofCOL8andCOL9swere regulated during fruit development (Fig.4) and in response to SA treatment during fruit senescence (Fig.5).To further understand the underlying mechanisms of COL-mediated fruit senescence,we performed yeast two-hybrid and BiFC assays to identify the interacting partners.Here,PpCOL8 showed strong interaction with PpMADS,a MADS-box protein.It is well documented that MADSbox proteins are key transcription factors in fruit ripening.The tomato MADS-box transcription factor,RIPENING INHIBITOR (RIN),was reported to regulate fruit ripening (Vrebalovet al.2002).BananaMADSs,MaMADS1andMaMADS2,are homologous to the tomato RINMADS ripening gene.Silencing either gene through antisense or RNA interference (RNAi) in banana delayed fruit ripening by suspending climacteric respiration and reducing the synthesis of ethylene,a well-known ripening hormone (Elitzuret al.2016).MaMADS1andMaMADS2transgenic plants also showed extended fruit shelf-life phenotypes,including color development and softening delay (Elitzuret al.2016).Additionally,Xieet al.(2014) demonstrated that a novel MADS-box gene (SlFYFL) could play a positive role in delaying fruit ripening and abscission in tomato plants.The ethylene content and the genes involved in ethylene biosynthesis and signaling pathways were down-regulated in35S:FYFLfruits (Xieet al.2014).Interestingly,SA treatments up-regulated the expression ofPpMADS.Thus,PpMDAScould play a remarkable role in increasing the shelf life of pear fruits.Moreover,PpCOL8may function in fruit senescence by interacting withPpMADSin the SA signaling pathway.The expression ofPpCOL9swas up-regulated by SA,but down-regulated during fruit senescence.Moreover,PpCOL9sdid not interact withPpMADS,suggesting thatPpCOL9smay be involved in delaying fruit senescence directlyviathe SA signaling pathway.Studies have shown that someCOLscan be involved in abiotic stress responses.For example,a rapeseedCOLgene (BnCOL2) was induced by PEG-6000,abscisic acid (ABA),and NaCl,and overexpression ofBnCOL2enhanced sensitivity to drought stress inArabidopsis.BnCOL2was mainly expressed in the cotyledons and leaves of rapeseed seedlings (Liuet al.2020).In banana,accumulation of theMaCOL1transcript was increased by abiotic and biotic stresses,such as chilling and pathogen infection (Chenet al.2012).As shown in Appendix F,the expression levels ofPpCOL8,PpCOL14,andPpMADSwere down-regulated in diseased sand pear fruit,suggesting thatPpCOLgenes might be involved in disease resistance to delay fruit senescence during pear storage.
