Circumscription of the East Asia clade(Apiaceae subfamily Apioideae)and the taxonomic placements of several problematic genera
2024-04-12JingZhouXinyuWngShilinZhouJunmiNiuJiruiYuZhnwnLiuStphnDowni
Jing Zhou ,Xinyu Wng ,Shilin Zhou ,Junmi Niu ,Jirui Yu ,Zhnwn Liu ,Stphn R.Downi
a School of Pharmaceutical Science and Yunnan Key Laboratory of Pharmacology for Natural Products,Kunming Medical University,1168 Western Chunrong Road,Yuhua Street,Chenggong New City,Kunming 650500,China
b Yunnan Academy of Forestry and Grassland,Kunming 650201,China
c Gaoligong Mountain,Forest Ecosystem,Observation and Research Station of Yunnan Province,Kunming 650201,China
d Yunnan Key Laboratory of Biodiversity of Gaoligong Mountain,Kunming 650201,China
e Department of Plant Biology,University of Illinois at Urbana-Champaign,Urbana,IL 61801,USA
f College of Modern Biomedical Industry,Kunming Medical University,Kunming 650500,China
Keywords: Physospermopsis Pimpinella Sinocarum Tongoloa Trachydium
ABSTRACT The East Asia (or Physospermopsis) clade was recognized in previous molecular phylogenetic investigations into the higher-level relationships of Apiaceae subfamily Apioideae.The composition of this clade,the phylogenetic relationships among its constituent taxa,and the placement of species previously determined to be problematic have yet to be resolved.Herein,nrDNA ITS sequences were obtained for 150 accessions of Apioideae,representing species whose distributions are in East Asia or genera having one or more species included within the East Asia clade.These data,along with published ITS sequences from other Apioideae (for 3678 accessions altogether),were subjected to maximum likelihood and Bayesian inference analyses.The results show that the East Asia clade contains representatives of 11 currently recognized genera: Hansenia,Hymenolaena,Keraymonia,Sinolimprichtia,Acronema,Hymenidium,Physospermopsis,Pimpinella,Sinocarum,Tongoloa,and Trachydium.However,the latter seven genera have members falling outside of the East Asia clade,including the generic types of all except Tongoloa.Within the clade,the species comprising these seven genera are widely intermingled,greatly increasing confusion among relationships than previously realized.The problematic species Physospermopsis cuneata is confirmed as falling within the East Asia clade,whereas P.rubrinervis allies with the generic type in tribe Pleurospermeae. Physospermopsis kingdon-wardii is confirmed as a member of the genus Physospermopsis,whereas the generic attributions of P.cuneata and Tongoloa stewardii remain unclear.Two species of Sinocarum (S.filicinum and S.wolffianum) are transferred into the genus Meeboldia.This is the most comprehensive molecular phylogenetic investigation of the East Asia clade to date,and while the results increase systematic understanding of the clade,they also highlight the need for further studies of one of the most taxonomically intractable groups in Apioideae.
1.Introduction
Apiaceae (Umbelliferae) are a large family of flowering plants,with 442 genera and 3575 species recognized (Christenhusz and Byng,2016).While most members of its largest subfamily Apioideae are readily identifiable because of their characteristic inflorescences and fruits,their higher-level relationships have traditionally been obscure (Constance,1971).A modern classification of Apioideae based on phylogenetic analyses of molecular data has elucidated the higher-level subdivisions within the subfamily and provided a framework for its classification (reviewed in Downie et al.,2001,2010).However,several major clades have not yet been treated formally,pending confirmation from further studies (Downie et al.,2010).
The East Asia (orPhysospermopsis) clade is one of these problematic groups.Calvi~no et al.(2006) recognized species ofPhysospermopsisH.Wolff,TrachydiumLindl.,TongoloaH.Wolff,SinolimprichtiaH.Wolff,NotopterygiumH.de Boissieu,HaplosphaeraHand.-Mazz.,andHanseniaTurcz.as comprising thePhysospermopsisclade.Included were species with an almost exclusively East Asian distribution.Zhou et al.(2008)thus suggested the alternative name “East Asia clade” and included species ofPimpinellaL.,SinocarumH.Wolff ex R.H.Shan &F.T.Pu,andVicatiaDC.within.Subsequent molecular phylogenetic studies supported its monophyly and expanded its circumscription to include representatives of 16 genera(Zhou et al.,2009,2020,2021;Downie et al.,2010;Mousavi et al.,2021;Wen et al.,2021).The East Asia clade is a sister group to tribe Komarovieae(Zhou et al.,2009),or arises from within it based on limited sampling and using the universal Angiosperms353 probe set (Clarkson et al.,2021).
Taxonomic relationships within the East Asia clade are extraordinarily complex,having species-rich genera whose boundaries are poorly defined and several monotypic genera.Taxonomic challenges surroundPimpinella,Sinocarum,Tongoloa,andTrachydium.Pimpinellaincludes~150 species that occur disjunctly in Africa,Europe,and Asia(Pimenov and Leonov,1993;Pu and Watson,2005).A fewPimpinellaspecies are also distributed in South America,with one species found in western North America (Pimenov and Leonov,1993).Members ofPimpinellacan be recognized by their perennial(rarely biennial or annual) herbaceous habit,stem bases without fibrous remnant sheaths,slightly laterally compressed fruits constricted at the commissure,and filiform ribs.Authors have arranged the species ofPimpinellainto various sections (de Candolle,1827;Bentham and Hooker,1867;Boissier,1875;Wolff,1910,1927;Shan and Sheh,1985a),and molecular phylogenetic studies have shown that the sectional classification ofPimpinellais highly artificial.Furthermore,Pimpinellais not resolved as a monophyletic group,with its members assigned to seven tribes and other major clades in Apioideae including the East Asia clade (Downie et al.,2010;Wang et al.,2014;Fereidounfar et al.,2016;Zhou et al.,2020).As the type species ofPimpinella(P.saxifragaL.) is placed within tribe Pimpinelleae,nomenclatural changes are required for thosePimpinellaspecies falling outside of Pimpinelleae.
Sinocarumis a small genus of approximately 20 species and exhibits a typical Sino-Himalayan distribution ranging from Nepal to SW China(Pu et al.,2005).Tongoloaalso occurs at high elevations in the Sino-Himalayan region,with 15 species recognized(Pan and Watson,2005a;Pimenov,2017).Both genera are poorly defined with diffuse generic boundaries,and relationship to putatively allied generaAcronemaEdgew.,Pimpinella,Physospermopsis,andTrachydiumhave been suggested.SinocarumandTongoloawere each determined to be non-monophyletic in their current circumscription (Downie et al.,2010;Zhou et al.,2020).Moreover,Sinocarumdisplayed conflicting placements in different phylogenetic studies(Zhou et al.,2008,2009,2020;Xiao et al.,2021).The proper circumscriptions ofSinocarumandTongoloaneed to be ascertained.
