OsNPF3.1,a nitrate,abscisic acid and gibberellin transporter gene,is essential for rice tillering and nitrogen utilization efficiency
2024-05-13JunnanHangBowenWuDiyangQiuGuoYangZhongmingFangMingyongZhang
Junnan Hang ,Bowen Wu ,Diyang Qiu ,Guo Yang ,Zhongming Fang# ,Mingyong Zhang
1 Institute of Rice Industry Technology Research/Key Laboratory of Functional Agriculture,Guizhou Provincial Department of Education/Key Laboratory of Plant Resource Conservation and Germplasm Innovation in Mountainous Region,Ministry of Education/Key Laboratory of Molecular Breeding for Grain and Oil Crops in Guizhou Province/College of Agricultural Sciences,Guizhou University,Guiyang 550025,China
2 Key Laboratory of South China Agricultural Plant Molecular Analysis and Genetic Improvement/Guangdong Provincial Key Laboratory of Applied Botany/South China Botanical Garden,Chinese Academy of Sciences,Guangzhou 510650,China
3 Institute of Fruit Tree Research,Guangdong Academy of Agricultural Sciences,Guangzhou 510640,China
Abstract Low-affinity nitrate transporter genes have been identified in subfamilies 4-8 of the rice nitrate transporter 1 (NRT1)/peptide transporter family (NPF),but the OsNPF3 subfamily responsible for nitrate and phytohormone transport and rice growth and development remains unknown.In this study,we described OsNPF3.1 as an essential nitrate and phytohormone transporter gene for rice tillering and nitrogen utilization efficiency (NUtE).OsNPF3.1 possesses four major haplotypes of its promoter sequence in 517 cultivars,and its expression is positively associated with tiller number.Its expression was higher in the basal part,culm,and leaf blade than in other parts of the plant,and was strongly induced by nitrate,abscisic acid (ABA) and gibberellin 3 (GA3) in the root and shoot of rice.Electrophysiological experiments demonstrated that OsNPF3.1 is a pH-dependent low-affinity nitrate transporter,with rice protoplast uptake assays showing it to be an ABA and GA3 transporter.OsNPF3.1 overexpression significantly promoted ABA accumulation in the roots and GA accumulation in the basal part of the plant which inhibited axillary bud outgrowth and rice tillering,especially at high nitrate concentrations.The NUtE of OsNPF3.1-overexpressing plants was enhanced under low and medium nitrate concentrations,whereas the NUtE of OsNPF3.1 clustered regularly interspaced short palindromic repeats (CRISPR) plants was increased under high nitrate concentrations.The results indicate that OsNPF3.1 transports nitrate and phytohormones in different rice tissues under different nitrate concentrations.The altered OsNPF3.1 expression improves NUtE in the OsNPF3.1-overexpressing and CRISPR lines at low and high nitrate concentrations,respectively.
Keywords: rice tillering,grain yield,phytohormone,nitrate,transporter,nitrogen utilization efficiency
1.lntroduction
Rice is an important crop for global food security (Weiet al.2022).Its grain yield is mainly determined by three factors: panicle number per plant,grain number per panicle,and thousand-grain weight (Wang and Li 2008).The panicle number per plant and grain number per panicle are important determinants of rice plant architecture,which in turn depend on the number of tillers produced by plants and the number of primary and secondary branches of the panicles (Xing and Zhang 2010).These traits are regulated by a complex gene regulatory network and influenced by external and internal environmental factors,such as nitrogen (N) and phytohormones (Wanget al.2018;Suet al.2021).
N is an essential element for the synthesis of proteins,nucleic acids,phospholipids,enzymes,and some growth hormones.When rice is deficient in N,the synthesis of these substances is hindered,rice growth is inhibited,and tillering is reduced (Xinet al.2021;Xuet al.2021).However,excessive N fertilizer application can have many adverse effects,such as reduced folding resistance,decreased yields,and increased occurrence of rice blast and grain blight (Zhanget al.2015;Sousaet al.2021).Therefore,the genetic basis of crop nitrogen use efficiency (NUE) needs to be elucidated,and crops that can produce high yields under low N conditions should be bred for sustainable agricultural development.
In plants,nitrate,ammonium,peptide,and amino acid transporter proteins work together to coordinate N uptake and utilization.Rice has 93 nitrate and peptide transporter family (NPF) members,but only a few of these family members have been described in terms of their functions (Liet al.2017).Within the rice NPF family,OsNPF2.2 is a vascular-specific transporter involved in nitrate unloading in the xylem and nitrate transport from the roots to the stems (Liet al.2015).OsNPF2.4 is a pHdependent low-affinity nitrate transporter functioning in the K+-coupled nitrate transport related to pollen fertility (Xiaet al.2015).OsNPF3.1regulates NUE and affects rice plant height,heading date,and thousand-grain weight (Yanget al.2022).OsNPF4.1(sp1) is highly expressed in panicles,controlling their size (Liet al.2009).OsNPF4.5participates in the mycorrhizal pathway of N acquisition (Wang S Set al.2020).OsNPF5.16 is a nitrate-inducible low-affinity nitrate transporter that is essential for rice tillering and yield (Wanget al.2022).OsNPF6.1 is a low-affinity nitrate transporter that affects NUE in rice under low N conditions (Tanget al.2019).Importantly,OsNPF6.5(OsNRT1.1B) inindicavariants enhance nitrate uptake and can increase the NUE in rice (Huet al.2015).In the OsNPF7 and OsNPF8 subfamilies,OsNPF7.1(Huanget al.2019),OsNPF7.2(Huet al.2016;Wang Jet al.2018),OsNPF7.3(Fanet al.2014;Fanget al.2017),OsNPF7.7(Huanget al.2018),andOsNPF8.20(Fanget al.2013) positively influence rice yield by regulating tillering,whileOsNPF7.4negatively affects rice yield (Huanget al.2019).OsNPF8.9 (OsNRT1) is a low-affinity nitrate transporter that mainly mediates nitrate uptake in rice roots (Linet al.2000).However,studies have yet to determine whether any members of subfamilies OsNPF1 and OsNPF3 are responsible for nitrate transport and grain yield in rice.
Gibberellin (GA) is another phytohormone that has vital regulatory effects on plant height and branching (Zhanget al.2020).GA3and GA4are transported inArabidopsis viathe low-affinity nitrate transporter AtNPF3.1 (Pikeet al.2014;Talet al.2016).Atnpf3.1mutants have impaired hypocotyl elongation and seed germination under low nitrate conditions (Davidet al.2016).Conversely,AtNPF3.1-overexpressing plants exhibit late germination,weak hypocotyl growth,and ssignificant decline in root and shoot growth (Talet al.2016).The glucosinolate transporter AtNPF2.10 (GTR1) can transport certain forms of GA in yeast andXenopusoocytes (Ishimaruet al.2017).Atnpf2.10mutant plants have reduced fertility under continuous light exposure,but their fertility can be restored by GA treatment (Saitoet al.2015).Moreover,the sucrose transporter members AtSWEET13 and AtSWEET14 inArabidopsishave been identified as GA transporters (Kannoet al.2016).Although phytohormones (ABA and GA) and their transporters are essential for multiple developmental aspects inArabidopsis,their transport mechanisms and functions in rice under N application remain largely unknown.
In this study,we found that OsNPF3.1,a rice NPF3 subfamily member,is a low-affinity nitrate transporter that exhibits ABA and GA3transport activity.Moreover,the ABA and GA concentrations accumulated significantly inOsNPF3.1-overexpressing lines under high nitrate concentrations compared with wild-type Zhonghua 11 (WT ZH11).Consequently,shorter axillary buds,fewer tillers,and lower nitrogen utilization efficiency (NUtE) were observed,whereas the opposite trends were found inOsNPF3.1CRISPR mutants.The tiller number per plant,filled grain number per plant,and NUtE of theOsNPF3.1CRISPR mutants in the field were significantly higher than those of WT ZH11.Therefore,OsNPF3.1is essential for rice tillering and NUtE by balancing the transport of nitrate,ABA and GA under different N levels.
2.Materials and methods
2.1.Sequence variation of OsNPF3.1
Single-nucleotide polymorphisms (SNPs) ofOsNPF3.1(LOC_Os06g15370) were obtained from the Rice VarMap 2.0 website (http://ricevarmap.ncpgr.cn/v2/).The rice plants of 533 accessions were grown during the season from April to October in the field at the Guiyang experimental field of Guizhou University,China with fertilizer (N-P-K=19%-7%-14%) application at 90 kg ha-1.The expression ofOsNPF3.1among five haplotypes (Hap1-Hap5) was detected by real time-PCR (RT-PCR) in the corresponding cultivars.The primers are listed in Appendix A.For association analysis,haplotypes with a frequency of 10% or more were selected,and their agronomic traits were measured,including tiller number per plant,weight of straw per plant,total weight per plant,grain yield per plant and NUtE.