Ethylene plays an important role in fruit ripening (Yueet al.2020).SA treatment can reduce the content of endogenous ethylene and delay the senescence of pear fruit (Xuet al.2023).Antioxidant enzymes are also closely related to fruit senescence.Melatonin treatment can enhance the activity of antioxidant enzymes in pear and sweet cherry,with the goal of eliminating the damage from malondialdehyde and delaying fruit senescence (Zhaoet al.2013;Zhaiet al.2018).MaCOL1 is a transcriptional activator involved in fruit ripening and stress responses (Chenet al.2012).SlCOL1 and SlBBX24 control tomato fruit size by regulating the expression levels of downstream genes (Cuiet al.2022).The overexpression ofMiCOL16AandMiCOL16Bimproved tolerance to drought and salt stress (Liuet al.2022).These results suggested that theCOLgenes play important roles in fruit development,ripening and stress tolerance.In this study,the antioxidant enzyme-related genesPpSOD1andPpPOD1were identified from the sand pear genome.Subcellular localization predictions showed that PpSOD1 and PpPOD1 were localized in the cytoplasm (data not shown).Compared with the control,overexpression of thePpCOL8gene in sand pear fruit increased ethylene production and MDA content,but down-regulated the activities of SOD and POD and the expression ofPpSOD1andPpPOD1,which indicated that overexpression of thePpCOL8gene promoted the senescence of sand pear fruit.During the ripening of sweet cherries,PaMADS7 directly bound to the promoter of thePaPG1gene and actively regulated its expression to promote fruit softening (Qiet al.2020).SlCMB1,a MADS-box protein,increased ethylene production and promoted fruit ripening by regulating ethylene synthesis and signal transduction in tomato (Zhanget al.2018).However,SlMBP8 (a MADSbox protein) can inhibit cell wall metabolism to delay the ripening of tomato fruit (Yinet al.2017).Therefore,different MADS-boxes play different,or even opposite,roles in regulating fruit ripening and senescence.Here,thePpMADSgene was overexpressed in sand pear fruit,which improved the antioxidant capacity of the fruit and played a role in delaying fruit senescence.Notably,the overexpression ofPpMADSreduced the production of ethylene,which may delay the senescence of sand pear fruit by regulating the genes related to ethylene synthesis and signal transduction.Transcription factors often function by interacting with other genes.For example,the interaction between VvMADS39 and VvAGAMOUS maintained the characteristics of grape fruit meristem and promoted the growth and development of grape fruit (Zhanget al.2022).The interaction between CiAGL9 and CiMADS43 regulated citrus leaf development (Yeet al.2021).However,the interaction protein of COL is rarely reported.Our study confirmed the interaction between PpCOL8 and PpMADS (Fig.7).The co-transformation ofPpCOL8andPpMADSreduced the production of ethylene and the content of MDA.Meanwhile,the activities of antioxidant enzymes (POD and SOD) were enhanced,which delayed the senescence of pear fruit (Fig.11).These results suggested that PpCOL8 may play an important role in fruit senescence by interacting with MADS-box protein PpMADSviathe SA signaling pathway.
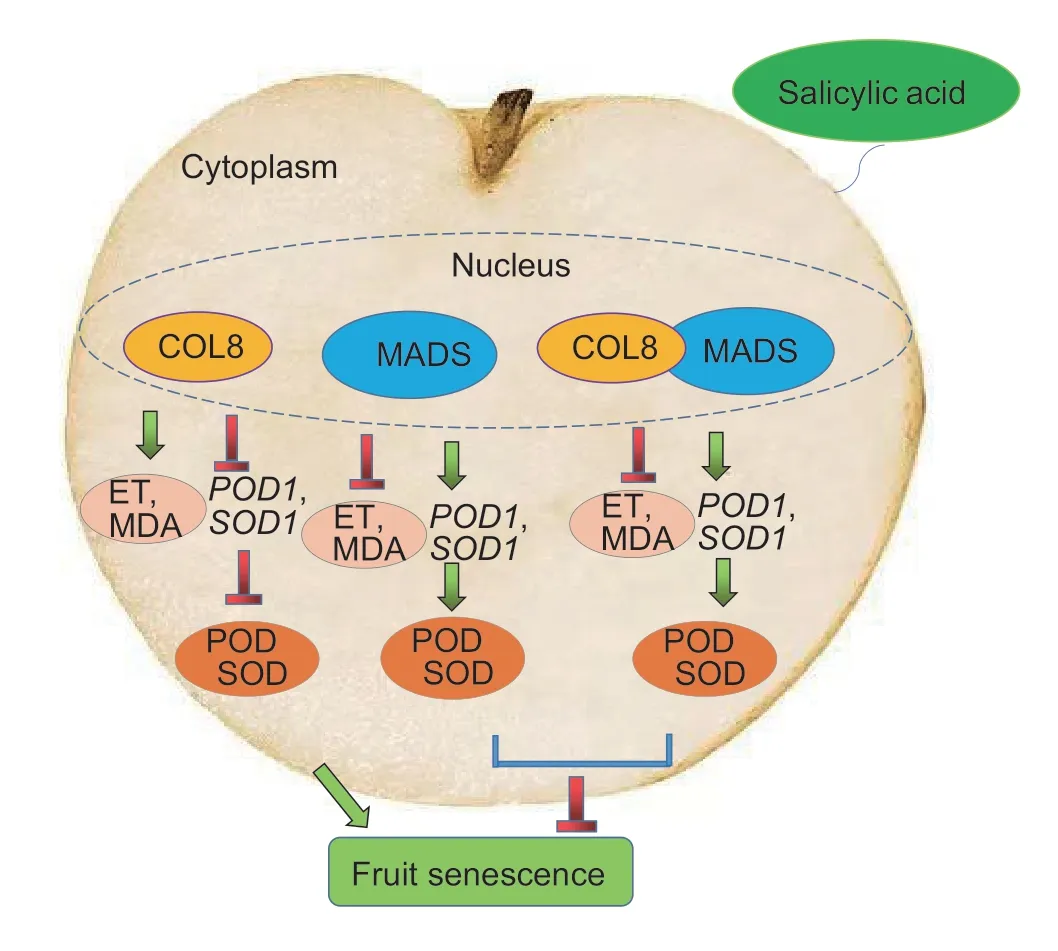