Trachydiumis another taxonomically complex genus,and its generic delimitation has historically been the subject of considerable debate.Since its establishment,those high montane species from the Sino-Himalayan region having a prostrate habit or shortened stems were described in,or transferred to,Trachydium(Pimenov and Kljuykov,2000).These include species previously attributable toAulacospermumLedeb.,ChamaesciadiumC.A.Mey.,ChamaesiumH.Wolff,LigusticumL.,Physospermopsis,PleurospermumHoffm.,Pseudotrachydium(Kljuykov,Pimenov &V.N.Tikhom.) Pimenov &Kljuykov,SchulziaSpreng.,andSinocarum.However,Pimenov &Kljuykov (2000) consideredTrachydiumto be a monotypic genus(T.royleiLindl.) and indicated that some of its species belong toSinocarumorTongoloa.Previous molecular results partly support such a viewpoint,as the four species examined to date occur throughout the East Asia clade and are allied withSinocarum,Tongoloa,andPhysospermopsis(Zhou et al.,2008,2009,2020).
No traditional classification of Apioideae has grouped together those genera that are presently included in the East Asia clade.The morphological diversity among its members is high,making it particularly difficult to identify clade-specific diagnostic characters.In this study,we (1) identify additional members of the East Asia clade by increasing the sampling of species whose distributions are in East Asia or genera having one or more species already included in the clade;(2) assess the phylogenetic relationships of those members comprising the East Asia clade,placing emphasis onSinocarum,Tongoloa,andTrachydium;and (3) confirm the phylogenetic placements of several species that have been treated variously (and perhaps erroneously) in previous molecular phylogenetic studies.The results obtained will be used to carry out nomenclatural transfers of some species and further clarify relationships within this taxonomically problematic group.
2.Materials and methods
2.1.Sampling
The nrDNA internal transcribed spacer(ITS)region was used for phylogenetic inference,as the numerous ITS sequences available in GenBank for Apiaceae subfamily Apioideae enabled an immediate consideration of these accessions for their possible inclusion in the East Asia clade.Comparative data for any specific chloroplast DNA locus does not yet exist across such a broad sampling of Apioideae.
To ascertain the limits of the East Asia clade,ITS sequence data were procured from 150 accessions of Apioideae (Table S1).These accessions included representation of species whose distributions in East Asia suggested possible close affinities with those taxa already included in the clade,or those genera whose species have already been unambiguously placed in the clade in earlier studies.Sampling of the generaTongoloa,Sinocarum,andTrachydiumwas more comprehensive than the other genera.For those nonmonophyletic genera whose species were previously determined to occur in several major clades,such asHymenidiumLindl.andPimpinella,those species that fell into the East Asia clade were resampled to confirm their phylogenetic positions.For those species having different ITS sequences in GenBank suggesting that at least one may be in error(i.e.,Physospermopsis rubrinervis(Franch.)C.Norman andP.cuneataH.Wolff),we collected new leaf material from the type localities.Where possible,each included species was represented by more than one accession to represent morphological,geographical,or possible infraspecific molecular variation.Apioideae ITS sequences were downloaded from GenBank and screened using the strategies described by Zhou et al.(2020) for possible inclusion in the East Asia clade.We only used those sequences released after March 2018,as sequences prior to that date were extracted in a previous study to ascertain the broad phylogenetic placements ofLigusticumspecies(Zhou et al.,2020).Thirtyeight separate ITS1 and ITS2 sequences from the previous matrix of Zhou et al.(2020) were replaced with the resequencing of the entire ITS region (ITS1-5.8S-ITS2).In summary,655 ITS sequences newly obtained from GenBank plus the 150 ITS sequences generated specifically for this study resulted in 805 sequences altogether.These were added to the matrix of Zhou et al.(2020) for a total of 3678 Apioideae ITS sequences.
2.2.DNA extraction
Total genomic DNAs were isolated from silica-gel dried materials or from herbarium specimens using a Plant Genomic DNA Kit(Tiangen Biotech,China)according to the manufacturer's protocol.The entire ITS region was amplified using the primers ITS4(5′-TCC TCC GCT TAT TGA TAT GC-3′) and ITS5 (5′-GGA AGT AAA AGT CGT AAC AAG G-3’;White et al.,1990) or N-nc18S10 (5′-AGG AGA AGT CGT AAC AAG-3′) and C26A (5′-GTT TCT TTT CCT CCG CT-3’;Wen and Zimmer,1996).If the PCR amplification failed,the region was amplified in two parts,using ITS-u1(5′-GGA AGK ARA AGT CGT AAC AAG G-3′)and ITS-u2(5′-GCG TTC AAA GAY TCG ATG RTT C-3′)for ITS1,and ITS-u3(5′-CAW CGA TGA AGA ACG YAG C-3′)and ITS-u4(5′-RGT TTC TTT TCC TCC GCT TA-3′) for ITS2 (Cheng et al.,2016).Procedures for amplification are the same as described in Zhou et al.(2008,2009).The PCR products were sequenced at the Sequencing Laboratory of the Institute of Botany using the same primers as for PCR on an ABI 3730 automated sequencer (Applied Biosystems,Foster City,California,U.S.A.).ITS sequence data acquired in this study were deposited in GenBank (Table S1).
2.3.Phylogenetic analysis
The sequences were assembled,aligned,and manually adjusted using the BioEdit sequence alignment editor (Hall,1999).Initially,we performed maximum likelihood (ML) analyses using RAxML v.8.0.0 (Stamatakis,2014) on XSEDE via the CIPRES Science Gateway (http://www.phylo.org/).The settings were rapid bootstrap analysis with 1000 replicates and utilizing the GTR+I+G substitution model.Three species of tribe Bupleureae were used to root the trees as the tribe occupies a sister-group relationship to all other apioid genera excluding its most early-diverging branches(Downie et al.,2001;Calvi~no et al.,2006).When the circumscription of the East Asia clade was delimited through this ML analysis,we selected only its accessions to conduct Bayesian inference (BI)with MrBayes v.3.2.5 (Ronquist and Huelsenbeck,2003;Ronquist et al.,2012).The BI trees were rooted with three members of tribe Komarovieae,as the tribe constitutes an immediate sister group to the East Asia clade (Zhou et al.,2009,2020).For the BI analyses,we determined the appropriate nucleotide substitution models using the corrected Akaike information criterion (AIC) in jModeltest v.2.1.7 (Posada,2008;Darriba et al.,2012).We performed the BI analysis using two independent runs of the Markov chain Monte Carlo (MCMC) for 10 million generations each and sampling one tree every 1000 generations.Analyses were run until the standard deviation of split frequencies was less than 0.001.Tracer was also used to graphically assess the convergence of runs,and to determine that the effective sampling size values exceeded 200.We discarded the first 25 % generations as burn-in,and the remaining trees were summarized into a 50% majority-rule consensus tree with posterior probabilities (PP).
3.Results
The aligned data matrix of 3678 ITS sequences contained 736 positions,with none excluded due to alignment ambiguities.Of these positions,63 were autapomorphic,63 were constant,and 610 were parsimony informative.Phylogenetic analysis of this data matrix resulted in a significant expansion of the East Asia clade from any previous circumscription.Two representatives from the genusAcronemaare newly recognized members of the clade and the numbers of species fromHymenidium,Tongoloa,andTrachydiumhave increased substantially.Except for the generaHansenia,HymenolaenaDC.,KeraymoniaFarille(including one accession),andSinolimprichtia(a monotypic genus),all other genera comprising the East Asia clade are polyphyletic and greatly intermingled.The nomenclatural types ofAcronema,Hymenidium,Physospermopsis,Pimpinella,Sinocarum,andTrachydiumfall outside of the East Asia clade.The ML tree containing all 3678 representatives of Apioideae is presented in Fig.S1,while a portion of this ML tree representing only the East Asia clade and its simplified phylogenetic tree showing relationships among groupings of taxa are presented in Figs.1 and 2A,respectively.In addition,we extracted portions of theAcronemaandSinodielsiaclades that contain taxa of interest,as shown in Figs.3 and 4,respectively.