2.2.Construction of OsNPF3.1 plasmid vector
To construct the overexpression vector forOsNPF3.1,a cDNA (1,794 bp) fragment containing theOsNPF3.1open reading frame (ORF) was inserted downstream of the35Spromoter of pCAMBIA1306 plasmid usingBamHI andXbaI to generate the p35S-OsNPF3.1plasmid vector.To construct theOsNPF3.1CRISPR vector,anOsNPF3.1target sequence was designed and added to the U3IPAT/U3OPST primer.A 450 bp fragment was amplified from target sequence 1 by PCR using ZRO589 with the U3 plasmid and theOsNPF3.1-U3IPAT2 primer.In the same manner,a 400 bp fragment from target sequence 2 was amplified by PCR usingOsNPF3.1-U3IPST and ZRO282 primers with U3 plasmids.The complete U3 fragment was amplified by a fusion PCR containing the U3 promoter-sgRNA.Finally,the complete U3 was inserted into the Per8-Cas9 vector usingKpnI andSacI and a cloning kit for the 850 bp fragment.To construct theOsNPF3.1promoter-GUS vector,a 2,267 bpOsNPF3.1promoter fragment was obtained using PCR amplification and inserted upstream of the β-glucuronidase (GUS) coding region of the pCAMBIA1391Z plasmid usingHindIII andEcoRI to generate the pOsNPF3.1-GUS plasmid vector,and the primers are listed in Appendix A.All the constructed plasmids were transformed intojaponicarice WT ZH11 by theAgrobacterium tumefaciens-mediated transformation method.
2.3.Field planting and transgenic detection
All transgenic lines ofOsNPF3.1and WT ZH11 were grown in the field at Guiyang and Sanya experimental fields of Guizhou University with fertilizer (N-P-K=19%-7%-14%) application at 270 kg ha-1.Tiller number per plant and filled grain number per plant were measured over three seasons from 2019 to 2021.The concentrations of total nitrogen in the surface water of paddy fields were about 1.5 mmol L-1.In general,30 rice plants were used for each experiment,and the planting density was 19.98 cm×19.98 cm.The T2transgenic lines with overexpression ofOsNPF3.1(OE) were identified using RT-PCR and CRISPR mutants ofOsNPF3.1(C) were identified by sequencing.The primers are listed in Appendix A.
2.4.GUS staining
To determine the expression pattern ofOsNPF3.1,histochemical GUS assays were conducted using various tissues ofOsNPF3.1promoter-GUS transgenic plants with GUS staining solution containing 100 mmol L-1sodium phosphate (pH 7.0),10 mmol L-1Na2EDTA,1 mmol L-1K3[Fe(CN)6],1 mmol L-1K4[Fe(CN)6],0.5% Triton X-100,20% methanol,and 1.5 mg mL-1X-gluc.Then the samples for GUS staining were vacuum infiltrated for 15 min and gently fixed in formalin-acetic acid (70% (v/v) ethanol (1:1:18)) at 4°C for 20-30 min.The samples were then incubated in the staining buffer at 37°C overnight.After the incubation in a solution of 80% (v/v) ethanol was cleared,the stained samples were observed using a stereomicroscope (Olympus,Tokyo,Japan).The GUSstained samples were transferred to a 50% ethanol:5% acetic acid:10% formaldehyde solution,dehydrated in a series of ethanol solutions (50-100%),and embedded in paraffin.Paraffin-embedded tissue sections (7 μm) were cut with a paraffin-slicing machine (Leica,Wetzlar,Germany) and observed under a light microscope (Zeiss,Oberkochen,Germany).
One miserable5 rainy day, one of my regular customers came in looking depressed6 and defeated. My co-worker and I asked what the problem was and if we could help, but the customer wouldn’t reveal any details. He just said he felt like crawling into bed, pulling the sheets up over his head, and staying there for a few years. I knew exactly how he felt.
2.5.Subcellular localization
The ORF ofOsNPF3.1was amplified and fused with green fluorescent protein (GFP) usingBglII andSpeI in the pCAMBIA1302 vector to generate theOsNPF3.1-GFPfusion plasmid.The constructed vector was transformed into WT ZH11 by theA.tumefaciens-mediated transformation method.Fluorescence was observed using a confocal laser scanning microscope (Nikon,Tokyo,Japan).The primers are listed in Appendix A.
2.6.Functional analysis of OsNPF3.1 in Xenopus laevis oocytes
OsNPF3.1andCHL1cDNAs were amplified separately into the pNB1 vector for expression in oocytes.Complementary RNA (cRNA) was transcribedin vitrousing a mMessage mMachine T7 Kit (Ambion,Austin,USA),and purified with a MEGA Clear Transcription Clean-up Kit (Invitrogen,USA).Oocytes were selected for injection with 50 ng cRNA or the same amount of water.Injected oocytes were incubated in Barth’s solution at 18°C for 2 days.The Barth’s solution contained 96 mmol L-1NaCl,2 mmol L-1KCl,1 mmol L-1MgCl2,1.8 mmol L-1CaCl2,and 10 mmol L-1HEPES,adjusted to pH 7.5 or pH 5.5 with Tris-base.Nitrate uptake assays to measure the nitrate uptake in oocytes were performed using15N-KNO3at 0.2 or 10 mmol L-1.
2.7.Protoplast ABA and GA3 uptake assays
Fluorescein isothiocyanate (FITC)-labeled ABA and GA3(ABA-FITC,GA3-FITC) were synthesized by Yuan Peptide Biotechnology Company,Nanjing,China.Rice protoplasts prepared from the basal part of transgenic seedlings fromOsNPF3.1and WT ZH11 were incubated in 1 mL W5 buffer (pH 5.6) with either FITC-labeled ABA or GA3for 4 h at 25°C in the dark.Then the protoplasts were washed five times to remove the free FITC-labeled ABA or GA3,and fluorescence was observed with a confocal laser scanning microscope (Nikon,Tokyo,Japan).
2.8.Seedling growth analysis with nitrate,ABA or GA3 treatment
To analyze the effects of different nitrate treatments on the growth and development of axillary buds ofOsNPF3.1transgenic plants,the WT ZH11,OE line and C line seedlings were cultured in nutrient solution with 0.2,2 or 10 mmol L-1NaNO3(pH=5.8) for 35 days.To analyze the effects of treatment with different concentrations of ABA or GA3on the growth and development ofOsNPF3.1transgenic plants under 2 mmol L-1NaNO3treatment,seedlings of the WT ZH11,OE lines and C lines were cultured in nutrient solutions with 0.5,1 or 5 μmol L-1ABA or GA3(pH=5.8) for 35 days.The nutrient solution was renewed every 7 days.These seedlings of the WT ZH11,OE lines and C lines were then used for axillary bud growth,biomass and plant height measurements.Axillary bud growth was photographed using a stereomicroscope (Olympus,Tokyo,Japan) with Image J software.The nutrient solutions for the above treatments were applied in a nitrogen-free culture solution with the following composition: 0.32 mmol L-1NaH2PO4,0.51 mmol L-1K2SO4,1 mmol L-1CaCl2,1.65 mmol L-1MgSO4,8.9 μmol L-1MnSO4,0.5 μmol L-1Na2MoO4,18.4 μmol L-1H3BO3,0.14 μmol L-1ZnSO4,0.16 μmol L-1CuSO4and 40 μmol L-1FeSO4.
2.9.Whole stage analysis with sand culture
To further analyze the effect of different nitrate treatments on the maturity stage ofOsNPF3.1transgenic plants,the WT ZH11,OE line and C line seedlings were cultured to maturity in sand pots containing 0,0.2,2 or 10 mmol L-1NaNO3(pH=5.8) nutrient solutions.A total of 4 L of nutrient solution was added every 7 days.The nutrient solutions for above treatments were applied in a nitrogen-free culture solution with the following composition: 0.32 mmol L-1NaH2PO4,0.51 mmol L-1K2SO4,1 mmol L-1CaCl2,1.65 mmol L-1MgSO4,8.9 μmol L-1MnSO4,0.5 μmol L-1Na2MoO4,18.4 μmol L-1H3BO3,0.14 μmol L-1ZnSO4,and 0.16 μmol L-1CuSO4and 40 μmol L-1FeSO4.
2.10.Gene expression analysis
Total RNA was extracted from the lateral root,basal part,leaf sheath,culm,leaf blade and young spikelet using TRIzol reagent (Vazyme,Nanjing,China).First strand cDNA was synthesized using M-MLV reverse transcriptase (TaKaRa,Shiga,Japan) from 2 μg of the total RNA.The first-strand cDNA was used as a template for RT-PCR after normalization using the riceActin1gene (AB047313).The RT-PCR was performed in a 10 μL reaction volume containing 1 μL of the cDNA solution,1 μL gene specific primers (10 μmol L-1),and 5 μL 2× SYBR PCR Mix (Vazyme,Nanjing,China) under the following conditions: 94°C for 2 min (1 cycle),94°C for 30 s,58°C for 30 s,and 72°C for 30 s (40 cycles),followed by 72°C for 1 min (1 cycle).The primers are listed in Appendix A.