Fig.11 Schematic model showing how salicylic acid (SA) regulates fruit senescence via COL8 interacting with MADSbox.ET,ethylene;MDA,malondialdehyde;POD,peroxidase;SOD,superoxide dismutase.
5.Conclusion
In this study,we identified 22CONSTANS-LIKEgenes (COLs) from sand pear (P.pyrifolia).Four of the 22COLs,PpCOL8,PpCOL9a,PpCOL9b,andPpCOL14,were reported in our previous study (Shiet al.2021).Based on the presence and absence of B-box and NLS domains,the PpCOLs were classified into three different groups.Tissue-specific expression analysis showed that the four pearCOLswere expressed in all tissues,but with different expression patterns,and regulated during fruit development.In addition,the transcripts ofPpCOL8were inhibited by SA treatment,but SA induced the expression ofPpCOL9aandPpCOL9b.However,the expression ofPpCOL14was not affected by SA treatment.In addition,PpCOL8 interacts with PpMADS.The transient overexpression ofPpCOL8andPpMADSinto sand pear fruits demonstrated that they antagonistically regulated fruit senescence.While overexpression ofPpCOL8promoted fruit senescence,overexpression ofPpMADSand co-transformation ofPpCOL8andPpMADSdelayed the senescence of sand pear fruits,which suggested thatPpCOL8plays a role in fruit senescence throughPpMADSand the SA signaling pathways.Interestingly,thePpCOL8,PpCOL14,andPpMADSexpression levels were down-regulated in diseased fruit.The results of this study revealed that PpCOL8 can play a crucial role in fruit senescence and disease resistance by interacting with the MADS-box protein,PpMADS,through the SA signaling pathway.The study also identified the target genes for regulating fruit senescence.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (32272654),the Natural Science Foundation of Hebei Province,China (C2023204016),the Hebei Province Introduced Overseas-Scholar Fund,China (C20220361),the S&T Program of Hebei,China (20326330D),and the Hebei Province Outstanding Youth Fund,China (2016,2019).
Declaration of competing interest
The authors declare that they have no conflict of interest.
Appendicesassociated with this paper are available on https://doi.org/10.1016/j.jia.2024.01.011
杂志排行
Journal of Integrative Agriculture的其它文章
- OsNPF3.1,a nitrate,abscisic acid and gibberellin transporter gene,is essential for rice tillering and nitrogen utilization efficiency
- Fine mapping and cloning of the sterility gene Bra2Ms in nonheading Chinese cabbage (Brassica rapa ssp.chinensis)
- Basal defense is enhanced in a wheat cultivar resistant to Fusarium head blight
- Optimized tillage methods increase mechanically transplanted rice yield and reduce the greenhouse gas emissions
- A phenology-based vegetation index for improving ratoon rice mapping using harmonized Landsat and Sentinel-2 data
- Combined application of organic fertilizer and chemical fertilizer alleviates the kernel position effect in summer maize by promoting post-silking nitrogen uptake and dry matter accumulation