3.1.Problematic sequences in GenBank
Simultaneous analysis of multiple ITS sequences from the same species indicated that some may be either misidentifications or resulting from contamination or improper laboratory practices because they did not fall as monophyletic in the phylogenetic trees.Sequences of some conspecific taxa differed considerably and occurred in different tribes and other major clades of Apioideae.Physospermopsis cuneatais confirmed as belonging to the East Asia clade.The placement of its two previously examined accessions(GenBank accession nos.EU236188 and EU236189) in tribe Pimpinelleae (Zhou et al.,2008,2009) must be attributable to identification errors.The accession ofPhysospermopsis rubrinerviscollected from its type locality (Luoping Mt.,Yunnan Province,China)allies with other representatives of the same species within the tribe Pleurospermeae;the accession ofP.rubrinervis(AF164836 and AF164861),previously attributable to the East Asia clade,is also a misidentification.Similar situations also occur forSinocarum coloratum(Diels) H.Wolff ex R.H.Shan &F.T.Pu andS.cruciatum(Franch.) H.Wolff ex R.H.Shan &F.T.Pu,where sequences of the same species fell into both theAcronemaclade(Fig.3)and the East Asia clade(Fig.1).After examination of their voucher materials,we considered thatSinocarum cruciatumaccession MN846686 andS.coloratumaccession MN846685 are correctly identified,whereasS.coloratumFJ385063 andS.cruciatumEU236209 in the East Asia clade andS.coloratumAY328927 andS.cruciatumAY038199,AY038213 and MH293141 in theAcronemaclade are not.Vicatia bipinnataR.H.Shan &F.T.Pu (EU236217) and its synonymous speciesMelanosciadium bipinnatum(R.H.Shan &F.T.Pu) Pimenov et Kljuykov(JX514721)fell into the East Asia clade(Fig.1)and tribe Selineae (Fig.S1),respectively.The voucher specimen ofV.bipinnatais actuallyHansenia forrestii(H.Wolff) Pimenov &Kljuykov,while the ITS sequence ofM.bipinnatum(JX514721) is identical to that ofPimpinella rhomboideavar.tenuilobaR.H.Shan&F.T.Pu.The newly obtainedMelanosciadium genuflexumPimenov&Kljuykov is placed in theSinodielsiaclade and is sister group to the clade ofTongoloa zhongdianensisS.L.Liou,Pterocyclus rivulorum(Diels) H.Wolff,andSinodielsia delavayi(Franch.) Pimenov &Kljuykov (Fig.4).Heptaptera anisopteraTutin (AY941273 and AY941301) andSpuriopimpinella brachycarpa(Kom.) Kitagawa(GQ379337) arise from within accessions ofSinolimprichtia alpinaH.Wolff andPimpinella rhomboideaDiels,respectively.However,other sequences ofH.anisopteraandS.brachycarpaare placed into theOpopanaxandAcronemaclades,so we excluded these two problematic genera from the East Asia clade.With nearly identical ITS sequences,Hymenidium lindleyanum(Klotzsch) Pimenov &Kljuykov (MZ191057) comprises a group withH.nanum(Rupr.)Pimenov&Kljuykov in the East Asia clade,while another accession ofH.lindleyanum(FJ469949 and FJ483488) falls into tribe Pleurospermeae (Fig.S1).

Fig.1.Portion of the large consensus tree (Fig.S1) obtained from maximum likelihood analysis of 3678 nrDNA internal transcribed spacer sequences from Apiaceae subfamily Apioideae showing only the East Asia clade(comprising 206 terminals).Support values ≥50%are provided.Clade designations are those defined in the text.The 106 newly sampled accessions in this portion of the tree are shown in bold.GenBank accession numbers of all sequences are provided;two numbers indicate sequences from separate ITS1 and ITS2 regions.*Indicates that the identifications of original materials from these accessions were confirmed and that these sequences were used in the Bayesian inference analysis;names in [] are the correct ones.**Indicates that the identifications of these sequences are problematic and thus these taxa were excluded from the Bayesian analysis.
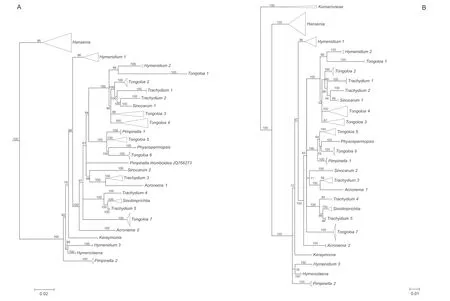
Fig.2.A.Simplified phylogenetic tree derived from maximum likelihood analysis of 3678 nrDNA internal transcribed spacer sequences from Apiaceae subfamily Apioideae showing the groupings of taxa in East Asia clade,with support values(≥50%)provided next to the branches.B.Simplified phylogenetic tree derived from Bayesian inference analysis of 195 nrDNA internal transcribed spacer sequences showing the group of taxa from the East Asia clade and outgroups,with Bayesian posterior probabilities(≥50%)provided next to the branches.

Fig.3.Portion of the large consensus tree (Fig.S1) obtained from maximum likelihood analysis of 3678 nrDNA internal transcribed spacer sequences from Apiaceae subfamily Apioideae showing only the accessions falling in the Acronema clade.Support values ≥50%are provided.The newly sampled accessions in this portion of the tree are shown in bold.GenBank accession numbers of all sequences are provided;two numbers indicate sequences from separate ITS1 and ITS2 regions.
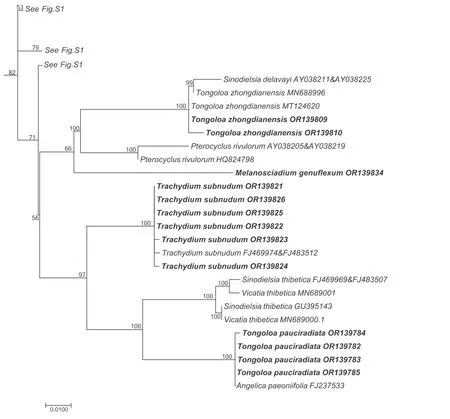
Fig.4.Portion of the large consensus tree (Fig.S1) obtained from maximum likelihood analysis of 3678 nrDNA internal transcribed spacer sequences from Apiaceae subfamily Apioideae showing only the accessions falling in the Sinodielsia clade.Support values ≥50% are provided.The newly sampled accessions in this portion of the tree are shown in bold.GenBank accession numbers of all sequences are provided;two numbers indicate sequences from separate ITS1 and ITS2 regions.