2.11.Determination of nitrate and total nitrogen contents
The free nitrate content was determined by the colorimetric method according to Caiet al.(2009).The total nitrogen content was determined using a Kjeldahl nitrogen analyzer (SKD-100,Peiou,China) and a semimicro Kjeldahl nitrogen method.NUtE was determined by the following equation: NUtE (%)=[Grain yield (g)/(Grain nitrogen content (g)+Straw nitrogen content (g)]×100
2.12.Nitrate uptake and translocation activity assay using
Rice seedlings grown in nutrient solution for 3 weeks were washed with 0.1 mmol L-1CaSO4for 1 min before transferring to fresh nutrient solution containing 0.25 or 5 mmol L-1NH415NO3with 99% atom excess of15N.The seedlings were washed again with 0.1 mmol L-1CaSO4for another 1 min after 30 min of incubation with the 0.25 or 5 mmol L-1NH415NO3solution,which was further washed with ddH2O five times before the shoot and root were sampled and dried at 80°C for the determination of their15N contents (Linet al.2008).
2.13.Determination of ABA and GA concentrations
The ABA and GA concentrations were measured by HPLC with a Waters e2695 High performance liquid chromatography system (e2695,Waters,USA).A 0.5 g rice sample was added to 5 mL of 80% 4°C methanol and ground overnight under dark conditions for extraction for 16 h.The extraction solution was centrifuged at 6,000 r min-1for 10 min.The supernatant was taken,and the filtrate residue was mixed at a volume ratio of 1:5.The supernatant was added to a 0.1 g crosslinked polyvinylpyrrolidone (PVPP) shaker for 20 min (150 r min-1) at room temperature for filtration.The filtrate was passed through a C18 solid-phase extraction column,and the effluent was dried under pressure at 40°C (150 r min-1).The evaporation flask was rinsed with 5 mL phosphoric acid buffer at pH 3.5.The aqueous phase was extracted three times with ethyl acetate.The aqueous phase was dried at 40°C,dissolved with 1 mL methanol,and then sampled by a 0.45 μm organic filter.The supernatant was collected and filtered through a filter membrane (0.45 μm).A total of 0.8 mL of each filtrate was analyzed using HPLC.
2.14.Statistical analysis
For statistical differences,thet-test was performed using SPSS 10 software (IBM,Inc.),and*,**,and***are used to indicate significant differences atP<0.05,P<0.01,andP<0.001,respectively.Duncan’s multiple range test was performed using SPSS 10 software (IBM,Inc.),and different letters indicate significant differences atP<0.05.Data are expressed as mean±standard deviation values.
3.Results
3.1.OsNPF3.1 promoter sequence is divergent between indica and japonica cultivars
Rice Variation Map v2.0,a database of rice genome variation (Chenet al.2014),was used for haplotype (Hap) analysis to investigate the sequences of theOsNPF3.1promoter region (2,700 bases upstream of the initiation codon ATG) in 533 cultivars worldwide.Four major Haps with 11 different SNPs were identified in 517 cultivars (Fig.1-A).Interestingly,Hap1 and Hap5 were mainly found inindicacultivars,and Hap2 and Hap4 were mainly present injaponicacultivars (Fig.1-A).The expression pattern ofOsNPF3.1was also detected in Hap1,Hap2,Hap4,and Hap5,revealing that the expression patterns ofjaponicahaplotypes (Hap2 and Hap4) differed from those ofindicahaplotypes (Hap1 and Hap5;Fig.1-B).Furthermore,our comparison of the tiller numbers ofindicaandjaponicahaplotypes showed thatindicahaplotypes produced more tillers thanjaponicahaplotypes (Fig.1-C).The grain yields per plant were significantly higher in cultivars with Hap1 and Hap5 than in cultivars with Hap2 and Hap4 (Fig.1-D).However,NUtE was significantly lower injaponicacultivars with Hap2 only than inindicawith Hap1 and Hap5 (Fig.1-E).Phylogenetic analysis showed that 31 haplotypes had two clusters,namely,a Hap1-containingindicacluster and Hap2-and Hap4-containingjaponicacluster (Appendix B).These results demonstrated the divergence in theOsNPF3.1promoter sequence between theindicaandjaponicacultivars.Additionally,eightindicaand eightjaponicacultivars were randomly selected to assess the association ofOsNPF3.1expression levels with the phenotypes of the different Haps.The results indicated that theOsNPF3.1expression levels,tiller number per plant,weight of straw per plant,total weight per plant,grain yield per plant,and NUtE in theindicacluster with Hap1 and Hap5 were higher than those in thejaponicacluster with Hap2 and Hap4 (Appendix C).Furthermore,we also sequenced the SNPs of the four haplotypes (Appendix D),and found that the expression ofOsNPF3.1in the roots and leaves ofindicaaccessions with Hap1 and Hap5 was higher than that ofjaponicaaccessions with Hap2 and Hap4 (Appendix D).Besides,the expression ofOsNPF3.1injaponicaaccessions with Hap2 is induced by GA3(Appendix D).Therefore,indicaaccessions with Hap1 and Hap5 had a higherOsNPF3.1expression and more tillers thanjaponicaaccessions with Hap2 and Hap4 under medium N concentration conditions,indicating thatOsNPF3.1expression was positively associated with tiller number.
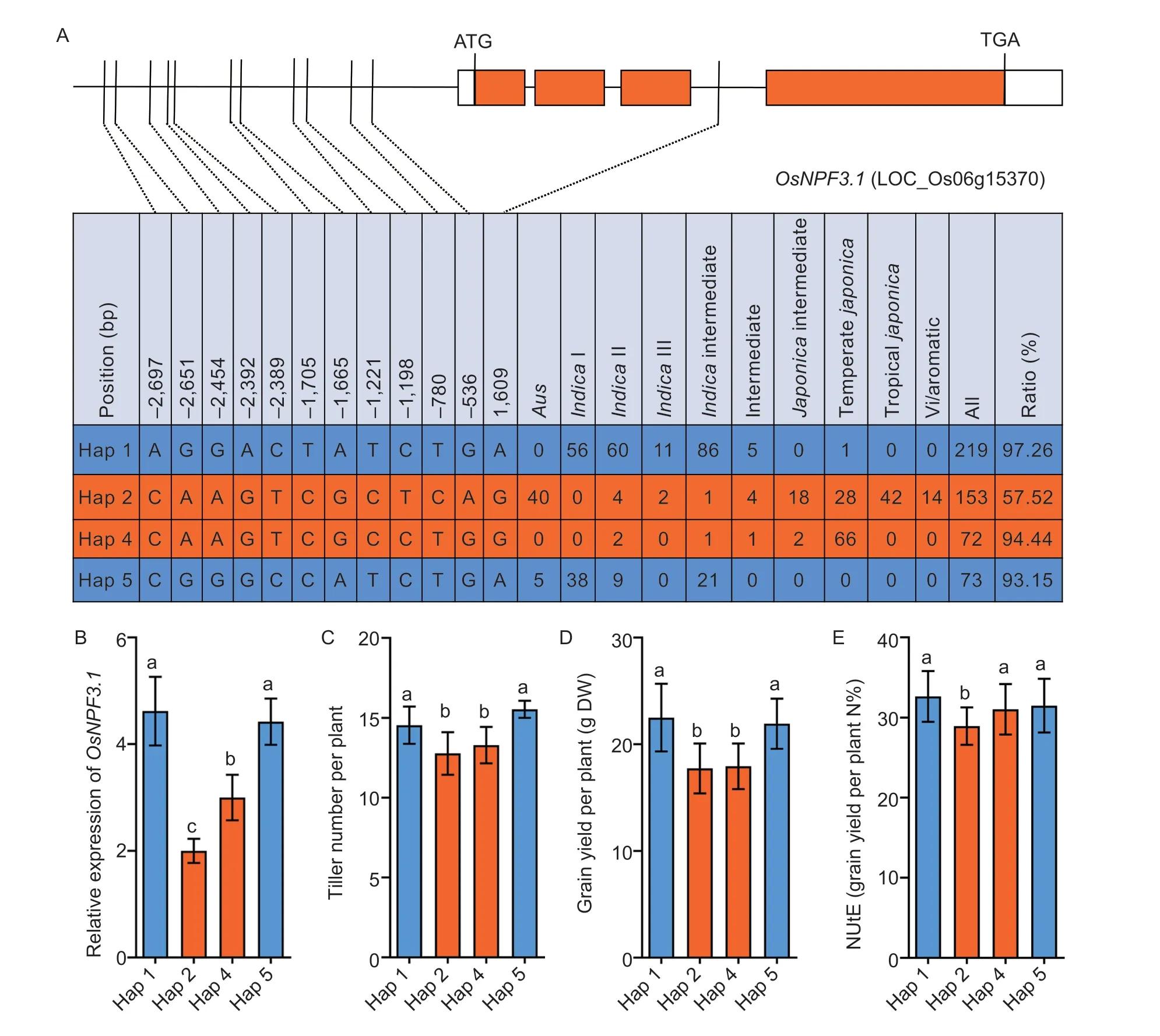
Fig.1 The expression level of OsNPF3.1 was positively correlated with rice phenotypes in different haplotypes.A,SNP divergence in the promoter sequence of OsNPF3.1 among 517 rice cultivars collected worldwide.The ratio represents the proportions of indica (Hap1 and Hap5) and japonica (Hap2 and Hap4) to each haplotype.B,relative expression of OsNPF3.1 in the basal part of rice Hap1-Hap5.C-E,means of tiller number per plant,grain yield per plant,and nitrogen utilization efficiency (NUtE) among all cultivars grouped by rice Hap1-Hap5.The 533 accessions of rice were grown at the Guiyang experimental field of Guizhou University,China under fertilizer (N-P-K=19%-7%-14%) treatment at 90 kg ha-1.Error bars indicate the SD (n=4).Different letters indicate significant differences at P<0.05.