The accessions ofPhysospermopsis kingdon-wardii(H.Wolff) C.Norman (AF164835,AF164860 and EU236190) do not ally.We further checked their voucher specimens(W810822 for EU236190,and E00052566 for AF164835 and AF164860),which should be identified asTrachydium soulieiH.de Boissieu andT.trifoliatumH.Wolff,respectively.The accessionSinolimprichtia alpina(GQ3 79336) is sister group to the clade comprising two accessions ofHymenidium lhasanumPimenov &Kljuykov,and distantly away from other accessions of the same species.With only one nucleotide difference between them,we consider that the species identified asS.alpina(GQ379336) is actuallyH.lhasanum.Three accessions ofPimpinella henryi Diels fall into both Pimpinelleae and the East Asia clade,with the two accessions in the East Asia clade not grouping together.Although we cannot confirm the identity of accessions with sequences EU236195 and MH710738,the one in the East Asia clade (JQ766274) arising from withinPimpinella rhomboideais no doubt a misidentification.Furthermore,Tongoloa napifera(H.Wolff) C.Norman (MT124605) does not ally with its synonymous speciesTrachydium napiferumH.Wolff,andPimpinella rhomboidea(JQ766273) forms an isolated lineage distantly away from its conspecific accessions.Other suspect sequences includeHymenidium amabile(Craib &W.W.Smith) Pimenov &Kljuykov(KP311487),Tongoloa elataH.Wolff(MH807993),Tongoloa silaifolia(H.de Boissieu) H.Wolff (EU236213),andTrachydium variabileH.Wolff (HQ342149 and HQ246206).In summary,11 genera are recognized herein as constituting the East Asia clade (Table 1).
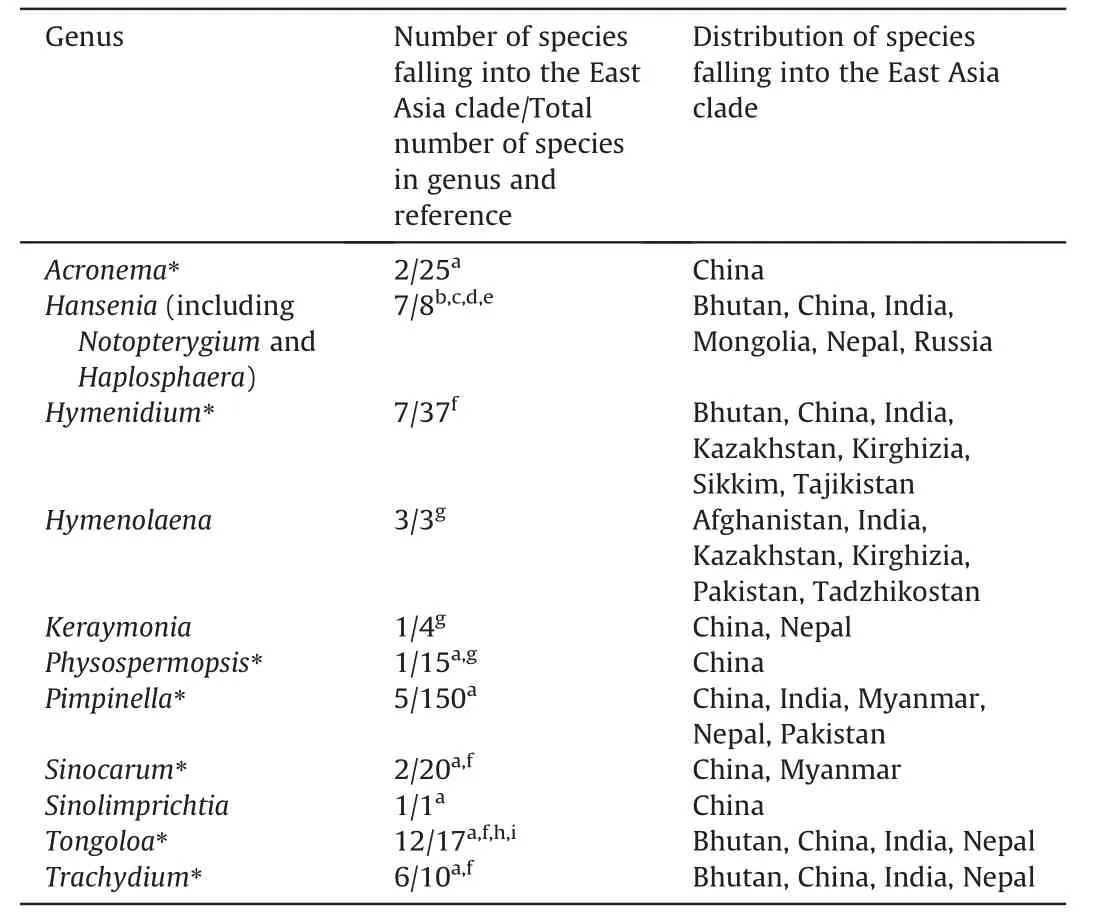
Table 1 Composition and distribution of the East Asia clade.Asterisks denote those genera that are not monophyletic based on the analyses presented herein.Distribution only involves species of the East Asia clade and does not include the undescribed species.
3.2.Phylogeny of the East Asia clade
The aligned 195 ITS sequences from taxa comprising the East Asia clade and three outgroup taxa (Komarovieae) resulted in a matrix of 620 positions,with none excluded because of alignment ambiguities.Of these positions,249 were parsimony informative,305 were constant,and 66 were autapomorphic.Gaps ranging in length from 1 to 3 bp were introduced to facilitate alignment.The mean percentage G+C content across all sequences was 54.8%.The BI results are generally congruent in topology with the portion of the ML tree containing the East Asia taxa;branch support values are indicated (Figs.2B and 5).
The seven species ofHanseniaconstituted a well-supported monophyletic group (BI=100),with each of its species also arising as monophyletic.Two species of the former genusHaplosphaera,Haplosphaera phaeaHand.-Mazz.(≡Hansenia phaea(Hand.-Mazz.) J.B.Tan &X.G.Ma) andH.himalayensisLudlow(≡Hansenia himalayensis(Ludlow) J.B.Tan &X.G.Ma),arise from within theHanseniaclade.The three species ofHymenolaenaalso allied as a strongly supported clade(BI=100)and is sister group toHymendium lhasanum.The four newly sampled species ofHymenidium[H.pulszkyi(Kanitz)Pimenov&Kljuykov,H.nubigenum(H.Wolff) Pimenov &Kljuykov,H.tsekuense(R.H.Shan) Pimenov &Kljuykov,andH.sp.] ally with their congenersH.amabileandH.nanumin cladeHymenidium1 (BI=98).Hymenidium virgatumPimenov&Kljuykov(cladeHymenidium2)andH.lhasanum(cladeHymenidium3) are separately placed in the East Asia clade and do not cluster with the aforementioned congeners.
With the exception ofTongoloa zhongdianensisandT.pauciradiataH.Wolff that are placed into theSinodielsiaclade(Fig.4),all other sampled accessions ofTongoloafell into the East Asia clade,albeit in seven different groups (Tongoloa1-7).Tongoloa tagongensisL.J.Gui &X.J.He (Tongoloa1),T.argutaL.J.Gui &X.J.He(Tongoloa2),T.rubronervisS.L.Liou (Tongoloa6),andT.gracilisH.Wolff(Tongoloa7)each comprises a separate,monophyletic group.Three species ofTongoloa[T.loloensis(Franch.)H.Wolff,T.stewardiiH.Wolff,andT.filicaudicisK.T.Fu] comprised a well-supported group (Tongoloa3;BI=87).Five Chinese endemic species ofTongoloa[Tongoloa elataH.Wolff,T.dunnii(H.de Boissieu) H.Wolff,T.silaifolia,T.taeniophylla(H.de Boissieu) H.Wolff,andT.sp.1]comprised theTongoloa4 group(BI=100).Four unknown species ofTongoloaally withT.tenuifoliaH.Wolff inTongoloa5 (BI=100).