3.2.Expression patterns in various tissues and protein subcellular localization of OsNPF3.1
To investigate the expression pattern ofOsNPF3.1,pOsNPF3.1-GUS transgenic plants were produced in the WT ZH11 background.GUS staining results showed the highestOsNPF3.1expression in the basal part (Fig.2-B),culm (Fig.2-D),and leaf blade (Fig.2-E),but it was also slightly expressed in lateral roots (Fig.2-A),leaf sheath (Fig.2-C),and young spikelet (Fig.2-F).RT-PCR analysis was performed to further verify the expression profile ofOsNPF3.1(Fig.2-G).The vascular tissues and parenchyma cells in the lateral root (Fig.2-H),basal part (Fig.2-I),leaf sheath (Fig.2-J),culm (Fig.2-K),leaf blade (Fig.2-L),and young spikelet (Fig.2-M) showed high GUS activities.RT-PCR (Appendices E and F) and GUS staining (Appendix E) were conducted using nitrate and phytohormone (GA3and ABA) treatments to characterize the regulation ofOsNPF3.1expression systematically.TheOsNPF3.1expression was strongly induced by either nitrate or GA3treatment in either the lateral root or basal part of rice (Appendix E).Intense GUS staining was also observed under nitrate or GA3treatment in the root and basal part (Appendix E).Moreover,theOsNPF3.1expression was strongly induced by different GA3treatments in either the root or shoot of rice.In addition,theOsNPF3.1expression decreased gradually with increasing concentrations of nitrate and ABA in either the root or shoot of rice (Appendix F).A 35S promoterdrivenGFPfused toOsNPF3.1was expressed in rice tissues to verify the subcellular localization of OsNPF3.1.The OsNPF3.1-GFP fusion protein was localized in the plasma membrane of rice root epidermis (Appendix G),which is consistent with a previous study (Yanget al.2022).These results suggested that OsNPF3.1 functions in membrane transport through rice roots and the basal parts to the aboveground parts.
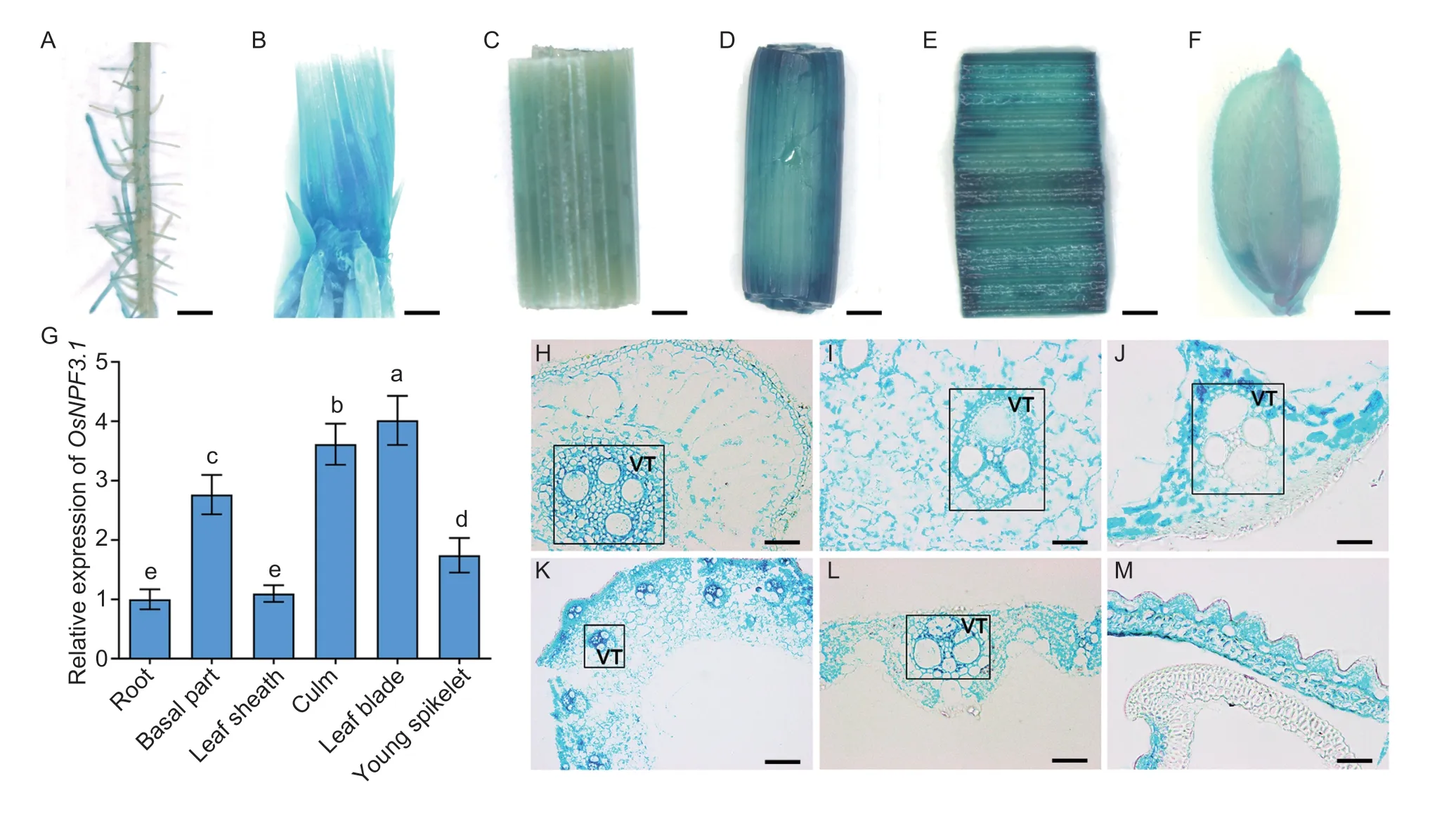
Fig.2 The expression pattern of OsNPF3.1.A-F,OsNPF3.1 promoter-GUS staining in the lateral root,basal part,leaf sheath,culm,leaf blade and young spikelet using the pOsNPF3.1-GUS-transgenic plants.Scale bars=5 mm.G,relative expression of OsNPF3.1 in different tissues of japonica rice wild-type Zhonghua 11 (WT ZH11) by RT-PCR.Error bars indicate the SD (n=4).Different letters indicate significant differences at P<0.05.H-M,transverse sections of lateral root,basal part,leaf sheath,culm,leaf blade and young spikelet using the pOsNPF3.1-GUS-transgenic plants.VT,vascular tissue.Scale bars=200 μm.
3.3.OsNPF3.1 is a pH-dependent low-affinity nitrate transporter
Thein vitro-synthesizedOsNPF3.1cRNA was microinjected intoXenopusoocytes to confirm that OsNPF3.1 protein transports nitrate through the plasma membrane.The nitrate transport activity of OsNPF3.1 was measured by the uptake of15NO3-at 0.2 and 10 mmol L-1.The well-characterized nitrate transporter CHL1 (AtNPF6.3 or AtNRT1.1) was used as a positive control.15NO3-uptake by oocytes injected withOsNPF3.1cRNA did not significantly increase in 0.2 mmol L-1K15NO3at pH 5.5 or 7.5.When oocytes with OsNPF3.1 cRNA were exposed to 10 mmol L-1K15NO3at pH 5.5 and 7.5,the15NO3-uptake was higher at pH 5.5 than at pH 7.5 (Fig.3).Therefore,OsNPF3.1 is a pH-dependent lowaffinity nitrate transporter.
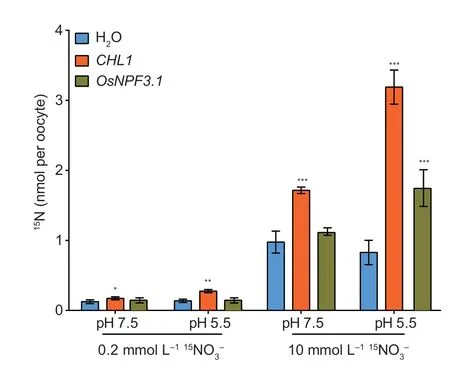
Fig.3 Functional analysis of OsNPF3.1 in Xenopus oocytes.The accumulation of 15N-labled nitrate by Xenopus oocytes is shown.Nitrate uptake activity was measured by OsNPF3.1-injected and CHL1-injected (as positive control) oocytes.Low-and high-affinity nitrate uptake activity was investigated by incubating the oocytes at pH 7.5 or pH 5.5 for 3 h.Error bars indicate the SD (n=5).*,** and *** indicate significant differences at P<0.05,P<0.01 and P<0.001,respectively.
3.4.CRlSPR/Cas9 mediated knockout mutant of OsNPF3.1 enhances NUtE by increasing the tiller number and filled grain number in rice
To reveal the biological function ofOsNPF3.1,we constructedOsNPF3.1-overexpressing (OE) lines in thejaponicarice WT ZH11 background and confirmed theOsNPF3.1expression levels in WT ZH11 and the OE lines (OE1,OE2 and OE3) through RT-PCR (Fig.4-A and C).We knocked out theOsNPF3.1sequence ofjaponicarice WT ZH11 by CRISPR technology and generated three independent CRISPR lines ofOsNPF3.1(C1,C2 and C3;Fig.4-A and D).To determine the effects of alteredOsNPF3.1expression on agronomic traits related to grain yield,we measured the tiller number per plant and filled grain number per plant of the OE and C lines in a paddy field (Fig.4-B,E and F).We found that the tiller number per plant (Fig.4-E) and filled grain number per plant (Fig.4-B and F) in the OE lines were less than those in the WT ZH11,but they increased significantly in the C lines compared with the WT ZH11 (Fig.4-B,E and F).To understand whetherOsNPF3.1affected NUtE,we measured the total N concentrations of these lines at the reproductive stage (Fig.4-G).We found that the CRISPR/Cas9 mediated knockout mutant ofOsNPF3.1enhanced NUtE,while the OE lines showed the opposite effect.
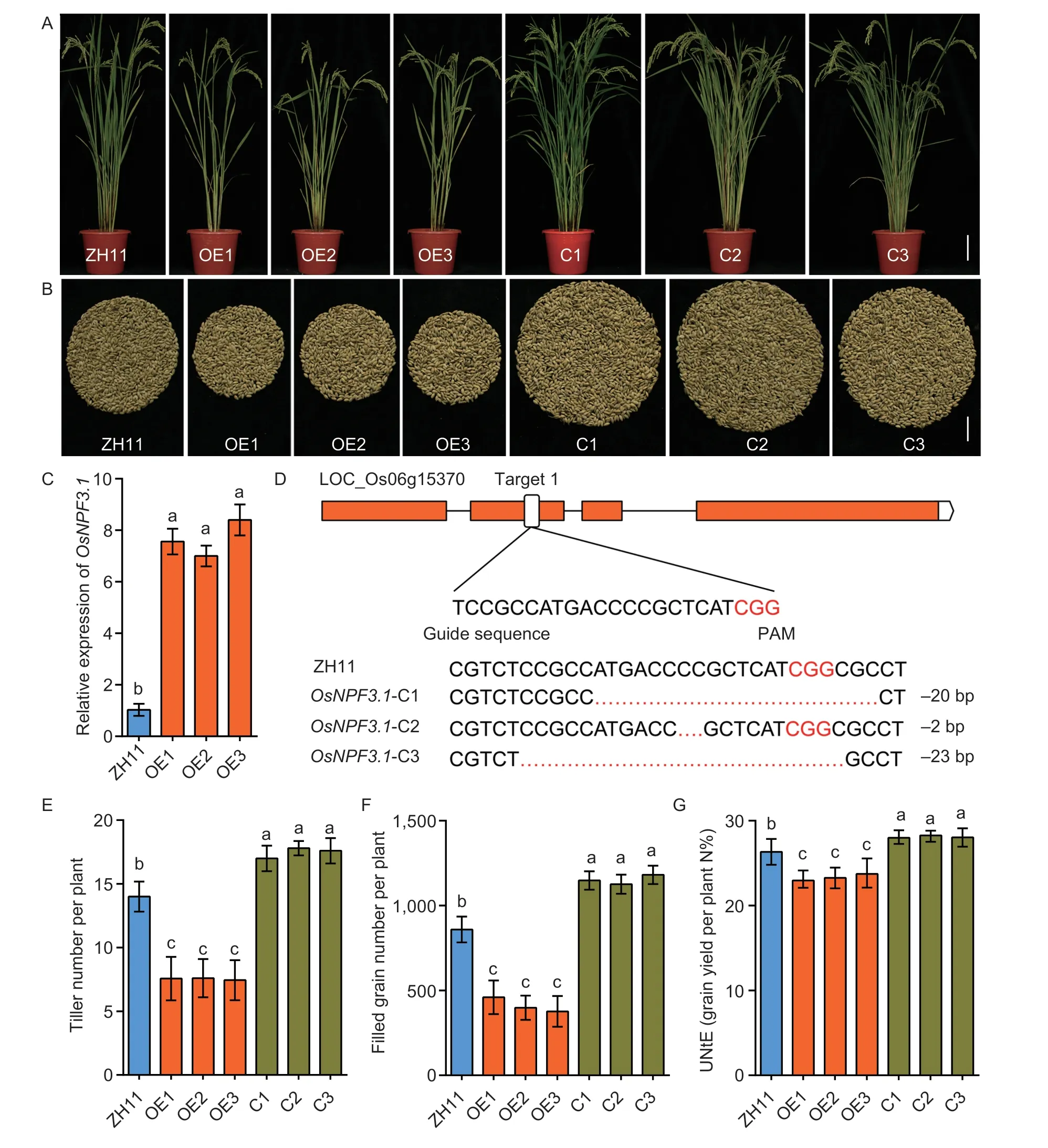
Fig.4 Phenotypes of the rice plants with altered expression of OsNPF3.1 grown in a paddy field.A,whole plant phenotypes of wild-type Zhonghua 11 (WT ZH11),OsNPF3.1-overexpressing lines (OE1-OE3) and OsNPF3.1 CRISPR lines (C1-C3).Scale bars=15 cm.B,filled grain numbers per plant of WT ZH11,OsNPF3.1-overexpressing lines (OE1-OE3) and OsNPF3.1 CRISPR lines (C1-C3).Scale bars=2 cm.C,relative expression of OsNPF3.1 in leaves of ZH11 and OsNPF3.1-overexpressing lines (OE1-OE3).D,gene knockout for OsNPF3.1 with CRISPR technology and sequencing results of base deletions of OsNPF3.1 CRISPR lines (C1-C3).E-G,means of tiller number per plant,filled grain number per plant and nitrogen utilization efficiency (NUtE) of WT ZH11,OsNPF3.1-overexpressing lines (OE1-OE3) and OsNPF3.1 CRISPR lines (C1-C3).All transgenic lines of OsNPF3.1 and WT ZH11 were grown in the field at Guiyang experiment field of Guizhou University under fertilizer (N-P-K=19%-7%-14%) treatment at 270 kg ha-1.Error bars indicate the SD for C (n=4) and for E-G (n=20).Different letters indicate significant differences at P<0.05.
3.5.OsNPF3.1 can mediate the transport of nitrate,ABA and GA3 and the outgrowth of axillary buds in rice
To further understand whether OsNPF3.1 mediates ABA and GA3transports similar to homolog AtNPF3.1 inArabidopsis(Talet al.2016),we performed assays for ABA and GA3uptake in protoplasts.We cultured rice protoplasts from the OE,C,and WT ZH11 lines with FITClabeled phytohormones,1 μmol L-1ABA-FITC (Fig.5-A) and 1 μmol L-1GA3-FITC (Fig.5-B),for 4 h.The ABA uptake assays indicated that the protoplast fluorescence signal of the OE lines was stronger than that of the WT ZH11 protoplasts,while the signal of the C lines was weaker than that of the WT ZH11 protoplasts (Fig.5-A and C).The GA3uptake assays indicated that the protoplast fluorescence signal of the OE lines was stronger than that of the WT ZH11 protoplasts,while the signal of the C lines was weaker than that of the WT ZH11 protoplasts (Fig.5-B and D).Interestingly,the fluorescence signal intensities of the GA3protoplasts were stronger than those of the ABA protoplasts of the corresponding materials in the OE,C and WT ZH11 lines (Fig.5-C and D).These results suggested thatOsNPF3.1can mediate ABA and GA3transmembrane transport,while it was previously reported that OsNPF3.1 imported GA4into oocytes (Talet al.2016).