The species ofTrachydiumalso comprise multiple groups in the East Asia clade (Trachydium1-5),while its nomenclatural type(T.roylei) andT.subnudumC.B.Clarke ex H.Wolff are included within tribe Pleurospermeae and theSinodielsiaclade,respectively(Figs.S1 and 4).All accessions ofTrachydium variabileally as monophyletic(Trachydium1;BI=100)and comprise a sister group to the clade ofTrachydium2 andSinocarum1.Trachydium2 comprises two accessions ofTrachydiumhaving identical ITS sequences(BI=100);their morphology is different from other species ofTrachydiumand therefore they likely represent a new species.Trachydium3 containsTrachydium simplicifoliumW.W.Smith,T.souleiH.de Boissieu,T.trifoliatum,andT.sp.3,of which the included accessions of all but the latter comprise a large polytomy having sequence divergence values of identity to 0.50%.Trachydium napiferum(Trachydium5) allies with the Chinese endemic genusSinolimprichtia,comprising a clade sister group to the clade ofT.involucellatumR.H.Shan &F.T.Pu andTrachydium sp.2 (Trachydium4).
The genusSinocarumis also not monophyletic,with accessions ofS.bellumPimenov &Kljuykov,S.coloratum,S.cruciatum,S.filicinumH.Wolff,S.muliensisL.J.Gui,Y.P.Xiao &X.J.He,S.vaginatumH.Wolff,S.wolffianum(Fedde ex H.Wolff)P.K.Mukh.&Constance,andS.sp.1 placed into theAcronemaclade(Fig.3)and away fromS.schizopetalum(Franch.)H.Wolff ex R.H.Shan&F.T.Pu andS.dolichopodum(Diels) H.Wolff ex R.H.Shan &F.T.Pu in the East Asia clade.The twoSinocarumspecies in the East Asia clade are placed into two separate monophyletic groups (Sinocarum1-2).Sinocarum dolichopodumallies withTongoloasp.MZ054152 (inSinocarum1),while the two included accessions ofSinocarum schizopetalum(Sinocarum2) fall as a sister group to the clade ofTrachydium3 andAcronema1 (BI=71).
The genusAcronemais referred to both theAcronemaclade and the East Asia clade.Two species occur in the East Asia clade.Acronema crassifoliumH.C.Wang,X.M.Zhou &Y.H.Wang (Acronema1)allies withTrachydium3,andA.chieniiR.H.Shan(Acronema2)occupies a separate lineage.Five species ofPimpinellafall in the East Asia clade and are divided into two distantly related groups(Pimpinella1 and 2).The first group comprises a large polytomy(with sequence divergence values ranging from identity to 0.17 %)and includesPimpinella acuminata(Edgew.)C.B.Clarke,P.purpurea(Franch.) H.de Boissieu,P.caudata(Franch.) H.Wolff,and one accession ofP.henryi.The second group consists solely ofPimpinella rhomboideaand comprises a trichotomy with all other species ofthe East Asia clade except for the genusHansenia.Lastly,Keraymonia cortiformisCauwet&S.B.Malla,the only included species of the genus,constitutes an isolated lineage without close relatives.
4.Discussion
4.1.Circumscription and distribution of the East Asia clade
The East Asia clade has previously been considered as comprising representatives of 16 genera(Downie et al.,2010;Zhou et al.,2020,2021).However,with the reduction ofHaplosphaera,Notopterygium,andLigusticum litangenseF.T.Pu toHansenia(Pimenov et al.,2008;Tan et al.,2020;Zhou et al.,2020),the exclusion of dubious sequencesS.brachycarpa,H.anisoptera,andV.bipinnatafrom the clade,and the examination of additional sequences available in GenBank for possible inclusion,the East Asia clade is herein circumscribed to include 11 genera(Table 1).Seven of these genera (Acronema,Hymenidium,Physospermopsis,Pimpinella,Sinocarum,Tongoloa,andTrachydium) are each not monophyletic,and all have members falling outside of the East Asia clade.Within the East Asia clade each of these genera exceptPhysospermopsiscomprises two or more lineages with its species intermingled with other East Asia taxa,resulting in relationships more complex than previously realized.ExcludingTongoloa,the nomenclatural types of six genera fall into other tribes and major clades of Apioideae indicating that substantial taxonomic rearrangements and new combinations will be inevitable.Upon expanded sampling of species distributed in East Asia and genera in which species have already been unambiguously placed in the clade,the number of species belonging toHymenidium,Tongoloa,andTrachydiumin the East Asia clade has increased greatly.
When the East Asia clade was first identified (asPhysospermopsisclade,Calvi~no et al.,2006),it included species almost exclusively distributed in East Asia.With the expansion of its circumscription herein,its distribution now extends to Central and South Asia,and even Asian Russia.Two species,namelyS.schizopetalumandPimpinella purpurea,have also been reported to occur in Southeast Asia(Pimenov,2017).All genera are native to Asia (the monotypic genusSinolimprichtiais endemic to China).With the exception ofHymenolaena,whose distribution is limited to Central and South Asia (Afghanistan,India,Kazakhstan,Kirghizia,Pakistan,and Tajikistan;Pimenov and Kljuykov,2000),all other genera have members distributed in China.The single species ofKeraymonia(K.cortiformis) is distributed in China and Nepal(Pimenov,2017).Six species ofHansenia[H.forbesii(H.de Boissieu)Pimenov&Kljuykov,H.forrestii,H.oviformis(R.H.Shan)Pimenov&Kljuykov,H.phaea,H.weberbaueriana(Fedde ex H.Wolff)Pimenov&Kljuykov,andH.trifoliolataQ.P.Jiang &X.J.He (not included in present analysis)] are confined to China,H.himalayensisis distributed in Bhutan,China,India and Nepal,and the typeH.mongolicaTurcz.occurs in Mongolia and Siberia,Russia (Sheh and Watson,2005;Pimenov et al.,2008;Pimenov,2017).Members of the remaining East Asia clade genera are distributed in Bhutan,China,India,Kazakhstan,Kirghizia,Myanmar,Nepal,Pakistan,Sikkim,and Tajikistan,with each genus having a species occurring in or endemic to China (Sheh et al.,2005;Pimenov,2017;Pimenov and Kljuykov,2000).
4.2.Morphological diversification and relationships within the East Asia clade
Unlike the tribes of Apioideae inferred from molecular data that can also be delimited using morphological or anatomical synapomorphies(e.g.,Bupleureae,Neves and Watson,2004;Oenantheae,Hardway et al.,2004;Chamaesieae,Zhou et al.,2009),the East Asia clade is morphologically heterogeneous.The differences among its members are extreme,exhibiting a great diversity of habit,life form,flower morphology,umbel structures,and fruit morphology and anatomy.The fruit presents variation in shape,degree and direction of mericarp compression(laterally compressed or slightly dorsally compressed),surface of mericarp(tuberculate or glabrous),modifications of ribs(winged or filiform),and shape of the ventral endosperm (flat or concave).We are not aware of any morphological features that can be used to unambiguously define the East Asia clade.