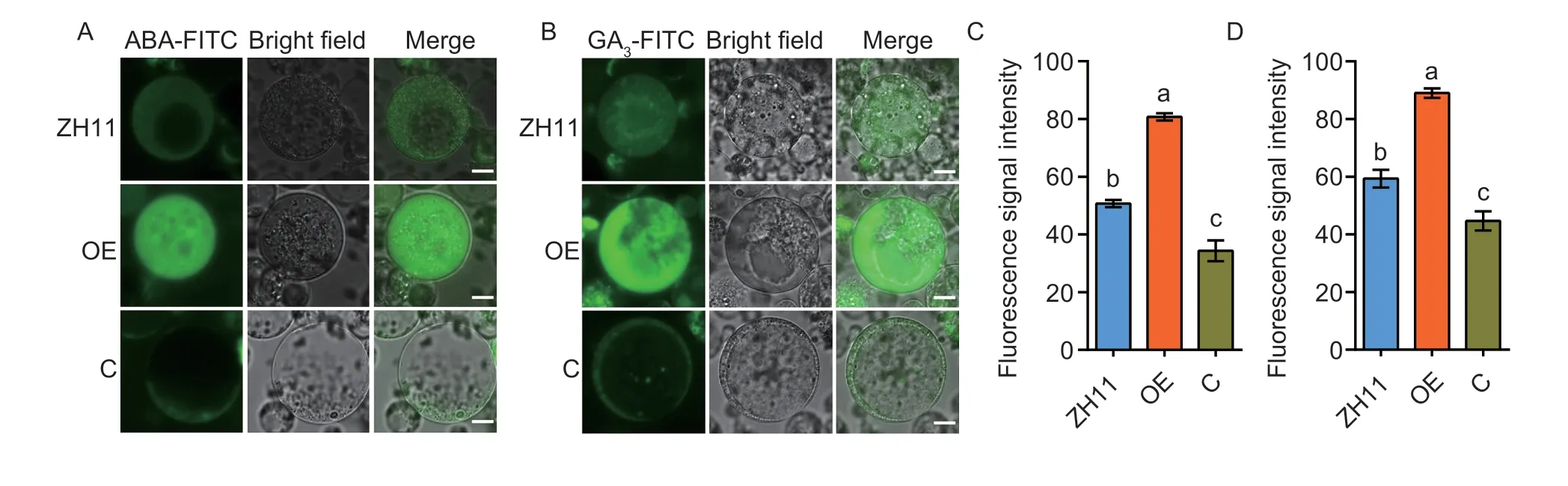
Fig.5 Protoplast abscisic acid (ABA) or gibberellin 3 (GA3) uptake assays among wild-type Zhonghua 11 (WT ZH11),OsNPF3.1-overexpressing line (OE) and OsNPF3.1 CRISPR line (C).Fluorescence was detected after culturing protoplasts with FITC-labeled ABA or GA3 for 4 h.A,green fluorescence images of the WT ZH11,OE and C lines under treatment with ABA-FITC.Scale bars=3 μm.B,green fluorescence images of the ZH11,OE and C lines under treatment with GA3-FITC.Scale bars=3 μm.C and D,detection of cell fluorescence signal intensities in A and B.Fluorescence intensities were normalized to the area of the respective cell by ImageJ software,and a total of 100 cells were statistically analyzed.Error bars indicate the SD (n=4).Different letters indicate significant differences at P<0.05.
To understand how OsNPF3.1 coordinates phytohormone and nitrate transport and plays a role in plant growth and development in rice,we cultured the OE lines,C lines,and WT ZH11 in different concentrations of nitrate nutrient solution or in ABA or GA3treatments for 35 days.Interestingly,the first axillary bud length of the OE lines was significantly greater than that of WT ZH11 at the low (0.2 mmol L-1) and medium (2 mmol L-1) nitrate concentrations (Fig.6-A and B,F).However,the first axillary bud outgrowth of the OE lines was significantly slower than that of WT ZH11 treated with high nitrate concentration (10 mmol L-1),medium nitrate concentration (2 mmol L-1)+medium ABA concentration (1 μmol L-1),and medium nitrate concentration (2 mmol L-1)+medium GA3concentration (1 μmol L-1;Fig.6-C,D,E,and F).Similarly,the second axillary bud outgrowth of the OE lines was significantly slower than that of WT ZH11 treated with 10 mmol L-1nitrate,2 mmol L-1nitrate+1 μmol L-1ABA,and 2 mmol L-1nitrate+1 μmol L-1GA3(Fig.6-C,D,E,and G).The first axillary bud of the C lines was significantly shorter than that of WT ZH11 at 0.2 mmol L-1nitrate but significantly longer than that of WT ZH11 at 10 mmol L-1nitrate and 2 mmol L-1nitrate+1 μmol L-1ABA (Fig.6-F).Furthermore,the second axillary bud of the C lines was significantly shorter than that of WT ZH11 at 0.2 mmol L-1nitrate but significantly longer than that of WT ZH11 at 2 mmol L-1nitrate+1 μmol L-1ABA and 2 mmol L-1nitrate+1 μmol L-1GA3(Fig.6-G).These results indicated that rice may produce more ABA and GA under high nitrate treatment,which can further inhibit the elongation of rice axillary buds after ABA and GA are directly transported by OsNPF3.1.
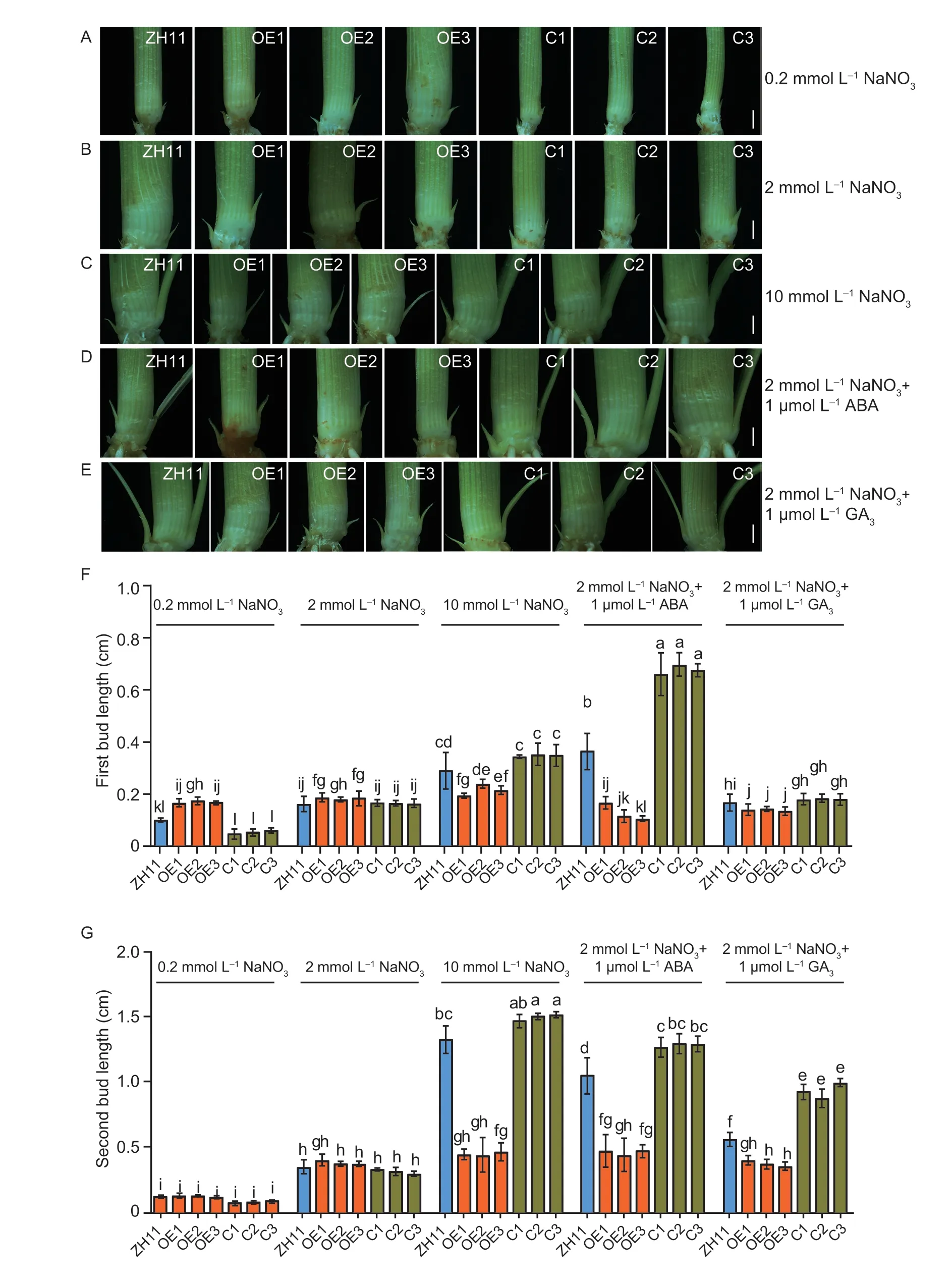
Fig.6 Axillary bud outgrowth of OsNPF3.1 transgenic plants under different concentrations of nitrate and phytohormones when the rice plants were grown for 35 days.A-E,phenotypes of axillary bud of wild-type Zhonghua 11 (WT ZH11),OsNPF3.1-overexpressing lines (OE1-OE3),and OsNPF3.1 CRISPR lines (C1-C3).ABA,abscisic acid;GA3,gibberellin 3.Scale bars=1 mm.F and G,means of first bud length and second bud length of WT ZH11,OE1-OE3,and C1-C3.Error bars indicate the SD (n=20).Different letters indicate significant differences at P<0.05.
In addition,the nitrate concentration in the aboveground part of the OE lines was significantly higher than that of WT ZH11 at 2 mmol L-1NaNO3;conversely,it was significantly lower in the C lines than in WT ZH11 (Appendix H).Furthermore,considering that OsNPF3.1 is a pH-dependent low-affinity nitrate transporter and expressed in the xylem parenchyma cells of vascular tissues,we suspected that OsNPF3.1 might mediate long-distance nitrate transport in rice.When supplied with 0.25 mmol L-1NH415NO3,the shoot/root15N concentration ratios were comparable in the WT ZH11,OE lines and C lines,with ratios of 0.179,0.184,and 0.176,respectively.However,when treated with 5 mmolthe ratios in the WT ZH11 and C lines were much lower (0.338 in WT ZH11 and 0.206 in C lines) than in in the OE lines (OE 0.432),reflecting the ability of the OE lines to facilitate the long-distance transport of nitrate from root to shoot (Appendix I).
Moreover,the biomass was higher in OE lines than in WT ZH11 at 0.2 mmol L-1nitrate and lower in OE lines than in WT ZH11 at 10 mmol L-1nitrate (Appendix J).By contrast,the biomass was lower in C lines than in WT ZH11 at 0.2 mmol L-1nitrate and 2 mmol L-1nitrate,and higher in C lines than in WT ZH11 at 10 mmol L-1nitrate (Appendix J).The biomasses of the WT ZH11,OE,and C lines were suppressed as the ABA or GA3concentration gradually increased (Appendices K and L).The OE lines were extremely sensitive to changes in the ABA or GA3concentration compared with the WT ZH11,whereas the C lines were insensitive to changes in the ABA or GA3concentration (Appendices K and L).Similarly,the plants of the OE lines withOsNPF3.1were significantly taller than those of WT ZH11 at 0.2 and 2 mmol L-1nitrate but were significantly shorter than those of WT ZH11 under different ABA or GA concentrations (Appendix M).Therefore,the OE lines withOsNPF3.1may transport more ABA or GA,thereby further inhibiting the bud elongation,biomass production,and plant height of rice.
3.6.OsNPF3.1 overexpression promotes ABA and GA accumulation
To further understand the effect ofOsNPF3.1on ABA and GA accumulation,we measured the ABA and GA concentrations in the roots and basal part under different nitrate concentrations and phytohormone (ABA and GA) treatments.The ABA concentrations were higher in the OE lines than in the WT ZH11 and C lines at 0.2 mmol L-1nitrate,2 mmol L-1nitrate,10 mmol L-1nitrate,and 2 mmol L-1nitrate+1 μmol L-1ABA in the roots and basal part (Fig.7).Surprisingly,the extremely accumulated ABA concentrations in the roots of the OE lines were similar under 10 mmol L-1nitrate and 2 mmol L-1nitrate+1 μmol L-1ABA conditions (Fig.7-A).The roots and basal part of the OE lines had very high ABA accumulation under 2 mmol L-1nitrate+1 μmol L-1ABA conditions (Fig.7).The GA concentration was higher in the OE lines than in WT ZH11 and the C lines under 2 mmol L-1nitrate+1 μmol L-1GA3in roots (Appendix N),while the GA concentrations were higher in the OE lines than in WT ZH11 and the C lines at 2 mmol L-1nitrate,10 mmol L-1nitrate,and 2 mmol L-1nitrate+1 μmol L-1GA3in the basal part (Appendix N).Therefore,OsNPF3.1had more significant effects on ABA accumulation in rice root and GA accumulation in the basal part.

Fig.7 Abscisic acid (ABA) concentration analysis of OsNPF3.1 transgenic plants under different concentrations of nitrate and medium concentrations of ABA when the rice plants were grown for 35 days.A and B,ABA concentrations in root,and the basal part of wild-type Zhonghua 11 (WT ZH11),OsNPF3.1-overexpressing lines (OE1-OE3),and OsNPF3.1 CRISPR lines (C1-C3).Error bars indicate the SD (n=5).Different letters indicate significant differences at P<0.05.
3.7.Altered OsNPF3.1 expression influences NUtE under different nitrate concentrations
To investigate the effect of the alteredOsNPF3.1expression on NUtE,we cultivated the WT ZH11,OE,and C lines in sand culture under different nitrate concentrations.Our results showed that plant height,biomass,and tiller number were higher in the OE lines than in WT ZH11 and the C lines for the 0.2 mmol L-1NaNO3(Fig.8-B;Appendix O) and 2 mmol L-1NaNO3(Fig.8-C;Appendix O) treatments.However,plant height,biomass,and tiller number were lower in the OE lines than in WT ZH11 at 10 mmol L-1NaNO3(Fig.8-D;Appendix O).Furthermore,NUtE did not differ among the WT ZH11,OE,and C lines at 0 mmol L-1NaNO3.The NUtE was higher in the OE lines than in WT ZH11 at 0.2 mmol L-1NaNO3and 2 mmol L-1NaNO3but lower in the C lines than in WT ZH11 only at 0.2 mmol L-1NaNO3(Fig.8-E).Interestingly,the total N concentration in the straw of the OE lines was lower than that of WT ZH11 at 0.2 and 2 mmol L-1NaNO3(Appendix P).Conversely,the total N concentration in the panicles of the OE lines was higher than that of WT ZH11 at 0.2 and 2 mmol L-1NaNO3(Appendix P).However,the total N concentration in the straw of the OE lines was significantly higher than that of WT ZH11 at 10 mmol L-1NaNO3,but significantly lower in the panicles of the OE lines than in WT ZH11 (Appendix P).However,an opposite trend was observed for the N concentration in the straw and panicles of the C lines compared with WT ZH11 (Appendix P).The total N content of the OE lines was higher than that of WT ZH11 at 0.2 and 2 mmol L-1NaNO3,while the total N content of the OE lines was lower than that of WT ZH11 at 10 mmol L-1NaNO3(Appendix P).These results showed that the nitrogen contents of the OE lines were significantly higher than those of WT ZH11 at low and medium nitrogen concentrations,while the nitrogen contents of the C lines were significantly higher than those of WT ZH11 at high nitrogen concentrations (Appendix P).Therefore,alteredOsNPF3.1expression was involved in NUtE regulation under different nitrate concentrations.In summary,our results showed thatOsNPF3.1is essential for nitrate transport,rice tillering and NUtE at low and medium nitrate concentrations,however,OsNPF3.1contributes to ABA and GA transport,and results in inhibition of rice tillering and NUtE at high nitrate concentrations (Fig.9).
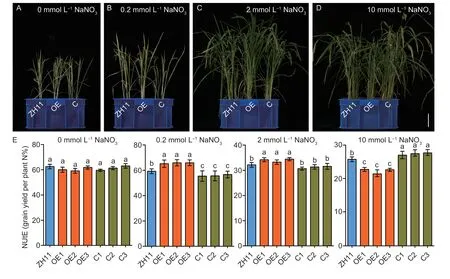
Fig.8 Phenotypic and nitrogen utilization efficiency (NUtE) analyses of OsNPF3.1 transgenic plants in sand culture under different nitrate concentrations.A-D,phenotypes of whole plants.Scale bars=10 cm.E,means of NUtE among OsNPF3.1-overexpressing (OE) lines,CRISPR (C) lines,and wild-type Zhonghua 11 (WT ZH11) in A-D.Error bars indicate the SD (n=4).Different letters indicate significant differences at P<0.05.
4.Discussion
4.1.OsNPF3.1 regulates the long-distance transport of nitrate
To efficiently utilize N sources in the environment,plants have developed two nitrate uptake systems,the low-and high-affinity nitrate transport systems (Glasset al.1992).Nitrate transporter 1 (NRT1)/peptide transporter (NPF) family members are mainly low-affinity nitrate transporters (Tsayet al.2007).In the rice NPF family,OsNPF2.2 (Liet al.2015),OsNPF2.4 (Xiaet al.2015),OsNPF4.5 (Wang S Set al.2020),OsNPF5.16 (Wanget al.2022),OsNPF6.1 (Tanget al.2019),OsNPF6.5 (Huet al.2015),OsNPF7.2 (Huet al.2016),and OsNPF8.9 (Linet al.2000) are low-affinity nitrate transporters.In the present study,15NO3-uptake assays using theXenopusoocyte system revealed thatOsNPF3.1in the NPF3 subfamily is a pH-dependent low-affinity nitrate transporter (Fig.3).Long distance nitrate transport to the aboveground part provides plants with essential mineral nutrients in their aerial parts.In rice,OsNPF2.4 functions in long-distance nitrate transport (Xiaet al.2015).OsNPF7.9 mediates nitrate loading into xylem vessels and long-distance transport to shoots (Guanet al.2022).In this study,OsNPF3.1was expressed in vascular tissues of the culm and leaf sheath (Fig.2),and the overexpression plants showed accumulation of nitrate in the shoot (Appendix H).Importantly,OsNPF3.1overexpression increased nitrate translocation from the roots to the shoots (Appendix I),indicating thatOsNPF3.1regulates the long-distance transport of nitrate.Moreover,AtNPF3.1,which is anOsNPF3.1homolog inArabidopsis,is a low-affinity nitrate transporter,suggesting thatOsNPF3.1is relatively evolutionarily conserved between monocotyledons and dicotyledons.