Of the many distinct groupings of taxa within the East Asia clade,four are equivalent to generally recognized genera(Hansenia,Sinolimprichtia,Hymenolaena,andKeraymonia).With the incorporation of species fromNotopterygiumandHaplosphaera,as well as the recognition of a new species,Hanseniais now represented by eight species (Pimenov et al.,2008;Tan et al.,2020;Jiang et al.,2022).It is monophyletic and diagnostic morphologically,with its(sometimes somewhat unequally) broad wings on all ribs.Sinolimprichtiais a monotypic genus endemic to China and is characterized by scaphoid petals and dark purple anthers (Pan and Watson,2005b).It falls as a sister group toTrachydium napiferumwithin the East Asia clade,which in turn is sister group toT.involucellatumandTrachydium sp.2.These three species ofTrachydiummay be referred toSinolimprichtia,but this remains to be confirmed.The genusHymenolaenaconsists of three distinctive species distributed in western Himalaya,Afghanistan,and Middle Asia.Its pinnate leaves and large,conspicuous,and almost entirely membranous bracteoles well delimit the genus (Pimenov and Kljuykov,2000);the group is also monophyletic in trees presented herein.Keraymoniacurrently comprises four species distributed in Nepal,Bhutan,and Xizang of China (Pimenov and Kljuykov,2000).The only species included in this study,K.cortiformis,forms an isolated lineage in the East Asia clade.The type species,Keraymonia nipaulensisCauwet-Marc &Farill,allies closely with members of tribe Scandiceae in analyses of chloroplast generbcLsequences (Saslis-Lagoudakis et al.,2012).The generic status ofK.cortiformismust be reconsidered.
Tongoloais highly polyphyletic,however.Two of its species(Tongoloa zhongdianensisandT.pauciradiata) ally with theSinodielsiaclade,while the remainder form seven separate clades within the East Asia clade (Table 2).Some authors have suggested thatT.zhongdianensisis conspecific withT.loloensis(Pan and Watson,2005a).In our analysis,T.zhongdianensisis paraphyletic,withSinodielsia delavayiarising from within.S.delavayiis the type ofSinodielsiaH.Wolff,which is conspecific withMeeboldia yunnanensis(H.Wolff) Constance&F.T.Pu;thus,it has been renamed asMeeboldia delavayi(Franch.)W.Gou&X.J.He(Gou et al.,2021).All species ofMeeboldiacluster together in theAcronemaclade.Tongoloa loloensis,which has been variously designated as a species ofCarum,PimpinellaorTrachydium(Franch,1894;Boissieu,1902;Hiroe,1958),allies strongly withTongoloa stewardii.Tongoloa pauciradiatais a poorly known species endemic to China and is not recorded in the Flora of China (Pan and Watson,2005a);Hiroe(1979) considered it to be a species ofPimpinella.In the present study,T.pauciradiataallies withAngelica paeoniifoliaR.H.Shan &C.Q.Yuan,both of which constitute a clade sister group toVicatia thibeticaH.de Boissieu (≡Sinodielsia thibetica(H.de Boissieu)Kljuykov &P.K.Mukh.).The ITS sequences of four accessions ofT.pauciradiataare identical to that ofA.paeoniifolia(FJ237533),while the sequences of our own accessions ofA.paeoniifolia(Zhou et al.,unpublished data),all falling in theSinodielsiaclade,do not match those of other conspecific members.A comparison of our material ofA.paeoniifolia(Z21-117 and Z21-124) with the types(PE00935513 and PE00617248) confirm that our collection is consistent with the types and protologue presented by Shan et al.(1980).Except for groupsTongoloa3,Tongoloa4,andTongoloa5,which each include two or more species ofTongoloa,the otherTongoloaspecies (including the type,T.gracilis) each constitute a separate clade.The morphological features characterizing the species comprisingTongoloa4 are subtle and include ultimate segments (length vs.width ratio),plant height (<30 cm vs.>30 cm),and rays (thick vs.slender).These features make species identifications extremely difficult.In addition,the morphological characters of five undescribed species (i.e.,T.sp.1,T.sp.2,T.sp.3,T.sp.4,andT.sp.MT124610)prevent them from being assigned to existingTongoloaspecies and describing them now as new species would only add to the confusion,given the polyphyletic nature of the genus.Overall,Tongoloais a rather critical genus,and its propercircumscription and taxonomy will be among the most difficult challenges remaining in Apioideae.

Table 2 Clade designation of Sinocarum,Tongoloa,and Trachydium species based on results of this study.Sinocarum filicinum and S.wolffianum are excluded from Sinocarum based on these same results.
Sinocarumis closely related to,and difficult to distinguish from,Acronemabecause the characters used to delimit these genera are variable and overlapping (Pu et al.,2005).Acronemais characterized by long-linear or long-aristate petals;however,some species have acute or obtuse petals (e.g.,A.yadongenseS.L.Liou andA.chinenseH.Wolff)that can also be seen inSinocarum.Both genera have species with ultimate linear segments making them indistinguishable (e.g.,Acronema schneideriH.Wolff,A.graminifolium(W.W.Smith) S.L.Liou &R.H.Shan,andSinocarum vaginatum).In the present study,species ofSinocarumandAcronemafall into both theAcronemaand East Asia clades(Table 2).In theAcronemaclade,their species are intermingled,indicative of their close relationship.The generic type ofSinocarum(S.coloratum)was synonymized withS.cruciatumby Pimenov (2017).However,S.coloratumallies withS.muliense,both of which comprise a clade sister group to the clade ofS.cruciatumandS.vaginatum.Upon examination of these specimens,we believe thatS.coloratumandS.cruciatumshould each be maintained as distinct species.Sinocarum bellumdoes not ally with its congeners.In a previous study(Xiao et al.,2021),S.bellumfell as sister group to allAcronemaspecies.Two other species ofSinocarumin theAcronemaclade,S.filicinumandS.wolffianum,form a sister group with three species ofMeeboldia.Their fruits are similar,being ovoid with filiform ribs,tapering toward the apex,slightly laterally compressed,and having a concave seed face (Wolff,1930;Xiao et al.,2021);all have recurved styles too (Wolff,1930;Shan and Sheh,1985b,Table 3).As a result,we transferS.filicinumandS.wolffianuminto the genusMeeboldia(see Taxonomic treatment section).The morphological characters ofS.dolichopodum,such as its long rhizome,leaf morphology,and divided bracteoles,are rather uncharacteristic ofSinocarum,suggesting that the species may be better placed elsewhere (Pu et al.,2005).The present results support such a treatment,as the species allies with several species ofTongoloaandTrachydium.The most characteristic feature ofS.schizopetalum(Franch.) H.Wolff is its petal apex being palmately 3-4-lobed.This species is sister group to theTrachydium3 group plusA.crassifolium.Its broad-ovoid mature fruits and subrhomboidal pollen are also different from thoseSinocarumspecies in theAcronemaclade (Xiao et al.,2021).A.crassifoliumis a newly described species characterized by thickly papery,ternate,abaxially dark purple leaves,terminal umbels with 8-13 rays,and no calyx teeth(Wang et al.,2013).Because the apex of its petals is linear(ca.1.7 mm long),it was described under the genusAcronema.However,the species falls within the East Asia clade,distantly awayfrom most of its congeners in theAcronemaclade.With its roots conic or tuberous,ultimate segments linear or linear-lanceolate,and petal bases long clawed and with an acute apex,Acronema chieniiis also separated from congeners and occupies an isolated position in the East Asia clade,further adding to the taxonomic complexities of the genus and the clade as a whole.