4.2.OsNPF3.1 mainly transports ABA and GAs at high nitrate concentrations
ABA and GA have been suggested to act as a longdistance signal that moves from the rootsviathe xylem to the shoot (Shabalaet al.2015;Binenbaumet al.2018).Efflux transporters would greatly enhance the loading of ABA into the xylem and the apoplast,thereby enabling a concentration to be reached to activate ABA signaling in other cells and organs (Boursiacet al.2013).The existence of GA efflux transporters is predicted in order for GA to effectively move locally,at both the tissue and cellular levels (Krameret al.2006).With respect to ABA uptake into cells,transporters would allow better access of ABA to intracellular receptors in the context of limited diffusion.Reports have shown that AtNPF4.6,as a protein that can mediate ABA uptake into cells for ABA distribution,was characterized as a low-affinity nitrate transporter (Kannoet al.2012).Moreover,numerous GA influx transporters have been identified.The glucosinolate transporter AtNPF2.10 can transport certain forms of GA (Ishimaruet al.2017).Importantly,AtNPF3.1 also transports ABA and GA3(Pikeet al.2014;Talet al.2016).Our uptake assays and HPLC analysis revealed that OsNPF3.1 is involved in ABA and GA3transport in protoplasts (Figs.5 and 7;Appendix N),which further verified that OsNPF3.1 imports GA4into oocytes (Talet al.2016).These results suggested that OsNPF3.1 transports not only nitrate but also ABA and GAs.OsNPF3.1overexpression promoted excessive ABA and GAs accumulation at higher nitrate concentrations (Fig.7;Appendix N).Similarly,the ABA transport activity of AtNPF4.6 was not inhibited by an excess amount of nitrate (Kannoet al.2013).In addition,overexpression ofAtNPF3.1enhanced the accumulation of GAs in all cells of the root (Talet al.2016).Likewise,the concentrations of GAs in roots ofOsNPF3.1CRISPR mutants decreased significantly at higher nitrate concentrations (Appendix N),which may lead to the elongation of their axillary buds.In addition,reports have shown thatAtNPF3.1expression is upregulated by the lack of nitrogen and by high levels of ABA,but downregulated in response to high levels of GA (Talet al.2016).In this study,OsNPF3.1expression was strongly increased by different GA3treatments,and decreased gradually with increasing concentrations of nitrate and ABA (Appendix F),indicating that the expression pattern ofOsNPF3.1under nitrate and GA treatment is similar to that ofAtNPF3.1inArabidopsis,but different under ABA treatment.Therefore,OsNPF3.1 mainly transports nitrate at low nitrate concentrations,and it mainly transports ABA and GAs at high nitrate concentrations.
4.3.Altered OsNPF3.1 expression differentially regulates rice tillering
Rice tillering is an important agronomic trait that determines the number of panicles and grain yield.In the present study,OsNPF3.1positively regulated axillary bud outgrowth and rice tillering under low and medium nitrate concentrations but negatively regulated axillary bud outgrowth and rice tillering under high nitrate concentrations (Fig.6;Appendix O).In the rice NPF family,onlyOsNPF7.4CRISPR lines promote rice tillering and enhance grain yield because the nitrate accumulation is reduced (Huanget al.2019).The overexpression of the GA catabolic GA2-oxidase gene increases the number of tillers or meristems in rice,tomato,turfgrass,and poplar,indicating that GA negatively regulates axillary bud formation (Agharkaret al.2007;Mauriatet al.2011;Huanget al.2021).Our results showed that GA inhibited the elongation of rice axillary buds under high nitrate and phytohormone treatments (Fig.6).Like previous reports on the inhibitory effects of ABA on rice axillary buds (Luoet al.2019),our study also showed that ABA inhibited the elongation of rice axillary buds (Fig.6).OsNPF3.1accelerated the inhibition of axillary bud elongation because of the transport of these two phytohormones (Fig.6).These results demonstrated that tillering inOsNPF3.1OE lines under high nitrate concentrations was inhibited because of the excessive ABA and GA3accumulation (Figs.6 and 7;Appendix N).In fact,the outgrowth of axillary buds was significantly influenced under different N concentrations,and the lengths of axillary buds were inhibited when the N concentration reached 15.0 mmol L-1(Wang R Net al.2020).Therefore,OsNPF3.1might affect ABA and GAs in rice tissues,which inhibits axillary bud outgrowth and rice tillering.
4.4.OsNPF3.1 regulates divergent NUtE under different nitrate concentrations
N is the primary factor limiting plant growth and yield (Sunet al.2014).The tillering,grain yield,and NUE ofjaponicarice were lower than those ofindica(Huet al.2015),indicating that improving NUE is the key to enhancing grain yield.Transporters for N acquisition are also essential for NUE (Tegeder 2014).The expression of N metabolism-related genes,especially nitrate transporter genes,is higher in super-high-yield rice in Yunnan (Zhonget al.2020).Although some efforts have been made in various plants (Liet al.2017),attempts to improve NUE in rice through nitrate transporter genes have been limited.In the rice NPF family,OsNPF7.3overexpression reduces NUtE in the presence of a high ammonium supply (Fanet al.2014).Furthermore,OsNPF7.7overexpression enhances NUtE from low to high N treatments (Huanget al.2018).OsNPF6.1overexpression also increases NUtE in rice under low N conditions (Tanget al.2019).Nitrate transporterOsNPF7.9mediates nitrate allocation and the divergent NUE betweenindicaandjaponicarice (Guanet al.2022).In addition,an amino acid mutation in theOsNPF3.1coding region was reported to cause different NUEs in wild and cultivated rice (Yanget al.2022).Furthermore,our study demonstrated thatOsNPF3.1contributes to NUtE with natural variations in its promoter (Fig.1-E),andOsNPF3.1overexpression could significantly promote NUtE under low and medium nitrate concentrations,while the CRISPR mutant ofOsNPF3.1could significantly promote NUtE under high nitrate concentrations (Fig.8).These results indicated thatOsNPF3.1differs fromOsNPF6.1,OsNPF7.3,OsNPF7.7andOsNPF7.9in terms of NUtE regulation,although these genes all belong to the NPF subfamily.Overall,the alteredOsNPF3.1expression improves NUtE in the overexpressing and CRISPR lines at low and high nitrate concentrations,respectively.
5.Conclusion
In this study,we demonstrated thatOsNPF3.1promoter sequences are divergent betweenindicaandjaponica,and OsNPF3.1 is a pH-dependent low-affinity nitrate transporter.In addition,OsNPF3.1transports phytohormones under different nitrate concentrations,andOsNPF3.1regulates the long-distance transport of nitrate.The alteredOsNPF3.1expression improves NUtE throughOsNPF3.1-overexpressing and CRISPR lines at low and high nitrate concentrations,respectively.
Acknowledgements
We thank Chen Y and Miller A at John Innes Centre,UK,for assistance with the functional analysis ofOsNPF3.1inXenopus laevisoocytes.This research was supported by the the Guizhou Provincial Excellent Young Talents Project of Science and Technology,China (YQK (2023) 002),the Guizhou Provincial Science and Technology Projects,China ((2022) Key 008),the Guizhou Provincial Science and Technology Support Plan,China ((2022) Key 026),the Key Laboratory of Molecular Breeding for Grain and Oil Crops in Guizhou Province,China ((2023) 008),and the Key Laboratory of Functional Agriculture of Guizhou Provincial Higher Education Institutions,China ((2023) 007).
Declaration of competing interest
The authors declare that they have no conflict of interest.
Appendicesassociated with this paper are available on https://doi.org/10.1016/j.jia.2023.04.024
杂志排行
Journal of Integrative Agriculture的其它文章
- Fine mapping and cloning of the sterility gene Bra2Ms in nonheading Chinese cabbage (Brassica rapa ssp.chinensis)
- Basal defense is enhanced in a wheat cultivar resistant to Fusarium head blight
- Optimized tillage methods increase mechanically transplanted rice yield and reduce the greenhouse gas emissions
- A phenology-based vegetation index for improving ratoon rice mapping using harmonized Landsat and Sentinel-2 data
- Combined application of organic fertilizer and chemical fertilizer alleviates the kernel position effect in summer maize by promoting post-silking nitrogen uptake and dry matter accumulation
- miR-24-3p promotes proliferation and inhibits apoptosis of porcine granulosa cells by targeting P27