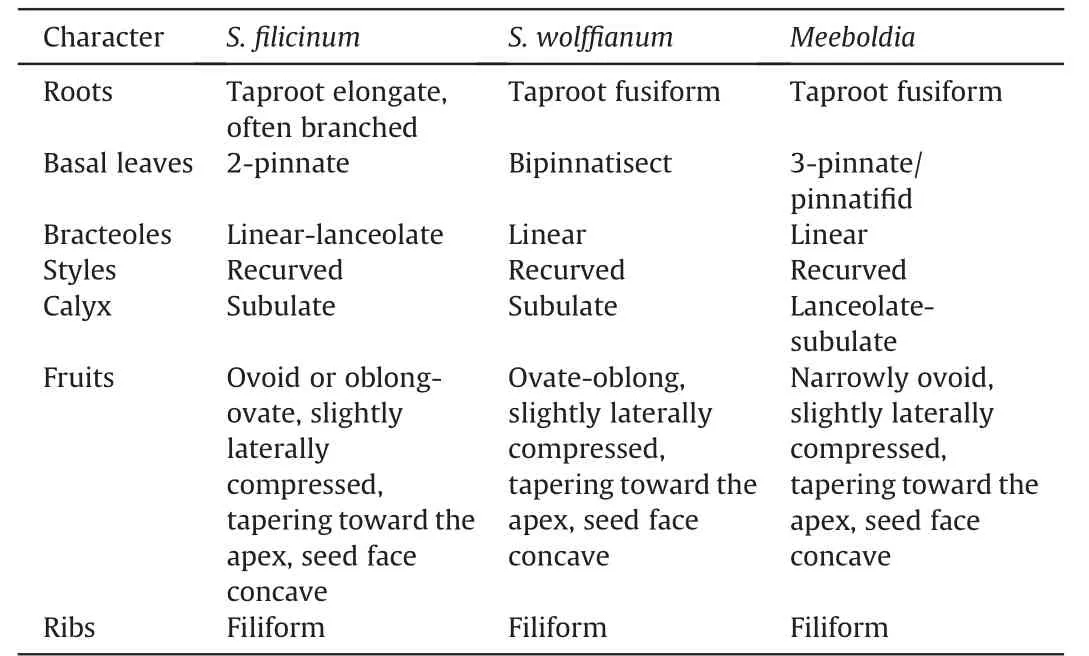
Table 3 Morphological comparisons of Sinocarum filicinum and S.wolffianum with Meeboldia.
Of the 37 species ofHymenidiumrecognized by Pimenov(2017),seven species (and one unknown species) occur in the East Asia clade as three separate lineages.H.amabileis widely distributed in China (Sichuan,Yunnan,and Xizang Provinces),Bhutan,and India(Pimenov,2017),and is easily identified by its showy leaf sheaths and bracteoles having its membranous and main veins dark purple.Hymenidium tsekuenseis a narrowly endemic species of northwestern China(Qinghai Province)and is part of thePleurospermum hookeriC.B.Clarke[=H.hookeri(C.B.Clarke)Pimenov&Kljuykov]complex (Pan and Watson,2005c).The main morphological differences betweenH.amabileandH.tsekuenseare that the latter is slender in habit and has fewer rays (Shan and Sheh,1979).In the present study,H.amabileis paraphyletic withH.tsekuensecontained within,whereasH.hookerifalls into theAcronemaclade.Thus,H.tsekuenseandH.amabilemay be regarded as conspecific or possibly close relatives having geographical variants.Hymenidium nubigenumis another poorly known taxon recorded from only a few collections,with its bracteoles margins white-membranous and ribs crisped-winged (Pan and Watson,2005c).The sample collected from its type locality (Que'er Mtn,Sichuan Province)yielded only a portion of its ITS sequence and this was enough to inform its phylogenetic position.H.nubigenumallies withH.nanum,as either a geographically distant species in the ML analysis (Fig.1) or a poorly supported sister group toH.tsekuenseandH.amabilein the BI analysis (Fig.5).BothH.virgatumandH.lhasanumare newly described species endemic to China(Pimenov and Kljuykov,2004).In the protologue,the authors consideredH.virgatumto be closely related to the generaSinolimprichtiaandTongoloa.In the present study,it is a sister group toTongoloa tagongensis.However,the morphological features ofH.virgatumandT.tagongensisare distinctly different.Hymenidium lhasanumforms a sister group to three species ofHymenolaena.Both taxa have clearly different morphologies,especially in the leaf blade (1-pinnate vs.bipinnatisect) and bracteoles (completely membranous vs.membranous-margined).

Fig.5.Majority-rule consensus tree for the East Asia clade and outgroups inferred from Bayesian inference analysis (195 terminals).Numbers above the branches are Bayesian posterior probabilities.The 106 newly sampled accessions are shown in bold.GenBank accession numbers of all sequences are provided;two numbers indicate sequences from separate ITS1 and ITS2 regions.Clades designations are described in the text.
Trachydiumhas been described as a storage-box genus,holding all taxa with shortened stems from the alpine Sino-Himalaya.Besides the two clades to whichTrachydiumhas been referred to previously (East Asia clade and Pleurospermeae;Downie et al.,2010),one species (Trachydium subnudum) is allied with theSinodielsiaclade (Table 2 and Fig.4).Trachydium subnudumis characterized by its scabrous leaf surfaces and densely tuberculate fruit,especially on its ribs.It occurs as a sister group to the clade ofTongoloa pauciradiataandV.thibetica.These three species share several characteristics,such as long rhizomes,carrot-like roots,and scabrous leaves,and all are distant from their respective generic types and congeners.The type species ofTrachydium(T.roylei) is clustered with several species ofHymenidiumin the tribe Pleurospermeae.Whether theseHymenidiumspecies are transferred intoTrachydiumor separated as a distinct genus remains to be seen.Species ofTrachydiumin the East Asia clade each comprise separate groups.The exception isTrachydium3,which includes four species ofTrachydium.Trachydium souliei,T.simplicifolium,andT.trifoliatumare morphologically distinct,but have almost identical ITS sequences.These species are distributed primarily in the Himalayas and their origins and low sequence divergence could be the result of rapid species radiation.
Pimpinella rhomboideais endemic to China and is characterized by its terminal leaflets being broad-ovate or rhombic.It has two varieties (P.rhomboideaandP.rhomboideavar.tenuiloba;Pu and Watson,2005) that do not cluster together.In the protologue,the authors considered the main morphological differences between the varieties to refer to the basal leaves(ternate-2-pinnate vs.2-ternate)and ultimate segments(1-1.5×0.5-1 cm vs.5-8×2-5 cm);fruit characters were not used (Shan and Pu,1989) for there are no ripe fruits on the type specimen (CDBI0172296).Pimpinella rhomboideavar.tenuilobawas synonymized withM.bipinnatum(≡V.bipinnata,Pimenov and Kljuykov,2006;Tan et al.,2015),which has close relationships to bothAngelica angelicifolia(Franch.) Kljuykov andA.yanyuanensis(F.T.Pu)J.Zhou of tribe Selineae.The generic type ofMelanosciadium,M.pimpinelloideumH.de Boissieu also belongs to Selineae,and it arises from withinAngelicaspecies.Thus,we consider that bothM.bipinnatum(=Pimpinella rhomboideavar.tenuiloba) andM.pimpinelloideumwill be transferred toAngelicapending confirmation from fruit characters.WhileM.genuflexumis away from congeners and falls into theSinodielsiaclade.Pimpinella rhomboideaoccupies an isolated position in the East Asia clade.The other four species ofPimpinellain the East Asia clade comprise a polytomy.Considering that members of the genusPimpinellaare scattered in no less than seven major clades of Apioideae,there is still a long way to go to resolve its nomenclatural and taxonomic problems.
4.3.Three species with ambiguous phylogenetic placements
Physospermopsis cuneatais a little-known species endemic to China.It is unusual within the genus because of its lack of conspicuous bracts and bracteoles(Pan and Watson,2005d).Based on fruit structure,general habit,and leaf dissection,Pimenov and Kljuykov (1999) transferred it into the genusSinodielsia,asS.cuneata(H.Wolff) Pimenov &Kljuykov.The results presented herein are consistent with our findings from prior studies in supporting its placement in the East Asia clade (Zhou et al.,2009,2020).In this clade,it comprises a sister group toTongoloa rubronervis,which in turn forms either a trichotomy withTongoloa5 andPimpinella1(Fig.1)or is a poorly supported sister group toTongoloa5 (Fig.5).The type ofSinodielsia[S.yunnanensisH.Wolff(≡M.yunnanensis)] falls into theAcronemaclade.Upon the results presented herein,the species may be better placed inTongoloa,or a new genus awaits description.
Physospermopsis kingdon-wardiiwas originally described as a species ofTrachydiumunder the name ofT.kingdon-wardiiH.Wolff(1929).It was later transferred into the genusPleurospermumasP.kingdon-wardii(H.Wolff)Hiroe(1979).Based on anatomical and micromorphological fruit characters,Pu and Liu(2005)considered it more appropriately placed inTrachydium.Pimenov (2017) classified it as synonymous withPhysospermopsis obtusiuscula(DC.) C.Norman,while other authors have argued thatP.kingdon-wardiidiffers fromP.obtusiusculain its reduced stem,dwarf plants,and small fruits with prominent and sinuate ribs(Xu et al.,2021).After careful comparison of the type specimens ofPhysospermopsis kingdon-wardii(E00000221)andP.obtusiuscula(K000697363),we believe that the treatment of Pimenov (2017) is justified.It is also supported by the present study,as accessions ofP.kingdon-wardii,with its entire or divided bracteoles and shortened stems(OR139727,OR139725,OR139724 and OR139726),andP.obtusiuscula(OR139722,OR139723 and OR139728),with its stems not shortened,comprised a polytomy.Herein,the generic attribution ofP.kingdon-wardiiis confirmed in the genusPhysospermopsis.
Tongoloa stewardiiis disjunctly distributed in Central and Southwest China and is distinguished from other species of the genus by its ovate to lanceolate-ovate ultimate segments and margins irregularly pinnate or coarsely serrate.Hiroe (1958)transferred the species toPimpinellaasP.stewardii(H.Wolff)Hiroe.Some authors considered it to be synonymous withTongoloa fortunatii(H.de Boissieu)Pimenov&Kljuykov(Pimenov and Kljuykov,1999;Pimenov,2017).We have examined type materials of both species and conclude thatT.stewardiiandT.fortunatiicannot be separated from each other.Herein,Tongoloa stewardiiis a sister group toT.loloensis,both of which fall into a large clade consisting ofTrachydium,Tongoloa,andSinocarum.
5.Conclusion
This is the most comprehensive study of the East Asia clade(Apiaceae subfamily Apioideae) carried out to date,specifically with regard to the number of new accessions examined,many of which are now included within the group.The clade is circumscribed to include representatives of 11 genera,seven (Acronema,Hymenidium,Physospermopsis,Pimpinella,Sinocarum,Tongoloa,andTrachydium)of which are not monophyletic and with some having their nomenclatural types falling outside of the group.Within the clade,the species comprising the polyphyletic genera are seriously intermingled,resulting in more confusing relationships than previously realized.The most perplexing taxonomic problems remaining in East Asia clade surround these polyphyletic genera,which are notoriously difficult to define.Thus,despite our attempt to clarify phylogenetic relationships of all genera comprising the East Asia clade,additional taxonomic study is warranted for the problematic species highlighted here.
6.Taxonomic treatment
Meeboldia filicina(H.Wolff) J.Zhou,comb.nov.
≡SinocarumfilicinumH.Wolff,1929,Repert.Spec.Nov.Regni Veg.27: 182,nom.inval.(Art.35.1).
≡Carum chinenseM.Hiroe,1979,Umbelliferae World: 872.
Type: CHINA.“Yunnan,eastern flank of the Tali Range,2540 m a.s.l.,Forrest 6863” (lectotype E! -barcode K00000440,designated by M.Farille in E;isolectotype K! -barcode K000 685663).
Meeboldia wolffiana(Fedde ex H.Wolff) J.Zhou,comb.nov.
≡Acronema wolffianumFedde ex H.Wolff,Repert.Spec.Nov.Regni Veg.27: 328.1930.
≡Sinocarum wolffianum(Fedde ex H.Wolff) P.K.Mukh.&Constance,1991,Edinburgh J.Bot.48(1): 42.
≡Pimpinella feddeanaM.Hiroe,1979,Umbelliferae World: 832.
Type: INDIA.“Himalaya,Sikkim,Jongri,12,000 ft,15.10.1875,Clarke 25796”(lectotype K!-barcode K000075372,designated by Mukherjee,Constance,1991: 42).
Author contributions
Jing Zhou:Writing-review&editing,Writing-original draft,Funding acquisition,Conceptualization.Zhenwen Liu: Writing -original draft,Conceptualization.Jia-Rui Yue: Investigation.Jun-Mei Niu: Formal analysis.Shi-Lin Zhou: Investigation.Xin-Yue Wang: Visualization,Data curation.Stephen R.Downie: Writing -review &editing
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (no.31960048 and 31872649),Yunnan Revitalization Talent Support Program (no.YNWRQNBJ-2019-208),the Department of Science and Technology of Yunnan Province (no.202201AT070118),the Hundred Talents Program of Kunming Medical University (no.60118260127),and the Gaoligong Mountain,Forest Ecosystem,Observation and Research Station of Yunnan Province (no.202205AM070006).We thank three anonymous reviewers for valuable,constructive comments that helped improve the manuscript.
Appendix A.Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2023.11.002.
杂志排行
植物多样性的其它文章
- Global patterns and ecological drivers of taxonomic and phylogenetic endemism in angiosperm genera
- Patterns and drivers of plant sexual systems in the dry-hot valley region of southwestern China
- Cryptic divergences and repeated hybridizations within the endangered “living fossil” dove tree (Davidia involucrata) revealed by whole genome resequencing
- Enhanced and asymmetric signatures of hybridization at climatic margins: Evidence from closely related dioecious fig species
- Cryptic diversity and rampant hybridization in annual gentians on the Qinghai-Tibet Plateau revealed by population genomic analysis
- Historical biogeography and evolutionary diversification of Lilium(Liliaceae): New insights from plastome phylogenomics
